Introduction
High fat diet and low fiber has become increasingly
widespread in the daily diet, which to some extent, accelerates the
incidence of colorectal cancer (1).
Colorectal cancer (CRC) is one of the most common malignant tumors
worldwide. Its incidence rate in China ranks fourth and its
mortality rate ranks third (2).
Therefore, the prevention and treatment of CRC have important
practical significance for improving quality of life of patients.
In its early stage, CRC mainly infiltrates locally while at its
advanced stage it is characterized by lymphatic spread, with
surgical resection being the only viable treatment option (3). However, the postoperative recurrence
rate of patients with CRC following surgical resection is
approximately 45.3–73.5%, which challenges the survival rate within
5 years of surgery (4). Therefore,
investigation into the pathogenesis of CRC is crucial. Caudal type
homeobox transcription factor 2 (CDX2) in the human body is a type
of transcription factor for regulating gene transcription and
expression in gastrointestinal embryo pathways (5–7).
CDX2 gene plays an important role in some relevant digestive
system cancers, including gastric and colon cancer (8). The downregulation of CDX2 gene
expression may lead to the loss of relevant differentiation
function and the increase of value-added ability in the process of
the occurrence of CRC, and it may also co-operate with other genes
to participate in regulating the occurrence of relevant tumors
(9). It is believed that the
methylation of DNA promoter to a certain extent can regulate the
expression of relevant genes (10).
However, the transcriptional regulation of many genes in the human
system specifically expressed in differentiation is de-methylated
(11), but it is methylated in
specific cells, not requiring the expression of genes.
Previous studies on CDX2 gene were primarily
focused on gastric and colon cancer. At present, few studies are
available on the function of CDX2 gene in CRC and on the
methylation of the promoter region of CDX2 gene in CRC. To
the best of our knowledge, the present study, for the first time,
examined the methylation of promoter region of CDX2 gene in
CRC to provide preliminary data on the correlation between the
methylation of the promoter regions of CDX2 and CRC. We also aimed
to determine strategies in the prevention and treatment of CRC in
advanced stage.
Patients and methods
Patients
In total 78 cases with CRC (tested by relevant
pathology) were selected at the Luoyang Central Hospital (Henan,
China) from May 2014 to April 2015. The patietns comprised 43 men
and 35 women, with an average age of 42±7.9 years. Additionally, a
total of 52 (31 men and 21women, with an average age of 45±9.3
years) participants who were diagnosed with colorectal polyps
served as controls (normal population). The study subjects were not
undergoing treatment in the form of surgery in vitro,
chemotherapy and radiotherapy.
Reagents
The following reagents were used: Mouse anti-human
CDX2 monoclonal antibody (Roche Diagnostics, Indianapolis, IN,
USA), goat anti-mouse HRP secondary antibody (Roche Diagnostics);
RT-polymerase chain reaction (PCR) kit (Takara Bio, Inc., Otsu,
Japan); mouse anti-human immunohistochemistry kit (Roche
Diagnostics); DBA staining kit (Thermo Fisher Scientific, Waltham,
MA, USA); animal cell genomic extraction kit (Axygen Scientific
Inc., Union City, CA, USA); CpG methylation enzyme (New England
Biolabs, Ipswich, MA, USA); MethylDetector bisulfite modification
kit (Active Motif, Carlsbad, CA, USA).
Detection of methylation
DNA extracted from the sample tissue was in
accordance with the instructions of the animal cell genomic
extraction kit, purchased from Axygen Scientific Inc. The
modification of DNA bisulfite was performed in accordance with the
instructions of the MethylDetector bisulfite modification kit
(Active Motif).
