Introduction
Acute lymphoblastic leukemia (ALL) is the most
common malignancy in the pediatric population. Over the past
decades, with the progress in risk classification schemes and
frontline protocol design, the overall survival (OS) in childhood
ALL has exceeded 80% with contemporary treatment regimens (1). However, 15–20% of children with ALL have
disease relapse (2). ALL is a
heterogeneous disease at the cytogenetic and genetic levels,
numerous acquired genetic abnormalities have been described in
patients with ALL, including chromosomal translocations,
aneuploidy, deletions, and amplifications (3). Certain genetic abnormalities, such as
breakpoint cluster region/Abelson murine leukemia viral oncogene
homolog 1 (BCR/ABL1) and ETS variant 6-Runt-related transcription
factor 1, are well-established prognostic factors (4). However, the identification and
understanding of novel genetic abnormalities remains important, in
order to optimize and refine the risk-stratification strategy and
individualized therapy, particularly for the high-risk and relapsed
patients.
ETS transcription factors are key in cell
development, differentiation, proliferation, apoptosis and tissue
remodeling (5). As a member of the
ETS family, the ETS-related gene (ERG) is preferentially and
strongly expressed in the immature B- and T-lymphoid lineages, in
addition to myeloid lineage cells (6). ERG has been demonstrated to act as a
proto-oncogene that can transform NIH3T3 cells and give rise to
tumors in nude mice (7). ERG gene
rearrangements have been identified in Ewing sarcoma and prostate
cancer (8,9). Previous studies suggest that ERG
overexpression is associated with inferior clinical outcome
(10,11) and is an independent prognostic factor
in cytogenetically normal acute myeloid leukemia (AML) (12–14).
Furthermore, in acute T cell-lymphoblastic leukemia (T-ALL)
patients, a high level of ERG expression has been associated with
poor relapse-free survival (RFS) (15,16). In
childhood AML, the impact of ERG expression level is somewhat
controversial: Pigazzi et al (17) suggested that high ERG expression was
an independent unfavorable prognostic marker for childhood myeloid
leukemia, whereas the study by Hermkens et al did not find
such correlation (18).
However, to the best of our knowledge, no studies
have yet investigated the prognostic correlation of ERG expression
in childhood ALL. The present study analyzed ERG mRNA expression
levels in 119 childhood patients with newly diagnosed ALL, and
aimed to validate their prognostic significance in pediatric ALL
patients.
Materials and methods
Patients and treatment
The expression levels of ERG in 119 bone marrow (BM)
samples from patients with newly diagnosed ALL (median age, 6
years; range, 1–14 years; 76 males and 43 females) were
retrospectively analyzed between January 2007 and December 2010 at
the Children's Hospital of Zhejiang University School of Medicine
(Hangzhou, China). The diagnostic criteria of ALL was based on
morphology, immunology, and cytogenetics, as described previously
(19). Overall, there were 91 B
cell-ALL (B-ALL) patients and 28 T-ALL patients. Furthermore, 36 BM
samples from children (median age, 6 years; range, 1–14 years; 23
males and 13 females) without history of malignancies were selected
and served as controls. Written informed consent was obtained from
the parents or guardians. All treatments were administered in
accordance with the Declaration of Helsinki and approved by the
ethics review board of the Children's Hospital of Zhejiang
University.
