Introduction
Renal cell carcinoma is one of the most common
malignant tumors of the urinary system. Its global incidence rate
has been increasing steadily every year and ~20–30% of patients
cannot be treated with radical operation because they have distant
metastases at the time of diagnosis (1). Renal cell carcinoma is known for being
insensitive to chemo- and radiotherapy. Sunitinib (sunitinib
malate) targeted therapy, represented by multi-target tyrosine
kinase inhibitors, has good clinical effects on treating advanced
renal cell carcinoma and has been applied as a first-line treatment
drug (2). However, previous findings
showed the effective rate to be only 60–75%, leaving many patients
to undergo ineffective treatment with added secondary adverse
reactions (3). A study has found that
individual single nucleotide polymorphisms (SNPs) can influence
signaling pathways such as MAPK/ERK/STAT3 and determine whether
sunitinib therapy will be effective against renal cell carcinoma
(4). The tumor in renal cell
carcinoma is generally highly vascular and notably expresses
vascular endothelial growth factors (VEGFs) and their receptors
(5).
The present study focused on specific SNPs, namely
VEGFR1, VEGFR2 and VEGFR3, which hypothetically have an impact on
the MAPK/ERK/STAT3 signaling pathways and influence sunitinib renal
cell carcinoma treatment, with the hope of contributing to a better
understanding of the pathogenesis of the cancer and allow for more
effective treatment adjustments.
Subjects and methods
Patients and methods
A total of 68 patients treated for advanced renal
cell carcinoma in our hospital from January, 2014 to July, 2015
participated in the present study. The following cases were
excluded: Patients with kidney failure, history of surgical and
chemoradiotherapy treatments, additional malignant tumors, severe
condition, predicted survival <12 months, sunitinib course of
<3 months, and incomplete data. The Ethics Committee of the
First People's Hospital of Yunnan Province approved the study and
the patients signed informed consent. There were 40 male and 28
female cases, aged between 46 and 72 years (with an average of
62.3±14.5 years); the disease had been present for 1–5 months (with
an average of 2.3±1.2 months); the Karnofski performance status
scoring ranged from 62 to 86 points (74.5±8.9 points in average);
and metastases were present in the following organs: 23 lung
metastases, 19 lymphatic metastases, 10 osseous metastases, 10
hepatic and adrenal metastases and 6 other metastases.
The dosage plan of sunitinib for each patient was 50
mg/day taken orally for 4 weeks, drug withdrawal for 2 weeks, and
then restarting the cycle for ≥3 months.
Assessment of response to therapy
The RECIST 1.0 standard was used to evaluate the
tumor lesions, dividing radiological results into: Complete
response (CR), partial response (PR), stable disease (SD) and
progression of disease (PD) and evaluating every 6 weeks (1 cycle).
Adverse effects were evaluated based on the NCI-CTC AE3.0 standard.
Progression-free survival (PFS) includes the time from the
beginning of treatment until PD or death and overall survival (OS)
is the time from the beginning of treatment to death.
Testing index and method
Polymerase chain reaction-restriction fragment
length polymorphisms (PCR-RFLPs) were used to determine candidate
nucleotide polymorphism loci (VEGFR1, VEGFR2 and VEGFR3), and
western blotting was performed to test the p-MAPK/ERK/STAT3 protein
expression levels.
Peripheral venous blood (5 ml) was drawn from each
patient after overnight fasting of ≥8 h and stored at −20°C. QIAamp
DNA kit was used to extract the DNA and protease K was used for
sample digestion. An equilibrated-phenol extraction method was used
to purify concentrated DNA; the genetic typing was accomplished in
a double-blind fashion by two people. The SNPs VEGFR1, VEGFR2 and
VEGFR3 were selected for sequencing. The screening standards were
as follows: Gene frequency for Chinese people >5%, linkage
disequilibrium principle of didymous SNP loci and related
coefficient r2 >0.8.