The methylation primer was designed according to the
supplier's protocol (12). The
primers are listed in Table I. The
reaction system of methylation-specific PCR was 25 µl, i.e.,
ddH2O 8.5 µl, DNA 2 µl, Premix Ex Taq DNA polymerase
mixture 12.5 µl, methylation- or nonmethylation-specific primer 2
µl. The PCR reaction conditions were: denaturation at 95°C for 3
min, annealing at 95°C for 30 sec, 58°C for 30 sec, and extension
72°C for 30 sec, for a total of 30 cycles. A final extension was
conducted at 72°C for 10 min, and final heat preservation was at
4°C. The PCR products was recovered and sequenced using gel
electrophoresis (Invitrogen Life Technologies, Carlsbad, CA,
USA).
 | Table I.Primer of methylation-specific
polymerase chain reaction. |
Table I.
Primer of methylation-specific
polymerase chain reaction.
| Primers | Length of products
(bp) |
|---|
| Methylation forward
primer |
|
|
5′-CGAAAATAAATCACTACGACG-3′ | 200 |
| Non-methylation
forward primer |
|
|
5′-ATTCAAAATAAAAATCACTACAACA-3′ | 204 |
| Common reverse
primer |
|
|
5′-AAAGGATATTGGAGAGTATTTTAG-3′ |
|
Fluorescence quantitative PCR
(qPCR)
RNA extraction was performed as described previously
(13). Fluorescence qPCR was
performed according to the manufacturer's instructions (Takara
fluorescent qPCR; Takara Bio, Inc.).
Enzyme-linked immune reaction
The procedure was performed according to the
instructions of ELISA kit. An assay buffer of PBST (Biosharp,
Hefei, China) was used to prepare dilutions of 1:25, and a standard
curve was then designed. Firstly, the sample to be measured was
diluted at a 1:100 ratio, and 100 µl of sample to be measured was
mixed with 50 µl of detection solution in each well. The samples
were kept for 2 h at 20°C, followed by the addition of TMB to
measure light absorption at 495 nm using a microplate reader
((Model 3550; Bio-Rad Labs, Hercules, CA, USA). The concentration
of CDX2 in every sample was measured according to the standard
curves.
Western blotting
The animal cell protein extraction kit (Roche
Diagnostics) was used to extract the total proteins. Western blot
analysis was performed as described previously (13). After protein quantitation using the
Lowery protein assay, equal amounts of proteins were separated by
SDS-PAGE and blotted onto nitrocellulose membranes by the semi-dry
blotting method using a three-buffer system. The membranes were
incubated with primary antibody at a dilution of 1:500 (anti-CDX2,
RayBiotech, Inc., Norcross, GA, USA; cat no.: 119-14329) overnight
at 4°C. The membrane was washed with TBST and incubated with a
peroxidase-conjugated secondary polyclonal goat-anti-mouse antibody
(1:1000) (Santa Cruz Biotechnology, Inc. CA, USA; cat no.:
sc-395763) for 1 h. Western blot film was scanned and the membrane
was stripped and reprobed with antibody against β-actin (1:1500)
(Santa Cruz Biotechnology, Inc.; cat no.: sc-8432) to confirm equal
sample loading
Immunohistochemical staining
The procedure was performed according to the mouse
anti-human immunohistochemistry kit (Roche Diagnostics). After
staining, the nucleus appeared yellow, which was considered
positive. The staining criteria were classified as: (−) negative,
unstained cells or stained very lightly and there was no noticeable
difference between the cell and the background; (+) extensively
weakly colored after staining, the color of cell was slightly
higher than that of the background; (++) extensively moderately
colored after staining, the color of cell was markedly higher than
that of the background; and (+++) extensively strong colored after
staining, where the cells were dark brown. In the study, all the
(+) (++) (+++) were considered positive. The final score was
evaluated by two experts independently, and the average score was
considered as final.
Statistics analysis
The SPSS 20.0 software (IBM SPSS, Armonk, NY, USA)
was used to process the obtained data. Measurement data were
presented as mean ± standard deviation and the measurement data was
tested using the χ2 test.