Patients received treatment with the modified
National Protocol of Childhood ALL in China 1997 at the hospital,
which has been described in our previous study (20). Based on the modified National Cancer
Institute criteria, patients were classified as low-risk (LR)
group, intermediate risk (IR) group or high-risk (HR) group
(20). Patients with one of the
following features were considered as IR: i) Age, >10 years old;
ii) initial white blood cell (WBC) counts >50×109/l
but <100×109/l; iii) central nervous system leukemia
and/or testicular leukemia at diagnosis; iv) T lineage ALL; v)
hypodiploidy (<45 chromosomes). Patients with aged <1 year
old, or initial WBC counts ≥100×109/l, or with t(9;22),
t(4;11) or BCR/ABL1, MLL/AF4 (the fusion of the MLL gene on
chromosome 11 and the AF4 gene on chromosome 4) fusion genes, or
poor response to prednisone or not achieving complete remission
(CR) on day 42 were considered HR. Otherwise, patients without any
features as described above were assigned to the LR group.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Mononuclear cells from BM samples were isolated by
Ficoll gradient centrifugation and cryopreserved in liquid
nitrogen. Total RNA was extracted using High Pure RNA Isolation Kit
(Roche Diagnostics GmbH, Mannheim, Germany) following the
manufacturer's protocols. First-strand cDNA was synthesized from
total RNA with reverse transcriptase and random primers, using the
ReverTra Ace qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan). Samples
were selected based on the quality and quantity of RNA.[optical
density (OD)260/OD280 ratio, 1.8–2.0].
qPCR was performed with a 96-well optic plate on a
StepOnePlus™ Real-time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), with GAPDH as an
endogenous control. Every sample was tested in triplicate. The
comparative cycle threshold (CT) method was used to determine the
relative expression levels of ERG to GAPDH, using the mean of ΔCT
from three replicates and expressed as 2µ(ΔCT)
(ΔCT=GAPDH-ERG), as previously described (10). Each reaction mixture consisted of 2 µl
cDNA, 12.5 µl SYBR Green PCR Master mix (Toyobo Co., Ltd.), 1 µl of
ERG primers (5 nmol/ml) or 1 µl of GAPDH primers (5 nmol/ml) and
deionized water to a total volume of 25 µl. The primers of ERG and
GAPDH were as described in previous studies (15,21), with
the sequences as follows: 5′-CACGAACGAGCGCAGAGTTA-3′ for ERG1;
5′-CTGCCGCACATGGTCTGTAC-3′ for ERG2; 5′-ATGGGGAAGGTGAAGGTCG-3′ for
GAPDH1; 5′-GGGTCATTGATGGCAACAATATC-3′ for GAPDH2. PCR amplification
was performed under the following conditions: First reaction, 94°C
for 5 min, followed by 30 cycles at 94°C for 30 sec, 55°C for 30
sec and 72°C for 1 min; second reaction, 94°C for 5 min, followed
by 35 cycles at 94°C for 30 sec, 58°C for 30 sec and 72°C for 1
min, using Taq DNA Polymerase from Toyobo (Osaka, Japan).
In addition, the Ikaros 6 (IK6) variant of Ikaros
family zinc finger protein 1 gene was detected by nested PCR using
primers described in a previous study (22). All PCR products were size-fractionated
by electrophoresis on a 1.5% agarose gel and stained with ethidium
bromide using the Image Lab™ Software of Gel Doc XR (Bio-Rad
Laboratories Inc., Hercules, CA, USA).
Statistical analysis
The median value of all measurements was selected to
define the low and high ERG expressing patients. Patients were
classified as having high ERG if they had expression values above
the median value of all patients and as low ERG if they had ERG
expression values below the median value. Comparisons of baseline
clinical variables across groups were conducted using the
χ2 test and the nonparametric Mann-Whitney U test for
categorical and continuous variables, respectively. Student's
t-test for independent samples was selected to perform mean
comparisons between distinct groups.
Estimated probabilities of RFS and OS were
calculated using the Kaplan-Meier method, and the log-rank test
evaluated differences between survival distributions. RFS was
determined from the date of achieving CR to relapse, any mortality
or last contact. OS was measured from the date of first diagnosis
until the date of mortality regardless of cause, censoring for
patients alive at last follow-up. The last follow-up was September
2014. Multivariable proportional hazards models were constructed
for RFS and OS, using a stepwise forward selection procedure. The
following covariates were included in the full model: Age (≤1 year,
≥10 years vs. 1–10 years), WBC counts (≥50×109/l vs.