The reagents for PCR-RFLP included TaqDNA
polymerase, dNTPs, and a DNA fragment length standard (PCR
markers), were purchased from Nanjing KeyGen Biotech Co., Ltd.,
(Nanjing, China); a restriction enzyme was purchased from Fermentas
(Glen Burnie, MD, USA). The Primer Premier 5.0 software was used
for primer design (Premier Biosoft International, Palo Alto, CA,
USA) and primers were synthesized by Shanghai Invitrogen
Biotechnology Co., Ltd. (Shanghai, China) (Table I). MJ-PTC200 type PCR amplification
apparatus was used with each test tube reaction containing 10 µl
(including 50 ng genome DNA in 5 µl, 2X Taq PCR Mix, 0.15 µl of 10
pmol/µl upstream and downstream primers, 4.2 µl ddH2O).
The PCR reaction protocol was programmed as: Initial
pre-denaturation step at 94°C for 5 min, followed by 34 cycles of a
denaturing step at 94°C for 30 sec, an annealing step at 59°C for
40 sec, and an extension 72°C for 45 sec, and a final extension at
72°C for l0 min. Then, 2% agarose gel electrophoresis was prepared
to observe the PCR product. The prepared enzymatic digestion system
included: 5 units of enzyme, l.0 µl 1X buffer, 3.5 µl
ddH2O, and 5 µl PCR reaction product. The mixtures were
incubated at 37°C thermostat overnight and the next morning a 3%
sepharose gel was electrophoresed to visualize the enzymatic
digestion results and identify the genotypes under a UV lamp.
 | Table I.Restriction enzyme used in the
PCR-RFLP method. |
Table I.
Restriction enzyme used in the
PCR-RFLP method.
| Gene name | Restriction
enzyme | Primers 5′-3′ |
|---|
| VEGFR1 |
|
|
|
rs664393 | SciI |
F-GACTAAACACCCCTCCAGCA |
|
|
|
R-TGTCAGCATTGTCCTTCTGC |
| VEGFR2 |
|
|
|
rsl870377 | AluI |
F-TTTCCTCCCTGGAAGTCCTC |
|
|
|
R-GGCTGCGTTGGAAGTTATTT |
|
rs7667298 | Hpy8I |
F-ATCCTTGGTCACTCCGGTTT |
|
|
|
R-TGCTGTGCTTTGGAAGTTCA |
| VEGFR3 |
|
|
|
rs448012 | SatI |
F-GAGGTTGACCACGTTGAGGT |
|
|
|
R-TTCAGAGCCGAGGGACCA |
|
rs72816988 | SsiI |
F-TGTGGGGGCTGTTCTGTATT |
|
|
|
R-ACCTCTGCTCCCTTCTCCTC |
For western blotting, RIPA cell lysis buffer and
PMSF (protease inhibitor) were used to extract proteins (both
reagents were from Shenneng Bocai Biotechnology Co., Ltd.,
Shanghai, China); the Bradford method was used to test the protein
sample concentrations (kit from R&D Systems, Inc., Minneapolis,
MN, USA); the samples were loaded onto 5X SDS-PAGE electrophoresis
gels (loading buffer from Beyotime Institute of Biotechnology,
Shanghai, China) and run; and Coomassie brilliant blue (from
Beijing Zhongshan Co., Beijing, China) was used to stain the gels.
After membrane transfer; the immunoreaction was set up. Briefly,
the PVDF membrane in a glass container was soaked with 5% blocking
buffer, and agitated at 25°C for 1 h. The primary mouse anti-human
monoclonal antibodies (Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) were added to the buffer at appropriate concentrations [p-MAPK
(cat. no. SC-6802) was added at a 1:2,000 concentration, p-ERK
antibody (cat. no. SC-154) at a 1:300 concentration, p-STAT3 (cat.
no. SC-7179) at a 1:1,000 concentration and β-actin (cat. no.