Results
Methylation status of CDX2 gene in
tissue of CRC and normal population
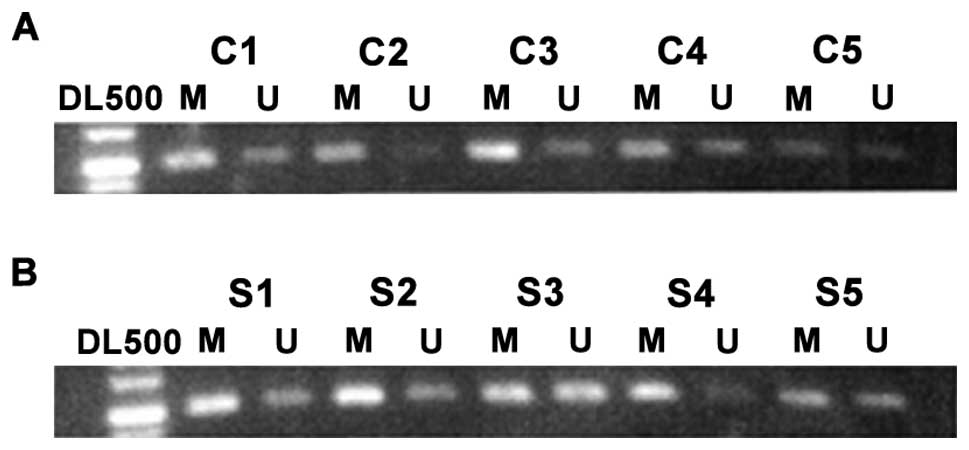
As shown in Fig. 1,
the controls had a lower degree of CDX2 promoter methylation, but
there was a high degree of methylation for CDX2 gene in the
lesion tissue of patients with CRC. Table II shows the rate of CDX2 promoter
methylation in the colorectal tissue of the normal population,
which was ~43.5%, but the rate in the lesion tissue of CRC was
~78.5%. The results were statistically significant.
 | Table II.Relative expression quantity of mRNA
of CDX2 in colorectal tissue in normal population and patients with