<50×109/l); risk (high vs. low + intermediate); ERG
expression (low vs. high), BM blasts (more than or equal to the
median vs. less than median), and BCR/ABL1 (presence vs.
absence).
All calculations were performed using the SPSS 17.0
software package (SPSS Inc., Chicago, IL, USA), and P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of ERG in childhood ALL
patients and control samples
ERG mRNA expression was retrospectively analyzed in
pretreatment BM samples of 119 patients with newly diagnosed ALL
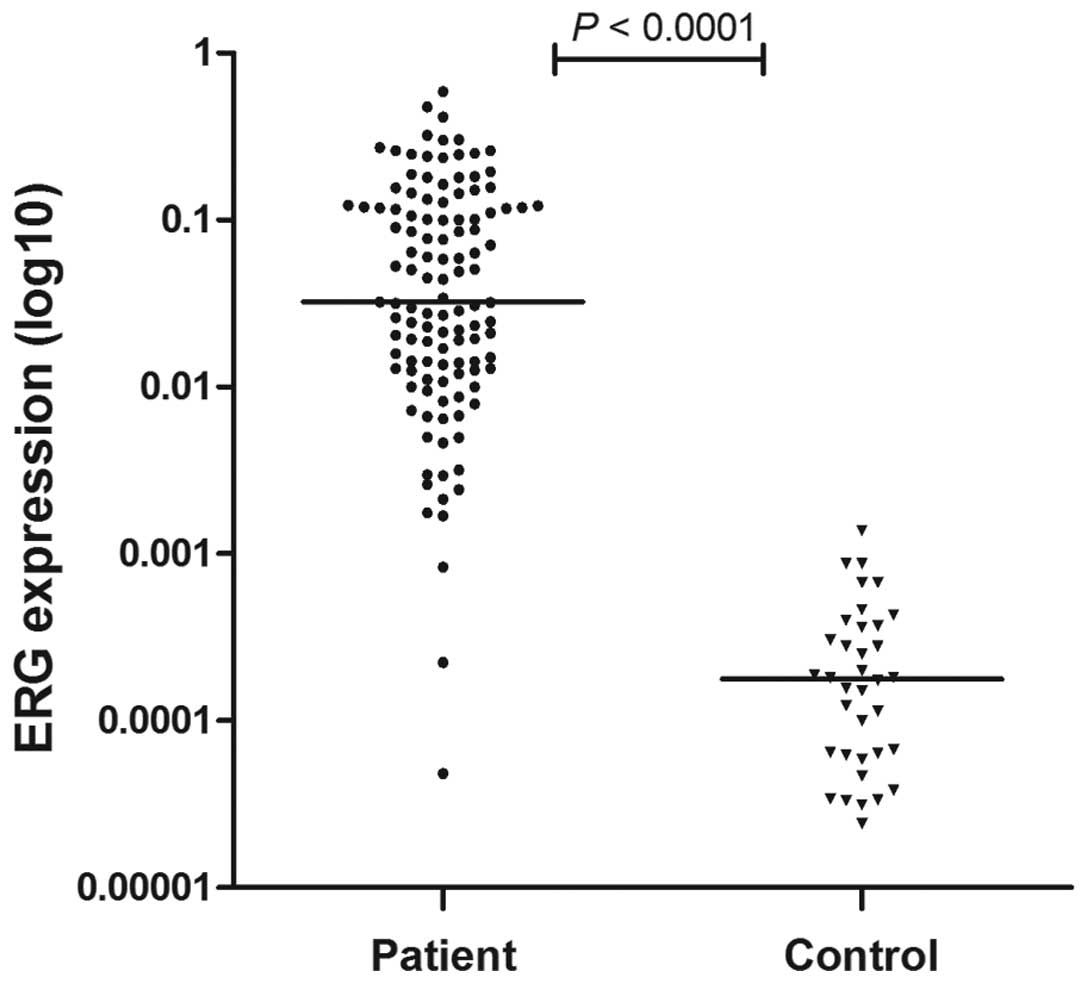
and BM samples of 36 controls. ALL patients demonstrated higher ERG
expression compared with controls (P<0.0001; Fig. 1).
