Introduction
Aberrant promoter hypermethylation contributes to
the transcriptional inactivation of a number of genes in various
malignant diseases (1). However, the
precise mechanisms underlying this aberration remain unclear. DNA
methylation is established by the catalytic activity of a family of
DNA methytransferases (DNMTs), which includes DNMT1, DNMT3A and
DNMT3B (2). The association between
DNMTs expression and promoter hypermethylation of the
tumor-suppressor gene (TSG) P15INA4B has been reported in
acute myeloid leukemia (3). In
another study on diffuse large B-cell lymphoma (DLBCL), the
overexpression of DNMT3B and DNMT1 proteins was significantly
correlated with the promoter hypermethylation of various genes,
including p16 and von Hippel-Lindau (4). However, a lack of association between
deregulated DNMTs expression and aberrant promoter methylation has
also been reported (5–8).
E-cadherin, the gene product of CDH1, is a
calcium-dependent cell adhesion molecule that is essential for
maintaining the integrity of cell-cell adhesions (9). Downregulation of E-cadherin has been
identified in numerous human cancers, and a loss of E-cadherin
function was demonstrated to be associated with CDH1
promoter hypermethylation and increased invasiveness and metastasis
in human tumors (9,10). However, in another study on DLBCL,
CDH1 promoter hypermethylation was observed not to be
correlated with the expression levels of DNMTs (4).
H-cadherin, the gene product of CDH13, is a
novel member of the cadherin family (11). H-cadherin has a unique feature, in
that it is devoid of a transmembrane domain, and is anchored to the
cell surface membrane via a glycosylphosphatidylinositol (GPI)
moiety instead. In addition, it also lacks a cytoplasmic domain
(11). CDH13 downregulation
has been associated with the tumorigenesis of multiple cancers
(11). CDH13 silencing due to
promoter hypermethylation and/or loss of heterozygosity is
associated with tumor progression in BCLs (12).
A disintegrin and metalloproteinase with
thrombospondin motif 18 (ADAMTS18) belongs to the ADAMTSs family, a
group of secreted proteases that control several cell functions
(13). Porter et al (14) demonstrated that there was a
downregulation of several members of the ADAMTS family,
including ADAMTS18, in human breast cancer compared with
non-neoplastic mammary tissues. In addition, silencing of
ADAMTS18 by methylation of promoter CpG islands has been
reported in multiple carcinoma cell lines, particularly in cell
lines derived from esophageal and nasopharyngeal carcinomas
(15).
Tumor cells are characterized by frequent deletions
of chromosomal regions that encode multiple TSGs (16). CDH1, CDH13 and ADAMTS18
are all located on chromosome 16q (16q22.1, 16q24.2 and 16q23.1,
respectively). Similar to epigenetic modifications, genetic changes
that are caused mainly by chromosomal loss of heterozygosity or
mutations within the genes have been demonstrated to be associated
with the upregulation of oncogenes or the downregulation of TSGs
(16,17).
To the best of our knowledge, the correlations among
the expression levels of CDH1, CDH13, ADAMTS18
and DNMTs, as well as their associations with the promoter
methylation status of CDH1, CDH1 and ADAMTS18
in human lymphoma, have not been analyzed to date. Therefore, in
the present study, the expression levels of CDH1,
CDH13 and ADAMTS18 were investigated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) in
human lymphoma. In addition, the frequency of methylation of the
CDH1, CDH13 and ADAMTS18 gene promoters was
examined using methylation-specific PCR (MSP). It was observed that
these chromosome 16-located TSGs are frequently methylated and
correlated, but they are not associated with the DNMTs
levels in human BCL.
Patients and methods
Patients
From the files of the Department of Hematology of
Gunma University Hospital (Gunma, Japan), cases of surgically
biopsied lymph nodes from patients that were collected between
December 2006 and July 2012 were obtained upon receiving
appropriate institutional review board approval from Gunma
University and patients' written informed consent. For all cases,
the diagnosis was based on a morphological and immunohistochemical
analysis according to the World Health Organization classification
(18). The criterion for the
selection of these cases was the availability of fresh-frozen
optimal cutting temperature compound-embedded tumor biopsy
specimens collected prior to any treatment. The present study
included 29 cases of DLBCL, 7 cases of mantle cell lymphoma and 16
samples of non-malignant lymphoid tissues (including necrotizing
lymphadenitis, reactive lymph nodes, immunoglobulin G4-associated
diseases, follicular hyperplasia and granulomatous
lymphadenitis).
Purification of genomic DNA (gDNA) and
total RNA
gDNA and total RNA were purified from each tumor
tissue using the AllPrep DNA/RNA Mini kit (Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocol.
RT-qPCR
Total RNA (≤1 µg) for qPCR assays was treated and
reverse transcribed using PrimeScript RT Reagent kit with gDNA
Eraser (Takara Biotechnology Co., Ltd., Dalian, China) according to
the protocol provided by the manufacturer. RT-qPCR was performed
using SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) on a 7300 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Complementary
DNA (1 µl) was mixed with gene-specific primers (listed in Table I), along with SYBR Green PCR Master
Mix in a final volume of 20 µl. PCR was performed under the
following conditions: Initial denaturation at 95°C for 10 min,
followed by 45 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 1 min. The relative level of
expression of DNMTs, CDH1, CDH13 and ADAMTS18 were
normalized using a reference gene (β-actin) and control cells
(HL60), and calculations were performed using the
2−∆∆Cq method (19).
 | Table I.Primers, amplicon sizes and accession
number of genes analyzed by RT-qPCR. |
Table I.