colorectal cancer (use GAPDH as internal control).a |
Table II.
Relative expression quantity of mRNA
of CDX2 in colorectal tissue in normal population and patients with
colorectal cancer (use GAPDH as internal control).a
|
|
| CDX2 gene | Promoter
methylation |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Group | Cases | Methylation | Non-methylation | χ2 | P-value |
|---|
| Normal
population | 63 | 27 | 36 | 5.8855 | 0.023b |
| Patients with
colorectal cancer | 78 | 61 | 17 |
|
|
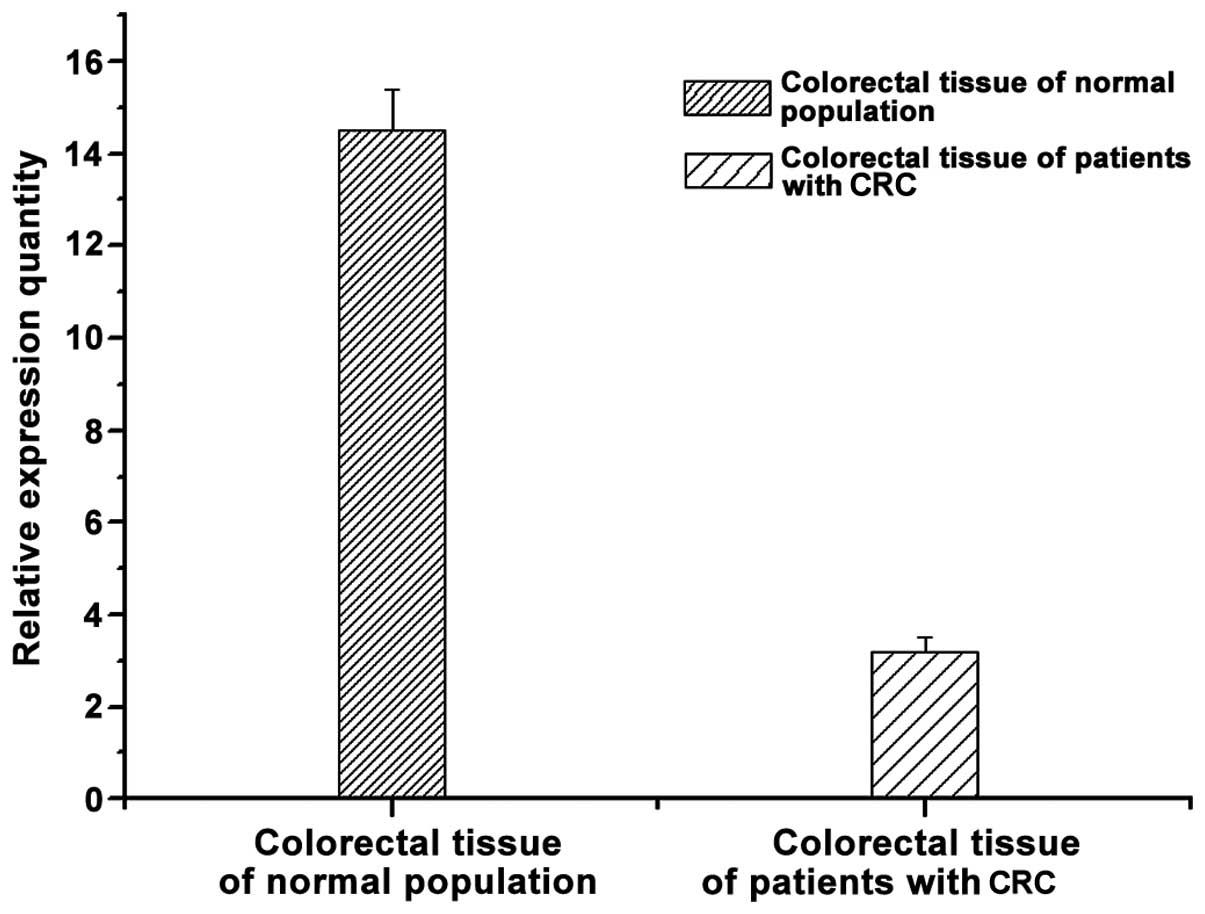
Fluorescence qPCR
By comparing the levels of CDX2 gene in
controls and patients with CRC, we found that the level of