ERG expression and correlation to
clinical features and molecular features
The clinical characteristics of 119 patients
included in the present study are summarized in Table I. Overall, patients with low ERG
expression exhibited higher pretreatment WBC counts (median,
32.2×109/l vs. 17.95×109/l; P=0.011),
compared with the high ERG expression group. In addition, low
expression of ERG was associated with higher percentage of T-ALL
(P=0.027). However, there was no significant association between
the ERG expression levels and the age, gender, platelet count, BM
blasts, peripheral blood blasts extramedullary involvement, the
risk group, Ik6 mutation, presence of BCR/ABL1 or MLL/AF4, and day
22 minimal residual disease.
 | Table I.Clinical characteristics of patients
with high and low ERG expression. |
Table I.
Clinical characteristics of patients
with high and low ERG expression.
| Characteristic | Low ERG (n=59) | High ERG (n=60) | P-value |
|---|
| Age, years |
|
| 0.432 |
|
Median | 7.5 | 5 |
|
|
Range | 1–14 | 1–14 |
|
| Sex, male, n (%) | 35 (59) | 41 (68) | 0.306 |
| WBC count,
×109/l |
|
| 0.011 |
|
Median | 32.20 | 17.95 |
|
|
Range | 2.4–552.9 | 0.8–760.0 |
|
| PLT count,
×109/l |
|
| 0.139 |
|
Median | 48 | 58 |
|
|
Range | 10–330 | 4–359 |
|
| BM blasts, % |
|
| 0.912 |
|
Median | 90 | 90 |
|
|
Range | 61–98 | 61–98 |
|
| PB blasts, % |
|
| 0.105 |
|
Median | 30 | 14.5 |
|
|
Range | 0–85 | 0–80 |
|
| Extramedullary
involvement, n (%) |
|
| 0.623 |
|
Yes | 1 (2) | 3 (5) |
|
| Immunophenotype, n
(%) |
|
| 0.027 |
|
B-ALL | 40 (68) | 51 (85) |
|
|
T-ALL | 19 (32) | 9 (15) |
|
| Risk, n (%) |
|
| 0.117 |
|
Low | 13 (22) | 22 (37) |
|
|
Intermediate | 15 (25) | 13 (22) |
|
|
High | 31 (53) | 25 (41) |
|
| Ik6, n (%) |
|
| 0.396 |
|
Mutated | 5 (8) | 8 (13) |
|
|
Unmuated | 54 (92) | 52 (87) |
|
| BCR/ABL1, n
(%) |
|
| 0.774 |
|
Presence | 5 (8) | 6 (10) |
|
|
Absence | 54 (92) | 54 (90) |
|
| MLL/AF4, n (%) |
|
| 1.000 |
|
Presence | 1 (2) | 1 (2) |
|
|
Absence | 58 (98) | 59 (98) |
|
| Day 22 MRD, n
(%) |
|
| 0.888 |
|
≥0.01% | 7 (12) | 6 (10) |
|
|
<0.01% | 52 (88) | 54 (90) |
|
In the subgroup analysis regarding the association
between ERG expression values and molecular genetics, no
significant difference was observed between patients with BCR/ABL1
compared with those patients without this fusion gene (P=0.273).
Similarly, there was no significant difference in ERG expression
levels between IK6 positive and negative groups (P=0.262).
Outcome and survival in childhood ALL
patients with respect to ERG expression
Compared with high ERG expressing patients, patients
with low ERG expression had a higher relapse rate (33 vs. 12%;
P=0.009; Table II). Patients with
low ERG expression exhibited a reduced prednisone response (79 vs.