SC-47778) at a 1:500 concentration] and were stored at 4°C
overnight. The following day, the PVDF membranes were washed three
times in PBST for 10 min. Blocking buffer was used to prepare
secondary antibody goat anti-mouse IgG marked by horseradish
peroxidase (Santa Cruz Biotechnology, Inc.; cat. no. SC-2054) at a
1:1,000 concentration, and the PVDF membranes were soaked at 37°C
on a shaking platform for 2 h. Finally, after washing the membranes
three times in PBST, the blots were visualized by chemiluminescence
(ECL fluorescence detection kit from Beyotime Institute of
Biotechnology), and the results were scanned into a computer
through a gel imager and analyzed using Quantity One 4.4.0
software. The experiments were done in triplicate and the values
obtained are averages of three experiments. After processing,
reported values represent the average of OD value ratios between
the target protein and the β-actin protein bands for
normalization.
Statistical analysis
Data were processed through SPSS 19.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Measurement data were
shown as mean value ± standard deviation and comparison among
groups was through t-test. Enumeration data are shown as cases or
percentages and comparison among groups were carried out using the
(correction) χ2 test. The logistic regression method was
used to analyze the correlation between SNPs and the effectiveness
of sunitinib-targeted therapy on renal cell carcinoma and the
step-back technique was used for screening. The comparison of PFS
and OS was analyzed by the Kaplan-Meier method; P<0.05 was
considered to indicate a statistically significant difference.
Results
SNP sequencing results
The elapsed time before the follow-up visits ranged
from 6 to 23 months, and the median time was 15 months. Of the 68
patients, there were 16 CR cases, 29 PR cases and 23 SD and PD
cases. Tested SNP mutation sites and mutation rates are shown in
Table II. Mutation rates of
rsl870377 and rs448012 loci in the CR+PR group are lower than those
in the SD+PD group while there are no differences for the other 3
loci.
 | Table II.SNP sequencing results. |
Table II.
SNP sequencing results.
| Gene type | Locus mutation | Gene frequency of
minor allele | Mutation rate [case
(%)] | CR+PR group
(n=45) | SD+PD group
(n=23) | χ2 | P-value |
|---|
| VEGFR1 |
|
|
|
|
|
|
|
|
rs664393 | G>A | 0.244 | 13 (19.1) | 9
(20.0) | 4
(17.4) | 0.000 | 1.000 |
| VEGFR2 |
|
|
|
|
|
|
|
|
rsl870377 | A>T | 0.467 | 31 (45.6) | 15 (33.3) | 16 (69.6) | 8.055 | 0.005 |
|
rs7667298 | C>T | 0.284 | 15 (22.1) | 8
(17.8) | 7
(30.4) | 1.418 | 0.234 |
| VEGFR3 |
|
|
|
|
|
|
|
|
rs448012 | C>G | 0.483 | 29 (42.6) | 14 (31.1) | 15 (65.2) | 7.239 | 0.007 |
|
rs72816988 | G>A | 0.079 | 7
(10.3) | 3 (6.7) | 4
(17.4) | 0.912 | 0.340 |
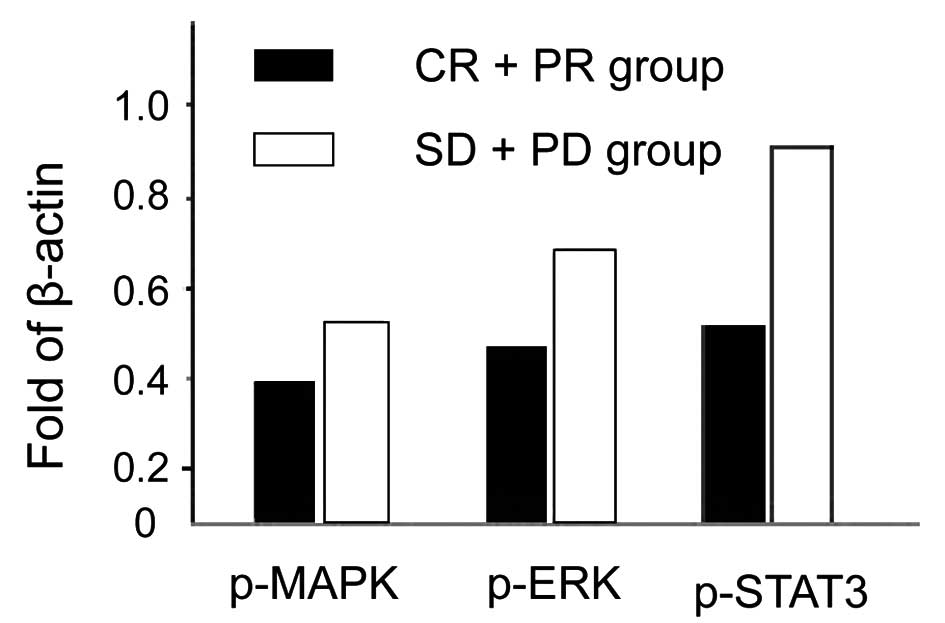
Comparison of p-MAPK/ERK/STAT3
expression levels
The expression levels for p-MAPK, p-ERK and p-STAT3
in the the CR+PR group are lower than those in the SD+PD group and
the differences are of statistical significance (P<0.05;
Fig. 1).