Primers, amplicon sizes and accession
number of genes analyzed by RT-qPCR.
| Target genes | Primer sequences
(5′-3′)a | Size (bp) | Accession
number |
|---|
| DNMT1 |
GCCAACGAGTCTGGCTTTGAG (sense) | 101 | NM001130823 |
|
|
GTGTCGATGGGACACAGGTGA (antisense) |
|
|
| DNMT3A |
ACCCGACTTCATAATGGTGCTTTC (sense) | 139 | NM022552 |
|
|
CGCCATCTGCAAGCTGTCTC (antisense) |
|
|
| DNMT3B |
TAAATACAAGGGCTGGAGTCTGCAC (sense) | 80 | NM006892 |
|
|
TGACGTCATCCCTGCTGAAATC (antisense) |
|
|
| CDH1 |
GAGTGCCAACTGGACCATTCAGTA (sense) | 86 | NM0043603 |
|
|
AGTCACCCACCTCTAAGGCCATC (antisense) |
|
|
| CDH13 |
GACATTGTCACTGTTGTGTCACCTG (sense) | 121 | NM0012573 |
|
|
CCGTGCCTGTTAATCCAACATC (antisense) |
|
|
|
ADAMTS18 |
AAGTGACATAAACGTGGTTGTGGTG (sense) | 89 | NM1993552 |
|
|
GAGACTGGTCTGCATGATGGTTG (antisense) |
|
|
| β-actin | TGGCACCCAGCACAATGAA
(sense) | 186 | NM_0011013 |
|
|
CTAAGTCATAGTCCGCCTAGAAGCA (antisense) |
|
|
Bisulfite modification and MSP
The extracted DNA was modified using the Methyl Easy
Xceed Rapid DNA Bisulphite Modification kit (Genetic Signatures,
Darlinghurst, Australia) according to the manufacturer's protocol.
MSP was performed in a total volume of 20 µl, containing 2 µl
sodium bisulfite- modified template DNA, using EpiTaq HS for
bisulfite-treated DNA (Takara Biotechnology Co., Ltd.). Each MSP
reaction was performed with the following conditions: Denaturation
at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C
for 30 sec, 30 sec at the specific annealing temperature for
CDH1 and CDH13 [53°C for methylated (M) and 57°C for
unmethylated (U) DNA] and ADAMTS18 (57°C for M and U DNA)
and extension at 72°C for 30 sec, followed by a final 4-min
extension at 72°C.
The primer sequences were as follows: CDH1
(CDH1UM sense, 5′-GGTTTGATTTGATTGTATTT-3′ and CDH1UM
antisense, 5′-AAATACATCCCTCACAAAT-3′; and CDH1M sense,
5′-GGTTCGATTCGATCGTATTC-3′ and CDH1M antisense,
5′-GAATACGTCCCTCGCAAAT-3′); CDH13 (CDH13UM sense,
5′-TTGTGGGGTTGTTTTTTGT-3′ and CDH13UM antisense,
5′-AACTTTTCATTCATACACACA-3′; and CDH13M sense,
5′-TCGCGGGGTTCGTTTTTCGC-3′ and CDH13M antisense,
5′-GACGTTTTCATTCATACACGCG-3′); and ADAMTS18
(ADAMTS18UM sense, 5′-AAATTGTAGTTTGGTAGGTTTGT-3′ and
ADAMTS18UM antisense, 5′-CAACTCCAAATAAAAACCACCA-3′; and
ADAMTS18M sense, 5′-TTGTATTCGGTAGGTTCGC-3′ and
ADAMTS18M antisense, 5′-ACTCCAAATAAAAACCGCCG-3′). Primer
sequences were described previously (15,20,21) and
were purchased from Eurofins MWG Operon, Inc. (Huntsville, AL,
USA). MSP products were visualized under ultraviolet illumination
following electrophoresis in 2% agarose gels containing GelRed
nucleic acid gel stain (Biotium, Fremont, CA, USA).
5-Aza-2′-deoxycytidine (5-aza-dC)
treatment
Established lymphoma cell lines [Raji (22) (Institute of Development, Aging and
Cancer, Tohoku University, Sendai, Japan), CTB-1 (23) and SLVL (24) (Riken, Saitama, Japan)] and one patient
primary DLBCL cell line were cultured in RPMI-1640 medium (Wako
Pure Chemical Industries, Ltd., Osaka, Japan) at 37°C in an
atmosphere of 5% CO2 and treated with the demethylating
agent 5-aza-dC (Wako Pure Chemical Industries, Ltd.). Cells
(5×104) were grown in the absence or presence of
5-aza-dC at a concentration of 3 µM. The demethylation treatment
was performed three times to verify the results.
Statistical analysis
Using the SPSS 20 software (IBM SPSS, Armonk, NY,
USA), differences in the mean rank values between groups were
analyzed with the Mann-Whitney U test. The significance of
associations between parameters were analyzed by Spearman's
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of DNMTs, CDH1, CDH13 and
ADAMTS18
The relative mRNA expression levels of DNMT1,
DNMT3A, DNMT3B, CDH1, CDH13 and
ADAMTS18 were detected by RT-qPCR in 36 cases of lymphoma
and in 16 samples of non-malignant lymphoid tissue. The mean rank
expression levels of DNMTs (1, 3A and 3B) in
the lymphomas were significantly higher (P=0.044, P=0.036 and
P=0.013) than those in the non-malignant tissues by 45.4, 45.9 and
60.4%, respectively (Fig. 1A-C). The
expression of CDH1 and ADAMTS18 were both
significantly (P<0.01) reduced in lymphomas by 36.0% with
respect to non-malignant tissues, while CDH13 expression was
non-significantly reduced by 14.4% in lymphomas compared with
non-malignant tissues (Fig.
1D-F).