91%; P=0.055; Table II). However, no
statistical significance was observed in CR rates between the low
ERG group and high ERG group.
 | Table II.Outcomes according to ERG
expression. |
Table II.
Outcomes according to ERG
expression.
| Outcome | Low ERG | High ERG | P-value |
|---|
| Induction
regimen |
|
| 0.055 |
|
Sensitive (%) | 44 (79) | 53 (91) |
|
| CR |
|
| 1.000 |
| No
(%) | 3 (5.4) | 6 (6.6) |
|
| Relapse |
|
| 0.009 |
| Yes
(%) | 18 (33) | 7 (12) |
|
| Relapse free
survival |
|
| 0.036 |
| Median,
months | 62.5 | 64 |
|
|
Relapse-free at 5-year, % | 58 | 79 |
|
| 95%
CI | 44–72 | 68–89 |
|
| Overall
survival |
|
| 0.620 |
| Median,
months | 71 | 65 |
|
| Alive
at 5-year, % | 71 | 78 |
|
| 95%
CI | 58–83 | 67–89 |
|
The median follow-up time was 67.5 months with a
range of 1–93 months. Following the exclusion of 9 patients who
received hematopoietic stem cell transplantation and 2 patients
whose exact survival data could not be obtained, the remaining 108
ALL patients were included for analyses of overall OS and 101 for
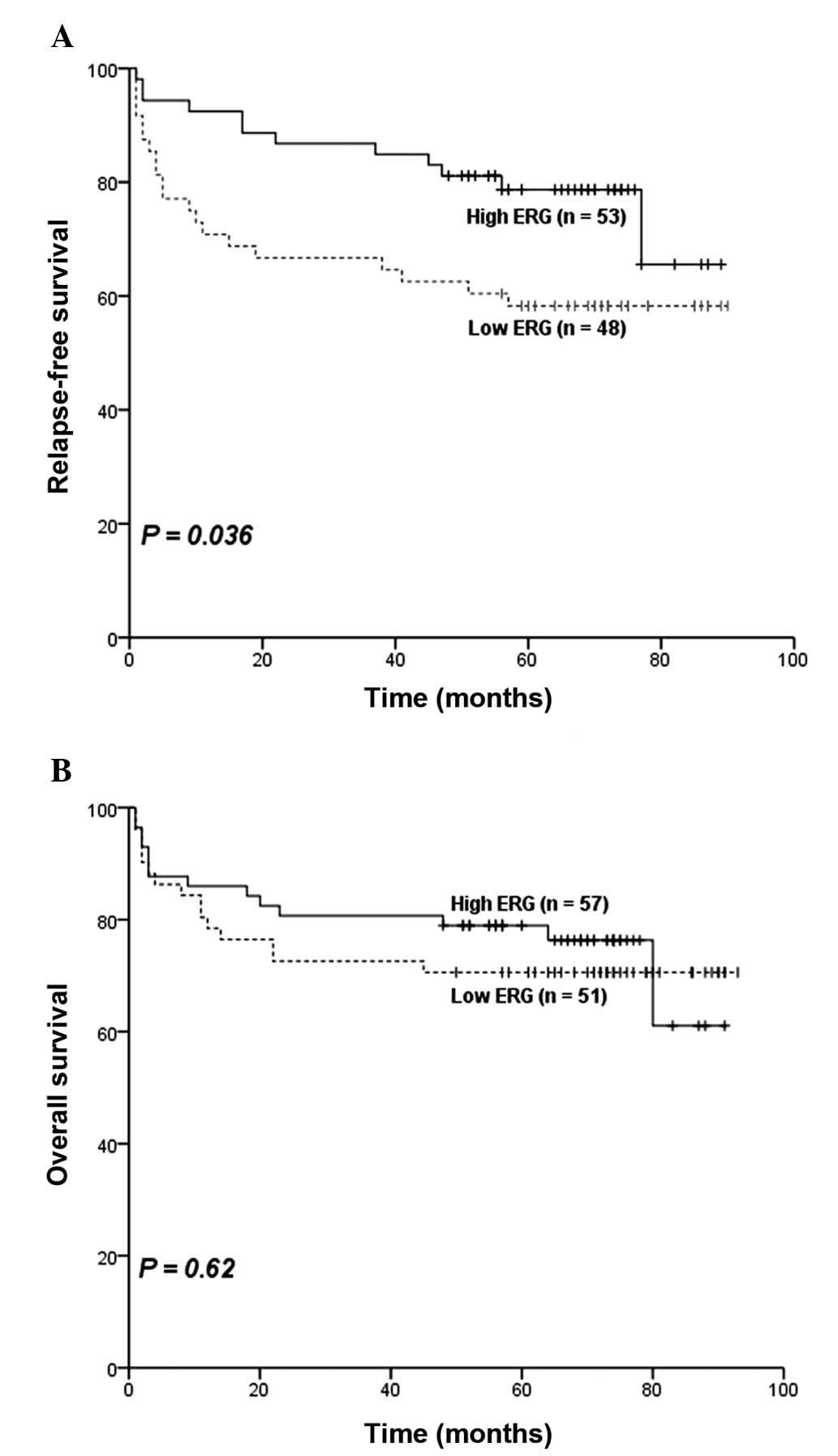
RFS. RFS was significantly different between the two groups
(P=0.036; Fig. 2A), patients in high
ERG group had an estimated 5-year relapse-free rate of 79% (95% CI,
68–89%; Table II) compared with 58%
(95% CI, 44–72%; Table II) in the
low ERG group. Furthermore, the estimated 5-year survival rate was
71% (95% CI, 58–83%; Table II) in
the low ERG group compared with 78% (95% CI, 67–89%; Table II) in the high ERG group, however, no
statistical difference was observed for differences in OS between
the two groups (P=0.62; Fig. 2B).