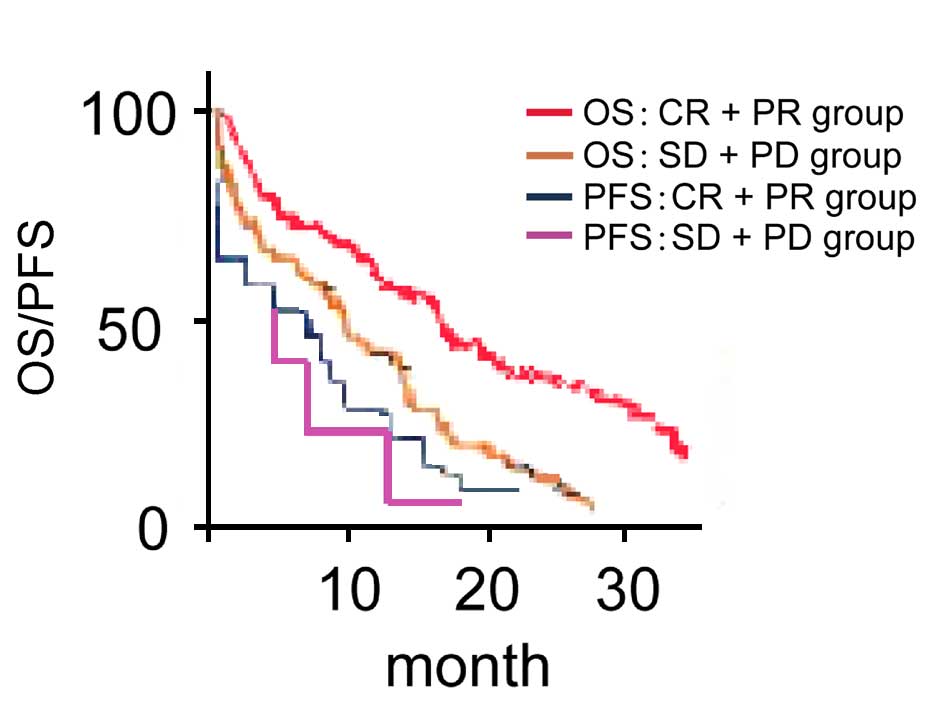
Comparison of PFS and OS in different
reaction groups
Median PFS and OS in CR+PR group are significantly
higher than those in SD+PD group (comparing 9.6 months with 6.2
months, χ2=8.924, P<0.001; comparing 17.5 months with
11.3 months, χ2=10.548, P<0.001; Fig. 2).