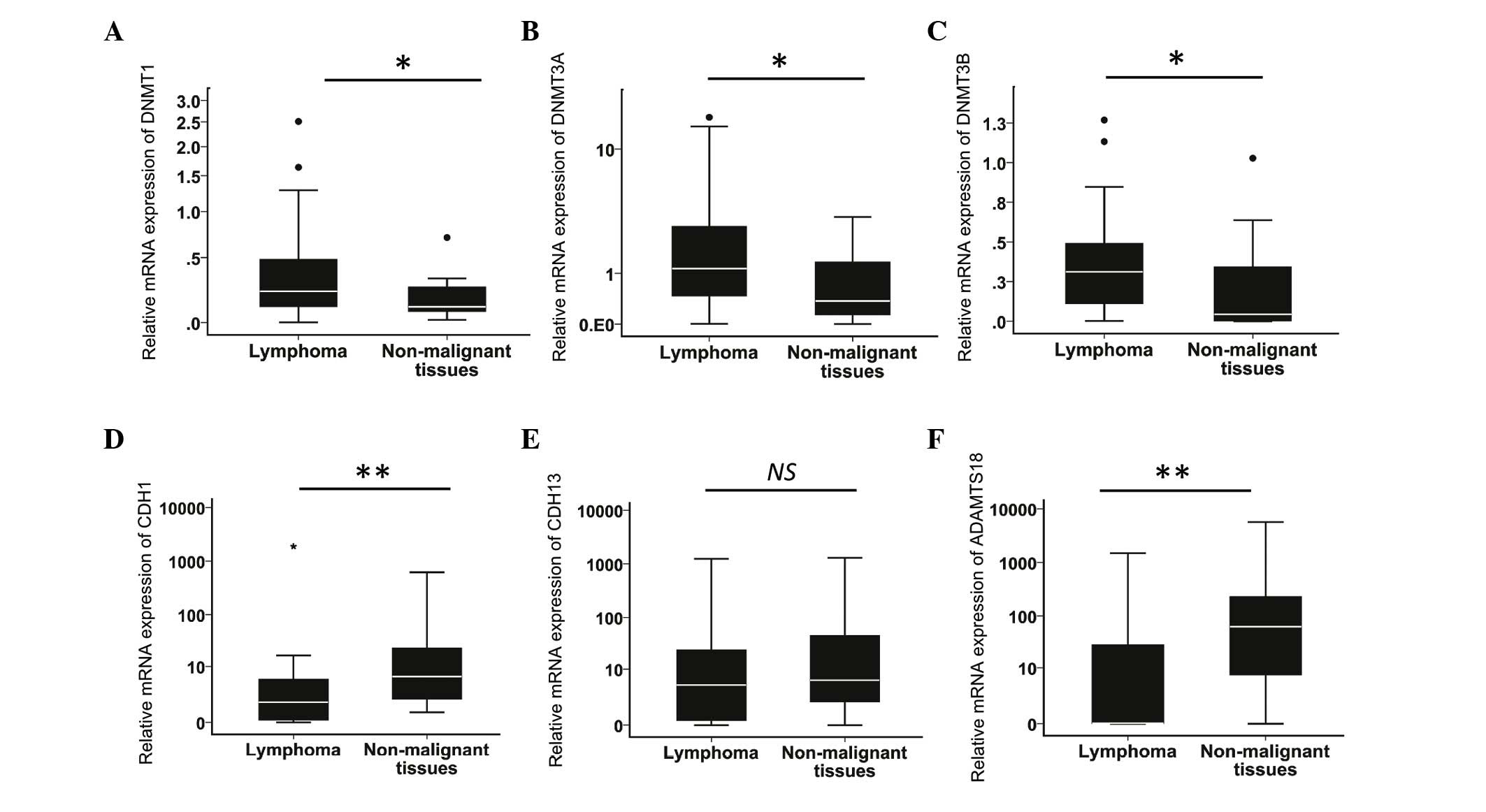 | Figure 1.Relative messenger RNA expression
levels of DNMT1, DNMT3A, DNMT3B, CDH1, CDH13
and ADAMTS18. The expression levels were determined by
reverse transcription-quantitative polymerase chain reaction in 36
cases of lymphoma (29 diffuse large B-cell lymphoma and 7 mantle
cell lymphoma) and 16 samples of non-malignant lymphoid tissues.
Relative (A) DNMT1, (B) DNMT3A, (C) DNMT3B,
(D) CDH1, (E) CDH13 and (F) ADAMTS18
expression levels (*P<0.05, **P<0.01 by the Mann-Whitney
U test). DNMT, DNA methyltransferase; CDH1,
E-cadherin; CDH13, H-cadherin; ADAMTS18, a
disintegrin and metalloproteinase with thrombospondin motifs 18;
NS, not significant. |
Methylation status of CDH1, CDH13 and
ADAMTS18, and their association with the mRNA expression levels of
DNMTs, CDH1, CDH13 and ADAMTS18
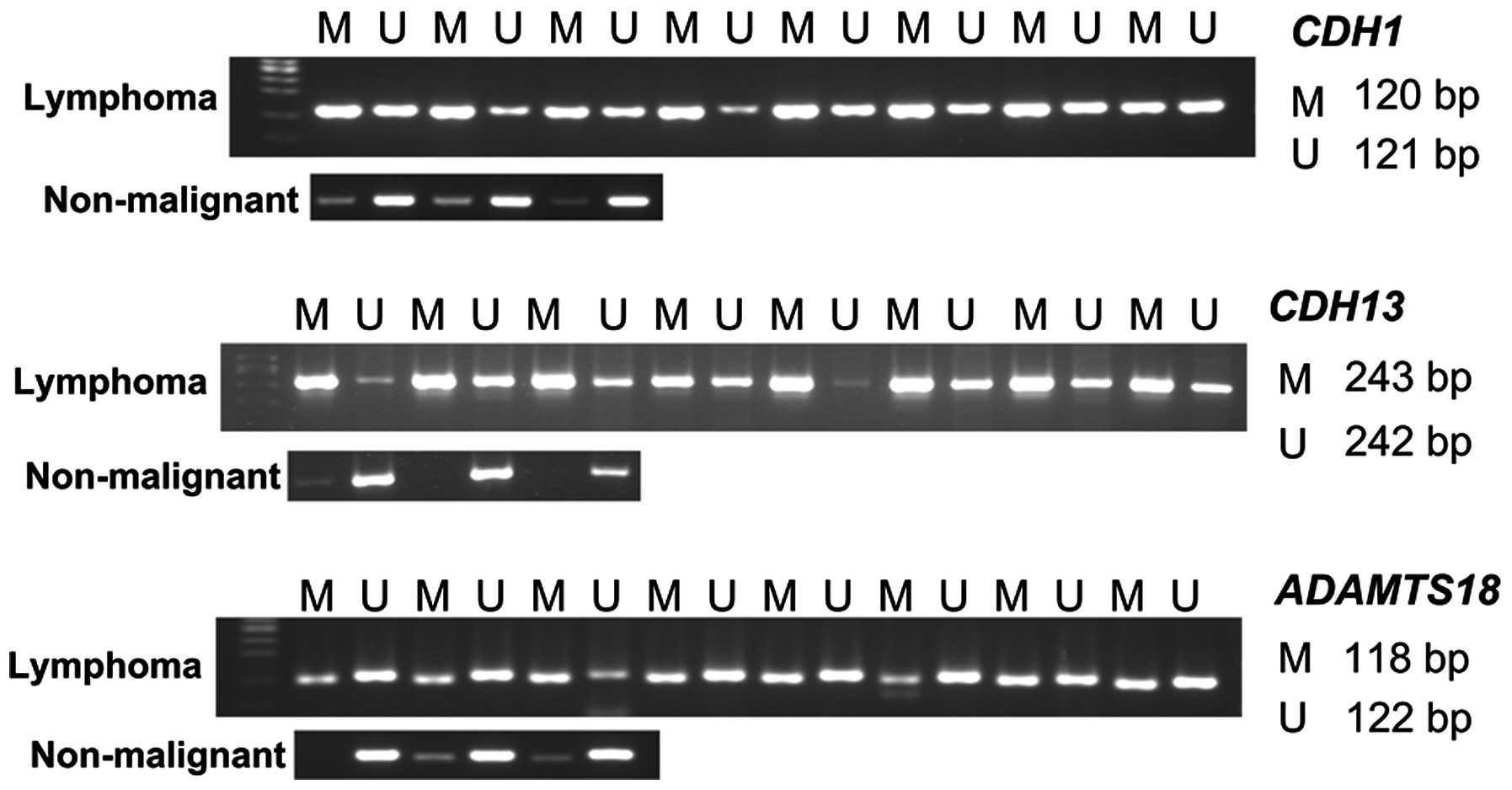
MSP was used (Fig. 2)
to investigate the promoter methylation status of CDH1,
CDH13 and ADAMT18 in 36 lymphoma and 16 non-malignant
lymphoid tissue samples. Promoter hypermethylation of CDH1,
CDH13 and ADAMTS18 was detected in 31/36 (86.1%),
33/36 (91.7%) and 28/36 (77.8%) lymphomas, respectively, and in
4/16 (25.0%), 8/16 (50.0%) and 5/16 (31.3%) non-malignant tissue
samples, respectively. The expression of CDH1 and
ADAMTS18 was significantly (P=0.008 and P=0.019,