CDX2 gene was higher in colorectal tissue of the normal
population and lower in colorectal tissue of the patients with CRC
(Fig. 2). The result indicates that
there may be a correlation between CDX2 gene and the disease
of CRC.
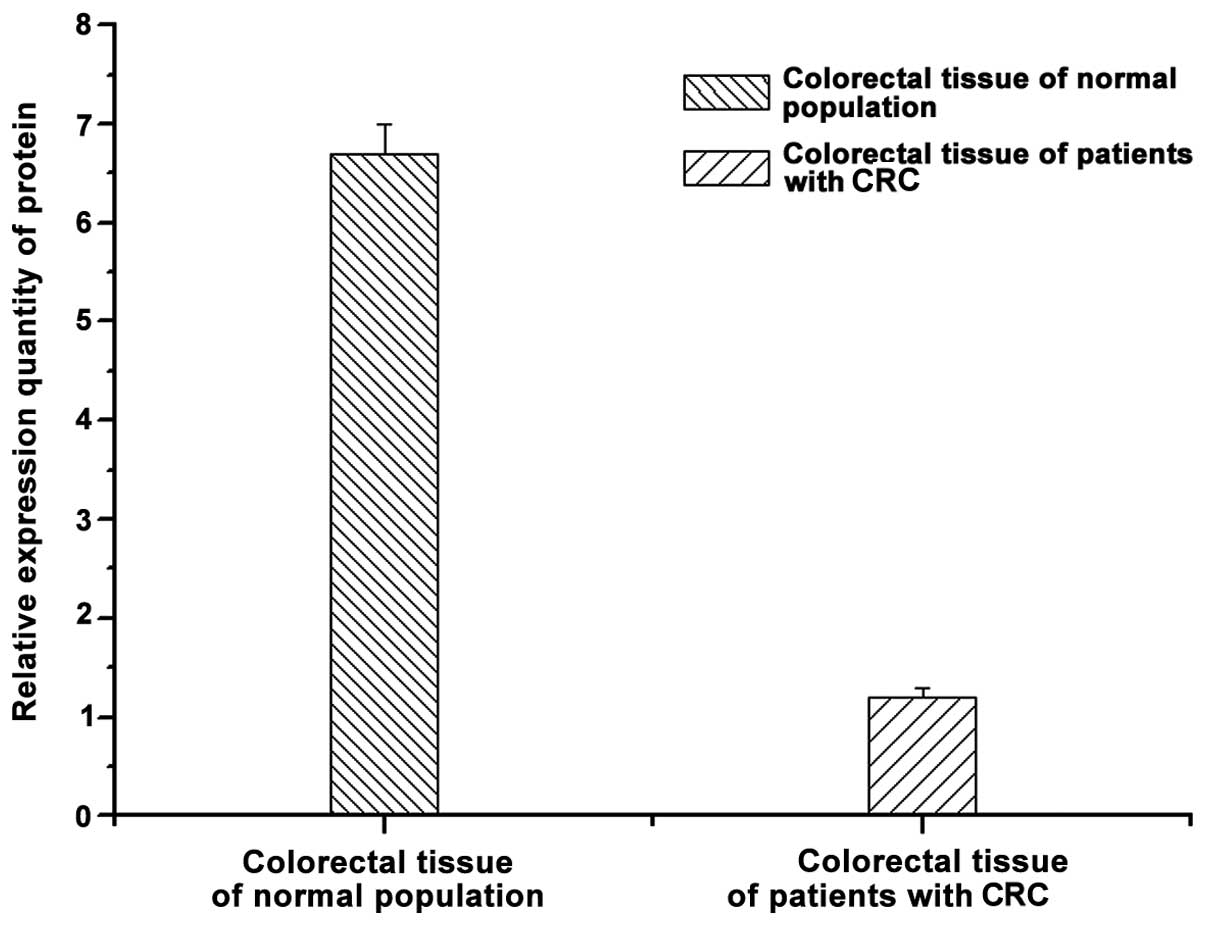
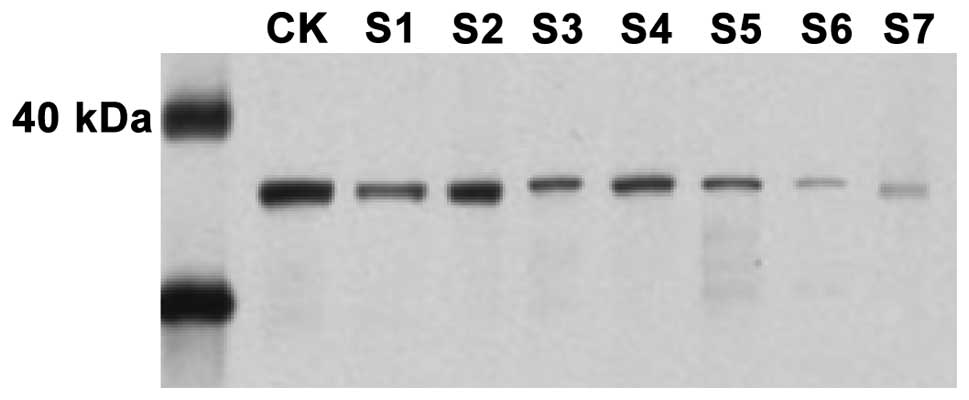
Enzyme-linked immune response and
western blotting
The protein in controls and patients with CRC were
determined by ELISA and western blotting (Figs. 3 and 4).
In comparison to CRC patients, the expression of CDX2 was higher in
the controls and a similar observation was made with the western
blotting and fluorescence qPCR results.