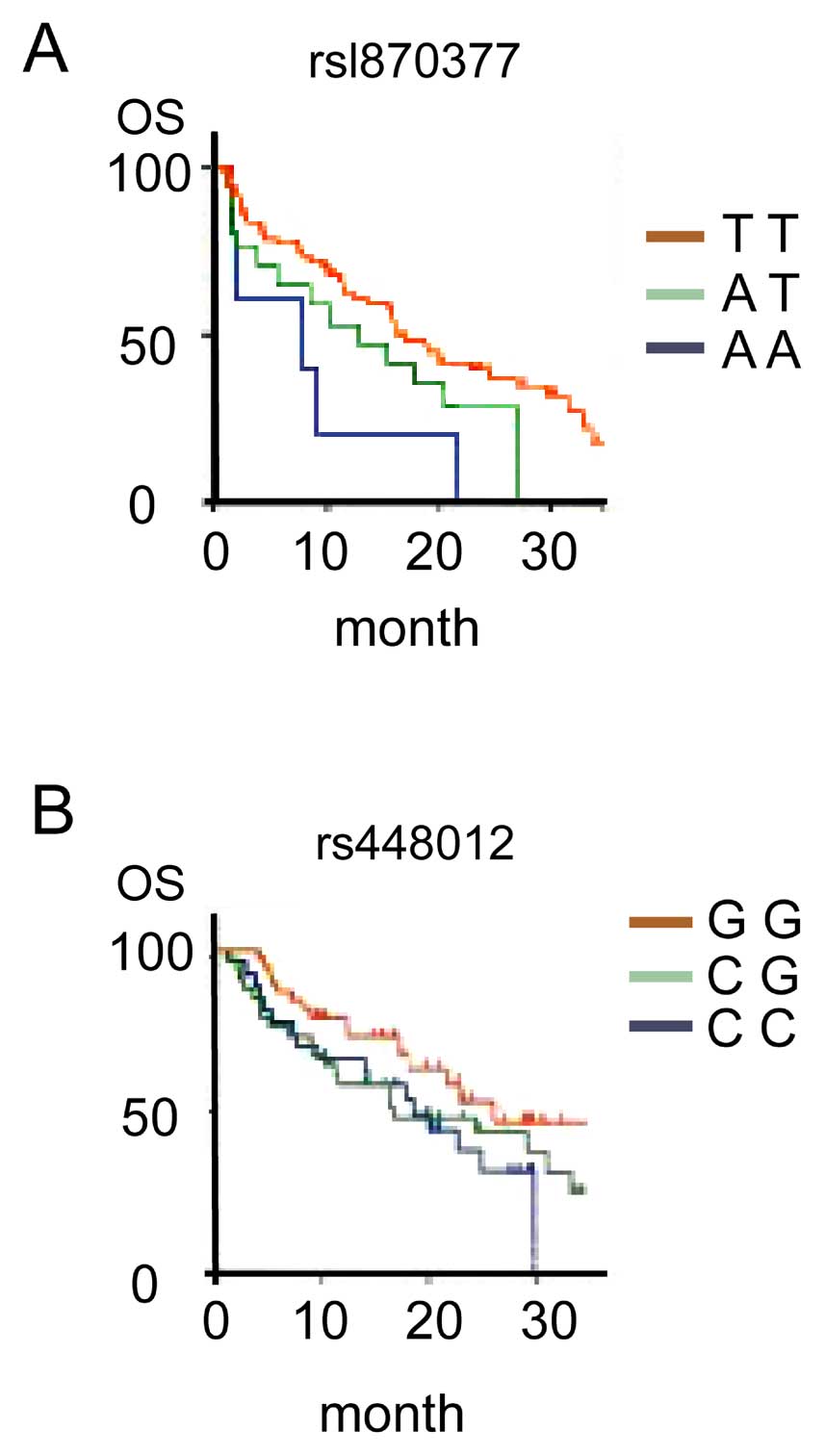
Association between SNPs and OS
The median OS of the rsl870377 genotype TT was
obviously higher than that for the genotype AA, and the median OS
of rs448012 genotype GG was higher than that for the genotype CC.
The differences are of statistical significance (comparing 15.8
months with 9.5 months, χ2=16.432, P<0.001; comparing
24.3 months with 17.2 months, χ2=12.623, P<0.001;
Fig. 3).
Correlation between SNP and
effectiveness analyzed by logistic regression method
Taking the abovementioned 5 SNPs as independent
variables and the effectiveness of CR+PR as the dependent variable,
it was concluded by a multi-factor logistic regression model that
rsl870377 and rs448012 are closely associated with the
effectiveness of sunitinib-targeted therapy on renal cell
carcinoma, and that A mutation of rsl870377 and C mutation of
rs448012 are independent risk factors that influence treatment
efficacy (Table III).
 | Table III.Analyzing correlation between SNP and
OS with logistic regression method. |
Table III.