respectively) reduced in samples with a hypermethylated promoter
compared with those with an unmethylated promoter, while
CDH13 expression displayed a non-significant reduction
(17.9%) in subjects with a methylated CDH13 promoter
(Fig. 3A). No associations were
identified between the increased levels of DNMTs mRNA and
the CDH1, CDH13 or ADAMTS18 promoter
hypermethylation (Fig. 3B-D).
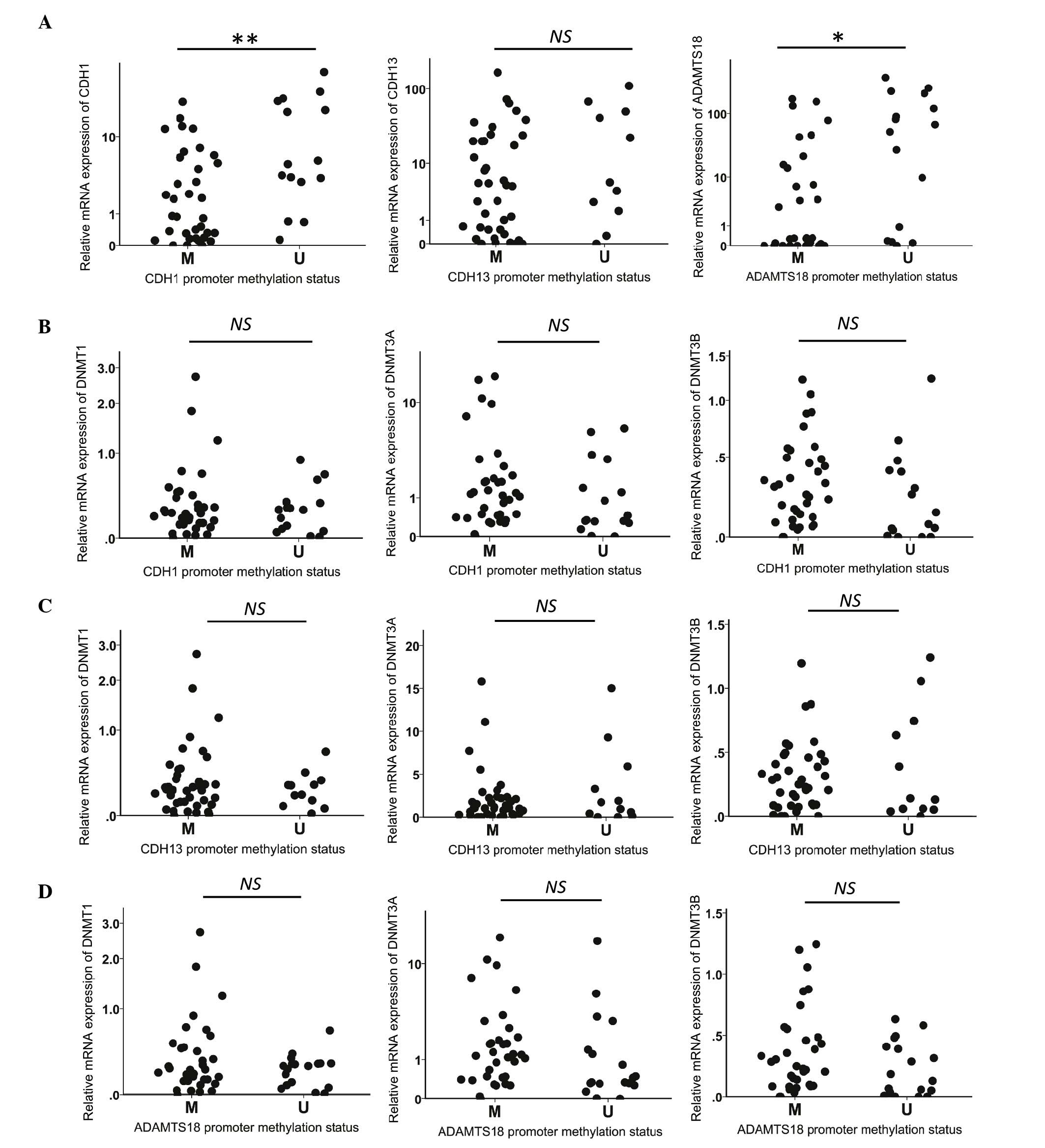 | Figure 3.(A) Relative CDH1, CDH13 and
ADAMTS18 mRNA expression levels and their corresponding
promoter methylation status. (B-D) Correlations among the
methylation status of (B) CDH1, (C) CDH13 and (D)
ADAMTS18 and the mRNA expression levels of DNMT1,
DNMT3A and DNMT3B (*P<0.05, **P<0.01 by the
Mann-Whitney U test). DNMT, DNA methyltransferase;
CDH1, E-cadherin; CDH13, H-cadherin; ADAMTS18,
a disintegrin and metalloproteinase with thrombospondin motifs 18;
NS, not significant; M, methylated; U, unmethylated; mRNA,
messenger RNA. |
To confirm that promoter methylation was responsible
for the silencing of CDH1, CDH13 and ADAMTS18
expression, methylated lymphoma cell lines were treated with
5-aza-dC, a demethylating agent, and CDH1, CDH13 and
ADAMTS18 expression was examined by RT-qPCR. Upon treatment
with 5-aza-dC, the promoter region of the CDH1, CDH13 and
ADAMTS18 genes exhibited hypomethylation, and their mRNA
expression levels were increased (Fig.