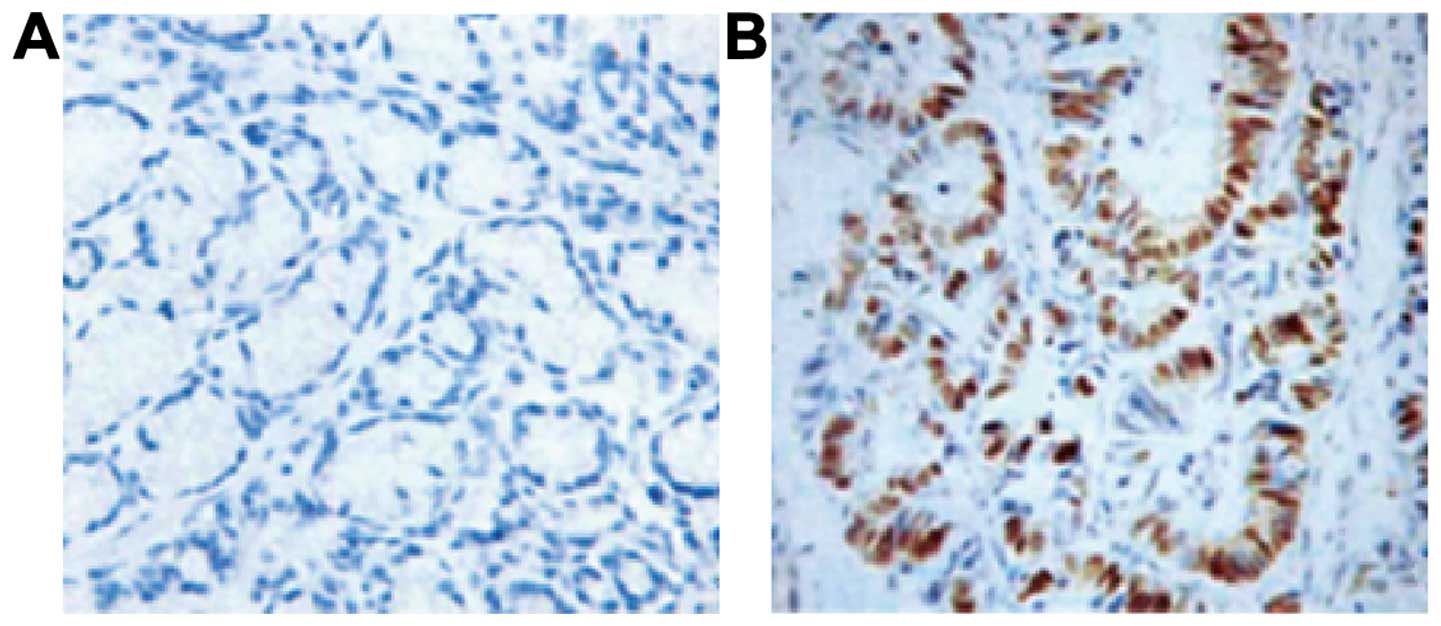
Immunohistochemistry
By comparing the results of colorectal tissue
staining in CRC patients and controls, we found that the positive
expression of CDX2 primarily exists in colonic epithelial goblet
cells (Fig. 5). Staining showed
brownish yellow granules. We observed that CDX2 protein mainly
exists in the colorectal tissue of the normal population and has a
lower expression in CRC tissue.
Discussion
The occurrence and aggravation of CRC involves
numerous factors and is not only influenced by physical conditions
but also by diet, and mental state (14). The increase of studies of genomics
with a focus on the occurrence of CRC have led to investigations at
gene level. CDX2, a tail-type homologous box gene family, is a type
of transcription factor of intestinal epithelial cell specificity
in the human body (15). CDX2
gene in human body is involved in the regulation of the expression
of intestine-specific genes, intestinal development, and the
differentiation and proliferation of intestinal epithelial cells
(16). CDX2 gene has also been
found to be involved in the occurrence of some tumors and cancers
in the human body (17). It has been
suggested that CDX2 gene has a higher expression in
intestinal metaplasia associated with chronic atrophic gastritis
and is expressed in gastric cancer (18). CDX2 may be associated with gastric
mucosal epithelial transformation and gastric mucosal
carcinogenesis (19–22). Previous findings have shown that
transfection of CDX2 cDNA, and human HT29 CRC cell line to express
CDX2 protein, indicated the oncogenic potential of the
abovementioned cells, and metastasis of related cells markedly
decreased while cell sensitivity for apoptosis significantly
increased (23).