Analyzing correlation between SNP and
OS with logistic regression method.
| SNPs | Β | Wald | P-value | OR | 95% CI |
|---|
| rs664393 | 0.321 | 1.302 |
0.616 | 0.427 | −0.325–2.302 |
| rsl870377 | 0.108 | 6.957 | <0.001 | 3.526 |
2.852–5.629 |
| rs7667298 | 0.254 | 2.625 |
0.938 | 0.854 |
0.232–3.935 |
| rs448012 | 0.163 | 7.854 | <0.001 | 4.113 |
3.593–5.942 |
| rs72816988 | 0.096 | 1.429 |
0.532 | 0.322 | −0.528–2.534 |
Discussion
Sunitinib is able to block vascular VEGF receptors,
platelet-derived growth factor receptors (PDGFR-α and PDGFR-β), the
stem cell factor receptor (C-kitR), FMS-like tyrosine kinase-3, the
type I colony stimulating factor receptor and the neurotrophin
receptor derived from neural glial cells, strongly inhibiting
proliferation of tumor cells and exerting an anti-angiogenesis
effect (6,7). In addition, sunitinib is able to inhibit
tumor cell proliferation directly by inhibiting several signal
transduction pathways such as Ras/Raf/MEK/ERK and MAPK/ERK/STAT3
(8,9);
directly inducing mechanisms such as tumor cell apoptosis (10), leading to a multi-targeted treatment
on advanced renal cell carcinoma. Sunitinib has been approved by
the FDA in the USA and by the European Commission to be used in the
treatment of advanced renal cell carcinoma.
Most molecular-targeted drug treatments have some
positive effects but bear low complete remission rates, which may
be due to SNPs (11). SNPs are able
to influence functions by modifying gene structures and may lead to
increased risks of developing tumors (12). It has been shown that the function of
genes related to angiogenesis such as VEGFA and VEGFR can be linked
to risks of renal carcinoma due to adjustments in the expression
levels of other genes (13). A study
found that the 460th gene polymorphism of VEGF is a risk factor for
renal carcinoma (14). Another study,
using a genome-wide association in European population, found three
loci related to susceptibility to renal carcinoma, one of which is
in the 2p21 region (rs7579899) and encodes the endothelial PAS
constitutive protein 1 (EPAS1, that is HIF-2α) (15). Nevertheless, related studies have not
found the same results in Chinese individuals (16), showing how genetic structures can
differ for different populations.
Among the population of patients with advanced renal
cell carcinoma in the present study, the mutation rates for VEGFR2
(rsl870377) and VEGFR3 (rs448012) were the highest. However, the
same mutation rates in the subgroup of CR+PR patients were lower
than those in the SD+PD subgroup. Also, the relative expression
levels of p-MAPK, p-ERK and p-STAT3 in the better response group
were significantly lower. Moreover, the median OS for the TT
genotype at rsl870377 was higher than that for the AA genotype, and
the median OS in the GG genotype for rs448012 was higher than that
for the CC genotype. Statistical analyses revealed the association
of these phenotypes to the effectiveness of treatment was
confirmed. It is clear from the present study that VEGFR SNPs can
control the MAPK/ERK/STAT3 signaling pathway protein expression and
thereby influence the effectiveness of sunitinib-targeted therapy.
This opens up possibilities for a new line of therapeutic
targets.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zimmermann K, Schmittel A, Steiner U,
Asemissen AM, Knoedler M, Thiel E, Miller K and Keilholz U:
Sunitinib treatment for patients with advanced clear-cell
renal-cell carcinoma after progression on sorafenib. Oncology.
76:350–354. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Motzer RJ, Hutson TE, Tomczak P,
Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik
C, Kim ST, et al: Sunitinib versus interferon alfa in metastatic
renal-cell carcinoma. N Engl J Med. 356:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamamoto K, Mizumoto A, Nishimura K, Uda
A, Mukai A, Yamashita K, Kume M, Makimoto H, Bito T, Nishigori C,
et al: Association of toxicity of sorafenib and sunitinib for human
keratinocytes with inhibition of signal transduction and activator
of transcription 3 (STAT3). PLoS One. 9:e1021102014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Finley DS, Pantuck AJ and Belldegrun AS:
Tumor biology and prognostic factors in renal cell carcinoma.