4).
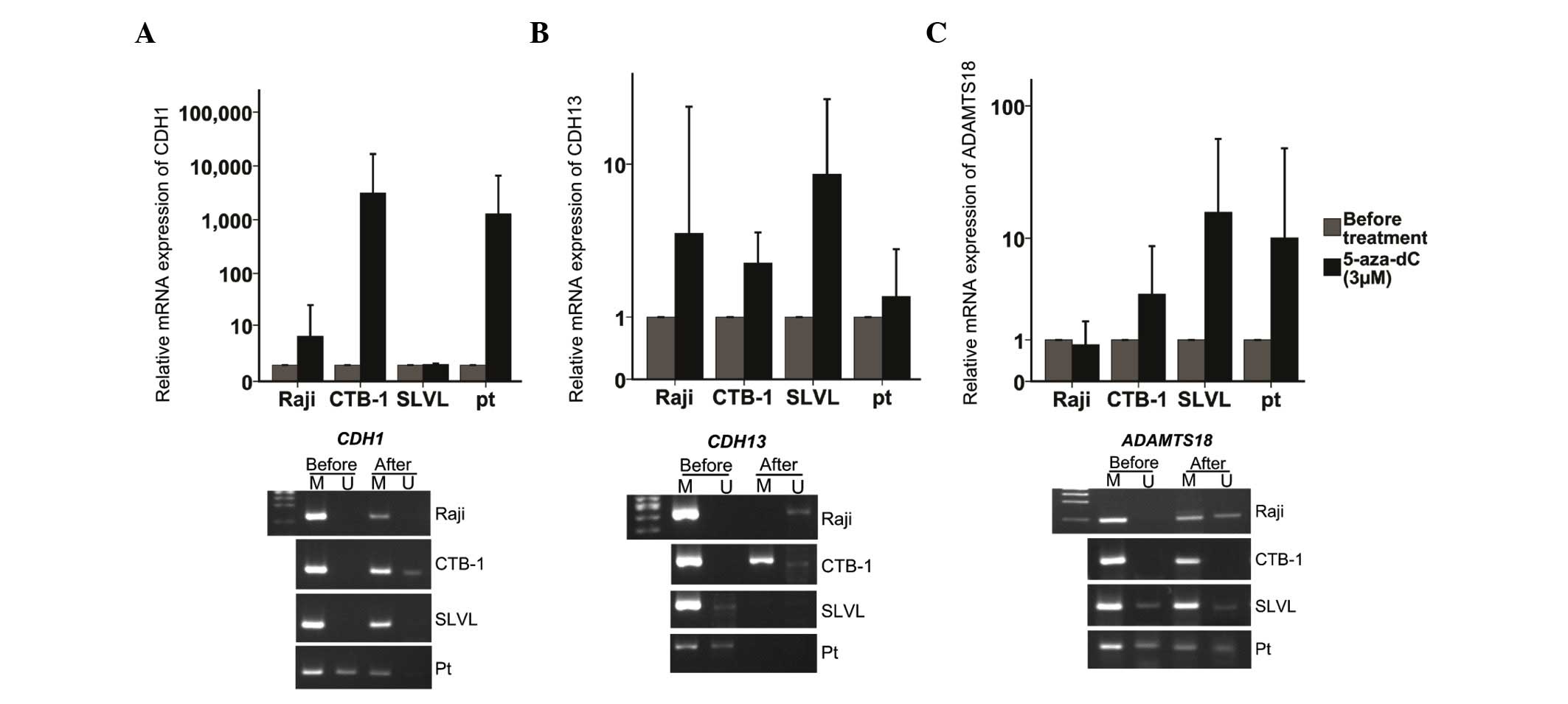 | Figure 4.Treatment of lymphoma cell lines with
5-aza-dC. (A) CDH1, (B) CDH13 and (C) ADAMTS18
messenger RNA expression levels and their corresponding promoter
methylation status prior and subsequent to the treatment of
lymphoma cell lines (Raji, CTB-1 and SLVL) and one primary cell
line from a patient with diffuse large B-cell lymphoma with
5-aza-dC at a concentration of 3 µM for three times. CDH1,
E-cadherin; CDH13, H-cadherin; ADAMTS18, a
disintegrin and metalloproteinase with thrombospondin motifs 18;
NS, not significant; M, methylated; U, unmethylated; pt, patient;
5-aza-dC, 5-aza-2′-deoxycytidine. Originally published in Blood
122: 4289, 2013. |
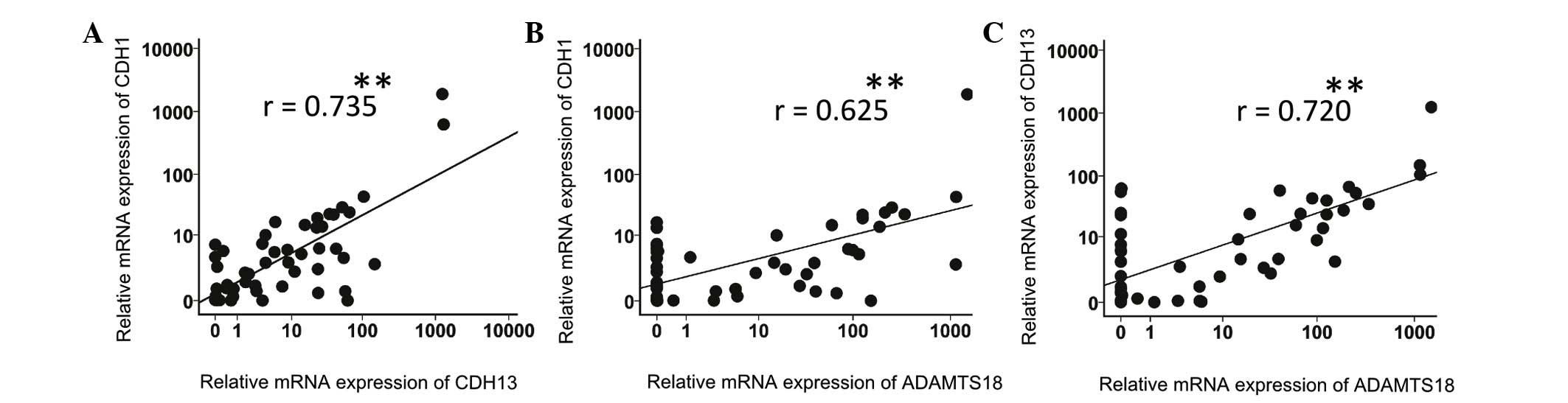
Associations between the relative mRNA
expression levels of CDH1, CDH13 and ADAMTS18
The correlations between the different tested
parameters were evaluated across the spectrum of both lymphoma and
non-malignant tissues. The CDH1 expression level exhibited a
significantly positive correlation with the CDH13 expression
level (r=0.735, P<0.01) (Fig.