In summary, to the best of our knowledge, for the
first time, we examined a correlation between methylation of the
promoter region of CDX2 in CRC and CRC. The results have shown that
in comparison to the normal population, the degree of methylation
of the promoter region of CDX2 in lesion tissue of patients with
CRC was higher than that of the normal population. The protein
expression in the control and lesion sections of CRC patients
showed that the expression level of CDX2 in the lesion section of
patients with CRC was lower. This finding suggested that there was
a certain correlation between CDX2 and CRC, or the decrease in the
degree of CDX2 gene promoter methylation to a certain
extent, promotes the risk of CRC.
Acknowledgements
The present study was supported by the Scientific
and Technologic Development Programme of Luoyang City (No.:
1401087A-3).
References
|
1
|
Gregoire R, Yeung KS, Stadler J, Stern HS,
Kashtan H, Neil G and Bruce WR: Effect of high-fat and low-fiber
meals on the cell proliferation activity of colorectal mucosa. Nutr
Cancer. 15:21–26. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu S, Zheng R, Zhang M, Zhang S, Sun X
and Chen W: Incidence and mortality of colorectal cancer in China,
2011. Chin J Cancer Res. 27:22–28. 2015.PubMed/NCBI
|
|
3
|
Dawson H, Galván JA, Helbling M, Muller
DE, Karamitopoulou E, Koelzer VH, Economou M, Hammer C, Lugli A and
Zlobec I: Possible role of Cdx2 in the serrated pathway of
colorectal cancer characterized by BRAF mutation, high-level CpG
Island methylator phenotype and mismatch repair-deficiency. Int J
Cancer. 134:2342–2351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang XT, Xie YB and Xiao Q: siRNA
targeting of Cdx2 inhibits growth of human gastric cancer MGC-803
cells. World J Gastroenterol. 18:1903–1914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baba Y, Nosho K, Shima K, Freed E, Irahara
N, Philips J, Meyerhardt JA, Hornick JL, Shivdasani RA, Fuchs CS,
et al: Relationship of CDX2 loss with molecular features and
prognosis in colorectal cancer. Clin Cancer Res. 15:4665–4673.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saandi T, Baraille F, Derbal-Wolfrom L,
Cattin AL, Benahmed F, Martin E, Cardot P, Duclos B, Ribeiro A,
Freund JN, et al: Regulation of the tumor suppressor homeogene Cdx2
by HNF4α in intestinal cancer. Oncogene. 32:3782–3788. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim SP, Park JW, Lee SH, Lim JH, Jang BC,
Lee SH, Jang IH, Freund JN, Suh SI, Mun KC, et al: Homeodomain
protein CDX2 regulates COX-2 expression in colorectal cancer.
Biochem Biophys Res Commun. 315:93–99. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawai H, Tomii K, Toyooka S, Yano M,
Murakami M, Tsukuda K and Shimizu N: Promoter methylation
downregulates CDX2 expression in colorectal carcinomas. Oncol Rep.
13:547–551. 2005.PubMed/NCBI
|
|
9
|
Brabletz T, Spaderna S, Kolb J, Hlubek F,
Faller G, Bruns CJ, Jung A, Nentwich J, Duluc I, Domon-Dell C, et
al: Down-regulation of the homeodomain factor Cdx2 in colorectal
cancer by collagen type I: An active role for the tumor environment
in malignant tumor progression. Cancer Res. 64:6973–6977. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li F, Wang Y, Zhang G, Zhou J, Yang L and
Guan H: Expression and methylation of DNA repair genes in lens
epithelium cells of age-related cataract. Mutat Res 766-767. 31–36.