Oncologist. 16:(Suppl 2). 4–13. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rock EP, Goodman V, Jiang JX, Mahjoob K,
Verbois SL, Morse D, Dagher R, Justice R and Pazdur R: Food and
Drug Administration drug approval summary: Sunitinib malate for the
treatment of gastrointestinal stromal tumor and advanced renal cell
carcinoma. Oncologist. 12:107–113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Motzer RJ, Michaelson MD, Redman BG, Hudes
GR, Wilding G, Figlin RA, Ginsberg MS, Kim ST, Baum CM, DePrimo SE,
et al: Activity of SU11248, a multitargeted inhibitor of vascular
endothelial growth factor receptor and platelet-derived growth
factor receptor, in patients with metastatic renal cell carcinoma.
J Clin Oncol. 24:16–24. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miyake H, Muramaki M, Imai S, Harada KI
and Fujisawa M: Changes in renal function of patients with
metastatic renal cell carcinoma during treatment with
molecular-targeted agents. Target Oncol. 11:329–335. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Korashy HM, Al-Suwayeh HA, Maayah ZH,
Ansari MA, Ahmad SF and Bakheet SA: Mitogen-activated protein
kinases pathways mediate the sunitinib-induced hypertrophy in rat
cardiomyocyte H9c2 cells. Cardiovasc Toxicol. 15:41–51. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi WH, Bian YH, Song XH and Wu JC:
Coadministration of sorafenib with adriamycin inhibits cell
proliferation in hepatocellular carcinoma cells HepG2. Prog Modern
Biomed. 24:4845–3848. 2011.
|
|
11
|
Diekstra MH, Belaustegui A, Swen JJ, Boven
E, Castellano D, Gelderblom H, Mathijssen RH, García-Donas J,
Rodríguez-Antona C, Rini BI, et al: Sunitinib-induced hypertension
in CYP3A4 rs4646437 A-allele carriers with metastatic renal cell
carcinoma. Pharmacogenomics J. 26:2–4. 2016.
|
|
12
|
Diekstra MH, Swen JJ, Boven E, Castellano
D, Gelderblom H, Mathijssen RH, Rodríguez-Antona C, García-Donas J,
Rini BI and Guchelaar HJ: CYP3A5 and ABCB1 polymorphisms as
predictors for sunitinib outcome in metastatic renal cell
carcinoma. Eur Urol. 68:621–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boers-Sonderen MJ, Desar IM, Fütterer JJ,
Mulder SF, De Geus-Oei LF, Mulders PF, Van Der Graaf WT, Oyen WJ
and Van Herpen CM: Biological Effects After Discontinuation of
VEGFR Inhibitors in Metastatic Renal Cell Cancer. Anticancer Res.
35:5601–5606. 2015.PubMed/NCBI
|
|
14
|
Bruyère F, Hovens CM, Marson MN, d'Arcier
BF, Costello AJ, Watier H, Linassier C and Ohresser M: VEGF
polymorphisms are associated with an increasing risk of developing
renal cell carcinoma. J Urol. 184:1273–1278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Purdue MP, Ye Y, Wang Z, Colt JS, Schwartz
KL, Davis FG, Rothman N, Chow WH, Wu X and Chanock SJ: A
genome-wide association study of renal cell carcinoma among African
Americans. Cancer Epidemiol Biomarkers Prev. 23:209–214. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Harten SK, Shukla D, Barod R, Hergovich A,
Balda MS, Matter K, Esteban MA and Maxwell PH: Regulation of renal
epithelial tight junctions by the von Hippel-Lindau tumor
suppressor gene involves occludin and claudin 1 and is independent
of E-cadherin. Mol Biol Cell. 20:1089–1101. 2009. View Article : Google Scholar : PubMed/NCBI
|