5A). Furthermore, the ADAMTS18 expression level was
positively correlated with the CDH1 and CDH13
expression levels (r=0.625, P<0.01; and r=0.720,
P<0.01, respectively) (Figs. 5B and
C).
Discussion
Promoter hypermethylation and loss of function of
CDH1, CDH13 and ADAMTS18 have been reported in
various cancers and cancer cell lines (10,11,13). In
the present study, the promoter methylation of CDH1, CDH13
and ADAMTS18, which are putative TSGs located on chromosome
16q (16q22.1, 16q24.2 and 16q23.1, respectively), was investigated
using MSP, and the expression levels of DNMTs, CDH1,
CDH13 and ADAMTS18 were examined using RT-qPCR to study
whether these parameters are correlated and associated with
DNMTs in human BCL.
Methylation of the CDH1 promoter CpG islands
and consequent loss of E-cadherin expression has been reported in
multiple tumor tissues (25–31). ADAMTS18 has been reported to be
downregulated through promoter methylation in esophageal and
nasopharyngeal cancer cell lines (15). In the present study, the relative mRNA
expression levels of CDH1 and ADAMTS18 were both
significantly reduced in lymphoma samples by 36.0% with respect to
non-malignant tissues. In addition, promoter hypermethylation of
CDH1 and ADAMTS18 was detected in 31/36 (86.1%) and
28/36 (77.8%) of lymphomas, respectively. Furthermore, the
reduction of CDH1 and ADAMTS18 was significantly
associated with their corresponding hypermethylated promoters when
compared with their unmethylated promoters. Therefore, the current
results suggest that the aberrant expression of CDH1 and
ADAMTS18 and their promoter hypermethylation may be
important in lymphomagenesis.
CDH13 downregulation due to promoter
hypermethylation has been observed in various cancers (11), including BCLs (12). In the present study, the relative mRNA
expression level of CDH13 in human lymphomas was reduced by
14.4% (not significant) compared with that in non-malignant
tissues. Furthermore, methylation of CDH13 was detected more
frequently in lymphoma (91.7%) than in non-malignant tissues
(50.0%), and there was a non-significant reduction in CDH13
expression in subjects with a methylated CDH13 promoter
(17.9%). No significant differences were observed among the
expression levels of DNMTs in subjects with a methylated
CDH13 promoter compared with those with unmethylated
promoters. These non-significant results may be due to the fact
that human clinical samples are heterogeneous, with cytological
diversity and different variants that may neutralize the aberrant
expression of genes.
Genes with transcriptional inactivation due to
methylation are sensitive to DNA methylation inhibitors and can
easily be reactivated (32). The
methylation inhibitor 5-aza-dC, can be incorporated during DNA
synthesis, and reduces the capacity for DNA methylation by DNMTs,
thus reversing the methylation status of the promoters of genes
(32,33). In the present study, methylated BCL
cell lines (Raji, CTB-1, SLVL) and a primary patient cell line were
treated with 5-aza-dC. Following demethylation treatment, the
expression levels of CDH1, CDH13 and ADAMTS18
mRNA were upregulated ≥2-fold, with corresponding complete or
partial promoter hypomethylation.
By contrast, in the current surgically-resected
tumor samples, not apparent significant correlation was identified
between the methylation status of TSGs and the expression levels of
DNMTs. These findings are in agreement with previous studies
that failed to demonstrate any significant correlation between
DNMTs expression and aberrant promoter methylation of the
tested genes (5,6,34,35). This finding can be explained by the
fact that the overexpression of DNMTs is considered to be
the primary mechanism responsible for the hypermethylation of TSGs
in cancer cells, while a gain of methylation could also be
secondary to the overexpression of transcriptional repressors or to
the loss of transcriptional activators, as well as the result of an
interallelic transfer of methylation via gene pairing (6,36). Another
study (8) revealed that transgenic
overexpression or complete depletion of DNMT3B did not
affect the methylation status of HCT-116 colon cancer cells.
Furthermore, in the present study, the expression levels of
DNMTs mRNAs were significantly higher in lymphomas than in
non-malignant tissue samples. This may be attributable to
differences in the cell proliferation rate between lymphomas and
non-malignant tissues (6). Taken
together, since tumor cell lines are developed from single cells,
and therefore consist of cells with a uniform genetic composition,
it is logical that the expression of genes displayed a clearer
association with their corresponding promoter's methylation status
than with that observed in the heterogeneous tumor clinical
samples.
CDH1 and CDH13 are important cell
adhesion molecules, and alterations in their structure and function
often cause a reduction in the adhesion between tumor cells, which
may cause the detachment of cells from the primary tumors and the
acquisition of invasive and metastatic properties (10,11).
Notably, in the present study, a significant positive correlation
between the expression levels of both CDH1 and CDH13
was observed. Regulation of VE-cadherin by N-cadherin was
previously described in non-malignant human umbilical vein
endothelial cells (37). It has also
been demonstrated that N-cadherin and VE-cadherin are co-expressed
in human breast cancer (38).