2014. View Article : Google Scholar
|
|
11
|
Zhang JF, Zhang JG, Kuai XL, Zhang H,
Jiang W, Ding WF, Li ZL, Zhu HJ and Mao ZB: Reactivation of the
homeotic tumor suppressor gene CDX2 by
5-aza-2′-deoxycytidine-induced demethylation inhibits cell
proliferation and induces caspase-independent apoptosis in gastric
cancer cells. Exp Ther Med. 5:735–741. 2013.PubMed/NCBI
|
|
12
|
Wojdacz TK, Hansen LL and Dobrovic A: A
new approach toprimer design for the control of PCR bias in
methylation studies. BMC Res Notes. 1:542008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Padda RS, Gkouvatsos K, Guido M, Mui J,
Vali H and Pantopoulos K: A high-fat diet modulates iron metabolism
but does not promote liver fibrosis in hemochromatotic Hjv¯/¯ mice.
Am J Physiol Gastrointest Liver Physiol. 308:G251–G261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sugimoto M, Yamaoka Y and Furuta T:
Influence of interleukin polymorphisms on development of gastric
cancer and peptic ulcer. World J Gastroenterol. 16:1188–1200. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gendron FP, Mongrain S, Laprise P, McMahon
S, Dubois CM, Blais M, Asselin C and Rivard N: The CDX2
transcription factor regulates furin expression during intestinal
epithelial cell differentiation. Am J Physiol Gastrointest Liver
Physiol. 290:G310–G318. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mutoh H, Hayakawa H, Sakamoto H and Sugano
K: Homeobox protein CDX2 reduces Cox-2 transcription by
inactivating the DNA-binding capacity of nuclear factor-kappaB. J
Gastroenterol. 42:719–729. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin R, Wang NN, Chu J and Wang X:
Expression and significance of homeodomain protein Cdx2 in gastric
carcinoma and precancerous lesions. World J Gastroenterol.
18:3296–3302. 2012.PubMed/NCBI
|
|
18
|
Bai YQ, Yamamoto H, Akiyama Y, Tanaka H,
Takizawa T, Koike M, Yagi O Kenji, Saitoh K, Takeshita K, Iwai T,
et al: Ectopic expression of homeodomain protein CDX2 in intestinal
metaplasia and carcinomas of the stomach. Cancer Lett. 176:47–55.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asano N, Imatani A, Watanabe T, Fushiya J,
Kondo Y, Jin X, Ara N, Uno K, Iijima K, Koike T, et al: Cdx2
expression and intestinal metaplasia induced by H. Pylori infection
of gastric cells is regulated by NOD1-Mediated innate immune
responses. Cancer Res. 76:1135–1145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mutoh H, Sakurai S, Satoh K, Tamada K,
Kita H, Osawa H, Tomiyama T, Sato Y, Yamamoto H, Isoda N, et al:
Development of gastric carcinoma from intestinal metaplasia in
Cdx2-transgenic mice. Cancer Res. 64:7740–7747. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mizoshita T, Tsukamoto T, Nakanishi H,
Inada K, Ogasawara N, Joh T, Itoh M, Yamamura Y and Tatematsu M:
Expression of Cdx2 and the phenotype of advanced gastric cancers:
Relationship with prognosis. J Cancer Res Clin Oncol. 129:727–734.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng J, Sun X, Wang W and Lu S:
Hypoxia-inducible factor-1α modulates the down-regulation of the
homeodomain protein CDX2 in colorectal cancer. Oncol Rep.
24:97–104. 2010.PubMed/NCBI
|
|
23
|
Soubeyran P, Mallo GV, Moucadel V, Dagorn
JC and Iovanna JL: Overexpression of Cdx1 and Cdx2 homeogenes
enhances expression of the HLA-I in HT-29 cells. Mol Cell Biol Res
Commun. 3:271–276. 2000. View Article : Google Scholar : PubMed/NCBI
|