N-cadherin controls the expression of VE-cadherin, and the latter
regulates the subcellular localization of N-cadherin by causing its
translocation from the cell surface (38). Since CDH13 lacks the transmembrane and
cytoplasmic regions of other cadherins, and instead uses a GPI
anchor and the interactions with specific ligands (11), we suggest that CDH1 may interact with
CDH13 and facilitate its signaling inside the cell.
In addition, the present study identified
significant positive correlations between the expression level of
ADAMTS18 and the expression levels of CDH1 and
CDH13. ADAMTSs, including ADAMTS18, are important in
mediating the degradation of extracellular matrix proteins, as well
as the ectodomain shedding of growth factors, growth factor
receptors and adhesion molecules (13). Interactions between metalloproteinases
and adhesion molecules have been reported (10,39,40). For
example, Maretzky et al (39)
demonstrated that ADAM10 contributes to E-cadherin shedding and to
cell proliferation by modulating β-catenin signaling through
E-cadherin shedding. Furthermore, ADAM15 catalyzes soluble
E-cadherin shedding, which in turn leads to its binding to the ErbB
receptor and to the stimulation of ErbB receptor signaling via
human epidermal growth factor (HER) 2 and HER3 in breast cancer
cells (40). Therefore, the positive
correlations between ADAMTS18 and CDH1 and
CDH13 mRNA in the current study may reflect the requirement
of ADAMTS for cadherin processing in the cells, and also
suggest the existence of a common factor regulating these genes,
which are all located on chromosome 16q, thus further emphasizing
the importance of this region.
Of note, as presented in Fig. 5B and C, the expression of
ADAMTS18 in replicate samples (16/52 samples, 30.8%) did not
display any correlation with the expression of either CDH1
or CDH13. The complete lack of ADAMTS18 expression
and the absence of correlation with CDH1, CDH13 or
DNMTs in these patients' samples may imply that there is a
loss of the ADAMTS18 gene locus.
Furthermore, to support our hypothesis that
chromosome 16 and cell surface membrane-located TSGs are correlated
with each other in human BCL, the present study examined the
expression of another TSG located on the same chromosome,
cyclin-dependent kinase inhibitor (CDKN) 2A (p16). It was
observed that the expression levels of our tested TSGs did not
exhibit any correlation with the CDKN2A expression level
(data not shown).
In conclusion, our findings demonstrated that the
expression levels of TSGs adjacently located at chromosome 16q
(CDH1, CDH13 and ADAMT18) are positively
correlated and frequently methylated, and that their methylation
status is not associated with the expression levels of DNMTs
in human lymphoma. Our findings suggest that aberrantly methylated
cell surface membrane TSGs located on chromosome 16q are correlated
and may be important role in the pathogenesis of human BCL.
Acknowledgements
The abstract was presented at the 55th American
Society of Hematology (ASH) Annual Meeting and Exposition December
7–10, 2013 in New Orleans, LA and published as abstract no. 21 in
Blood 122: 4289, 2013. The authors would like to thank Miss Rumiko
Koitabashi (Department of Medicine and Clinical Sciences, Graduate
School of Medicine, Gunma University, Gunma, Japan) for supporting
the present study by collecting the patients' samples. The current
study was supported in part by the National Cancer Research and
Development Fund (Tokyo, Japan, grant no. 23-A-17).
References
|
1
|
Akhavan-Niaki H and Samadani AA: DNA
methylation and cancer development: Molecular mechanism. Cell
Biochem Biophys. 67:501–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bestor TH: The DNA methyltransferases of
mammals. Hum Mol Genet. 9:2395–2402. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mizuno S, Chijiwa T, Okamura T, Akashi K,
Fukumaki Y, Niho Y and Sasaki H: Expression of DNA
methyltransferases DNMT1, 3A and 3B in normal hematopoiesis and in
acute and chronic myelogenous leukemia. Blood. 97:1172–1179. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amara K, Ziadi S, Hachana M, Soltani N,
Korbi S and Trimeche M: DNA methyltransferase DNMT3b protein
overexpression as a prognostic factor in patients with diffuse
large B-cell lymphomas. Cancer Sci. 101:1722–1730. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oue N, Kuraoka K, Kuniyasu H, Yokozaki H,
Wakikawa A, Matsusaki K and Yasui W: DNA methylation status of
hMLH1, p16 (INK4a) and CDH1 is not associated with mRNA expression
levels of DNA methyltransferase and DNA demethylase in gastric
carcinomas. Oncol Rep. 8:1085–1089. 2001.PubMed/NCBI
|
|
6
|
Sato M, Horio Y, Sekido Y, Minna JD,
Shimokata K and Hasegawa Y: The expression of DNA
methyltransferases and methyl-CpG-binding proteins is not
associated with the methylation status of p14(ARF), p16(INK4a) and
RASSF1A in human lung cancer cell lines. Oncogene. 21:4822–4829.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park HJ, Yu E and Shim YH: DNA
methyltransferase expression and DNA hypermethylation in human
hepatocellular carcinoma. Cancer Lett. 233:271–278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hagemann S, Kuck D, Stresemann C, Prinz F,
Brueckner B, Mund C, Mumberg D and Sommer A: Antiproliferative
effects of DNA methyltransferase 3B depletion are not associated
with DNA demethylation. PloS One. 7:e361252012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Strathdee G: Epigenetic versus genetic
alterations in the inactivation of E-cadherin. Semin Cancer Biol.
12:373–379. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodriguez FJ, Lewis-Tuffin LJ and
Anastasiadis PZ: E-cadherin's dark side: Possible role in tumor
progression. Biochim Biophys Acta. 1826:23–31. 2012.PubMed/NCBI
|
|
11
|
Andreeva AV and Kutuzov MA: Cadherin 13 in
cancer. Genes Chromosomes Cancer. 49:775–790. 2010.PubMed/NCBI
|
|
12
|
Ogama Y, Ouchida M, Yoshino T, Ito S,
Takimoto H, Shiote Y, Ishimaru F, Harada M, Tanimoto M and Shimizu
K: Prevalent hyper-methylation of the CDH13 gene promoter in
malignant B cell lymphomas. Int J Oncol. 25:685–691.
2004.PubMed/NCBI
|
|
13
|
Rocks N, Paulissen G, El Hour M, Quesada
F, Crahay C, Gueders M, Foidart JM, Noel A and Cataldo D: Emerging
roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie.
90:369–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Porter S, Scott SD, Sassoon EM, Williams
MR, Jones JL, Girling AC, Ball RY and Edwards DR: Dysregulated
expression of adamalysin-thrombospondin genes in human breast
carcinoma. Clin Cancer Res. 10:2429–2440. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin H, Wang X, Ying J, Wong AH, Li H, Lee
KY, Srivastava G, Chan AT, Yeo W, Ma BB, et al: Epigenetic
identification of ADAMTS18 as a novel 16q23.1 tumor suppressor
frequently silenced in esophageal, nasopharyngeal and multiple
other carcinomas. Oncogene. 26:7490–7498. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knudson AG: Two genetic hits (more or
less) to cancer. Nat Rev Cancer. 1:157–162. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
18
|
Swerdlow SH, Campo E, Harri NL, Jaffe ES,
Pileri SA, Stain H, Thiele J and Vardiman JW: WHO Classification of
Tumours of Haematopoietic and Lymphoid Tissues. 2. 4th. IARC Press;
Lyon: 2008
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sakai M, Hibi K, Koshikawa K, Inoue S,
Takeda S, Kaneko T and Nakao A: Frequent promoter methylation and
gene silencing of CDH13 in pancreatic cancer. Cancer Sci.
95:588–591. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Asiaf A, Ahmad ST, Aziz SA, Malik AA,
Rasool Z, Masood A and Zargar MA: Loss of expression and aberrant
methylation of the CDH1 (E-cadherin) gene in breast cancer patients
from Kashmir. Asian Pac J Cancer Prev. 15:6397–6403. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karpova MB, Schoumans J, Ernberg I, Henter
JI, Nordenskjold M and Fadeel B: Raji revisited: Cytogenetics of
the original Burkitt's lymphoma cell line. Leukemia. 19:159–161.
2005.PubMed/NCBI
|
|
23
|
Uchida Y, Miyazawa K, Yaguchi M, Gotoh A,
Iwase O, Ohyashiki K and Toyama K: Establishment of a novel
B-lymphoma cell line, CTB-1, with strong Pas antigen expression
having chromosomal translocation (14;22). Int J Oncol.
10:1103–1107. 1997.PubMed/NCBI
|
|
24
|
Inokuchi K, Abo J, Takahashi H, Miyake K,
Inokuchi S, Dan K and Nomura T: Establishment and characterization
of a villous lymphoma cell line from splenic B-cell lymphoma. Leuk
Res. 19:817–822. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeong DH, Youm MY, Kim YN, Lee KB, Sung
MS, Yoon HK and Kim KT: Promoter methylation of p16, DAPK, CDH1 and
TIMP-3 genes in cervical cancer: Correlation with clinicopathologic
characteristics. Int J Gynecol Cancer. 16:1234–1240. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harbst K, Staaf J, Måsbäck A, Olsson H,
Ingvar C, Vallon-Christersson J, Ringnér M, Borg A and Jönsson G:
Multiple metastases from cutaneous malignant melanoma patients may
display heterogeneous genomic and epigenomic patterns. Melanoma
Res. 20:381–391. 2010.PubMed/NCBI
|
|
27
|
Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ and
Park JY: Aberrant methylation of E-cadherin and H-cadherin genes in
nonsmall cell lung cancer and its relation to clinicopathologic
features. Cancer. 110:2785–2792. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsumura T, Makino R and Mitamura K:
Frequent down-regulation of E-cadherin by genetic and epigenetic
changes in the malignant progression of hepatocellular carcinomas.
Clin Cancer Res. 7:594–599. 2001.PubMed/NCBI
|
|
29
|
Barber M, Murrell A, Ito Y, Maia AT,
Hyland S, Oliveira C, Save V, Carneiro F, Paterson AL, Grehan N, et
al: Mechanisms and sequelae of E-cadherin silencing in hereditary
diffuse gastric cancer. J Pathol. 216:295–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ling ZQ, Li P, Ge MH, Zhao X, Hu FJ, Fang
XH, Dong ZM and Mao WM: Hypermethylation-modulated down-regulation
of CDH1 expression contributes to the progression of esophageal
cancer. Int J Mol Med. 27:625–635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qian ZR, Sano T, Yoshimoto K, Asa SL,
Yamada S, Mizusawa N and Kudo E: Tumor-specific downregulation and
methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes
correlate with aggressiveness of human pituitary adenomas. Mod
Pathol. 20:1269–1277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ling ZQ, Sugihara H, Tatsuta T, Mukaisho K
and Hattori T: Optimization of comparative expressed sequence
hybridization for genome-wide expression profiling at chromosome
level. Cancer Genet Cytogenet. 175:144–153. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murgo AJ: Innovative approaches to the
clinical development of DNA methylation inhibitors as epigenetic
remodeling drugs. Semin Oncol. 32:458–464. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Danenberg PV and Laird PW: CpG island hypermethylation in human
colorectal tumors is not associated with DNA methyltransferase
overexpression. Cancer Res. 59:2302–2306. 1999.PubMed/NCBI
|
|
35
|
Saito Y, Kanai Y, Sakamoto M, Saito H,
Ishii H and Hirohashi S: Expression of mRNA for DNA
methyltransferases and methyl-CpG-binding proteins and DNA
methylation status on CpG islands and pericentromeric satellite
regions during human hepatocarcinogenesis. Hepatology. 33:561–568.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tycko B: Epigenetic gene silencing in
cancer. J Clin Invest. 105:401–407. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luo Y and Radice GL: N-cadherin acts
upstream of VE-cadherin in controlling vascular morphogenesis. J
Cell Biol. 169:29–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rezaei M, Friedrich K, Wielockx B,
Kuzmanov A, Kettelhake A, Labelle M, Schnittler H, Baretton G and
Breier G: Interplay between neural-cadherin and vascular
endothelial-cadherin in breast cancer progression. Breast Cancer
Res. 14:R1542012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maretzky T, Reiss K, Ludwig A, Buchholz J,
Scholz F, Proksch E, de Strooper B, Hartmann D and Saftig P: ADAM10
mediates E-cadherin shedding and regulates epithelial cell-cell
adhesion, migration, and beta-catenin translocation. Proc Natl Acad
Sci USA. 102:9182–9187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Najy AJ, Day KC and Day ML: The ectodomain
shedding of E-cadherin by ADAM15 supports ErbB receptor activation.
J Biol Chem. 283:18393–18401. 2008. View Article : Google Scholar : PubMed/NCBI
|