Considering the difference of therapy in LR, IR and
HR patients, survival analyses for OS and RFS based on ERG
expression were also conducted in the LR, IR and HR subgroups
respectively. However, the results did not indicate any survival
difference between the high ERG group and the low ERG group (data
not shown) in any subgroup.
Multivariate analysis for clinical
outcome in childhood ALL
A multivariate analysis including ERG expression,
age, WBC counts, risk, BM blast and BCR/ABL1 was also performed to
determine the prognostic importance of ERG expression on RFS and
OS. Subsequent to adjusting for the impact of other factors, ERG
expression remained an independent prognostic factor for RFS
(Table III). In the Cox
proportional hazards model, BM blast (more than or equal to the
median vs. less than the median) and BCR/ABL1 (presence vs.
absence) were independent prognostic factors for RFS (Table III). In addition, WBC counts
(≥50×109/l vs. <50×109/l) and BM blast
(more than or equal to the median vs. less than the median), were
independent prognostic factors for OS (Table III).
 | Table III.Multivariate analyses for RFS and OS
in pediatric ALL patients. |
Table III.
Multivariate analyses for RFS and OS
in pediatric ALL patients.
|
| RFS | OS |
|---|
|
|
|
|
|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, ≤1 year, ≥10
years vs. 1–10 years | 1.67 | 0.82–3.42 | 0.157 | 1.83 | 0.87–3.84 | 0.109 |
| WBC,
≥50×109/l vs. <50×109/l | 1.75 | 0.86–3.59 | 0.125 | 2.19 | 1.05–4.57 | 0.036 |
| Risk, high vs. low
+ intermediate | 1.77 | 0.88–3.55 | 0.108 | 1.72 | 0.83–3.59 | 0.146 |
| ERG expression, low
vs. high | 2.17 | 1.05–4.48 | 0.036 | 1.21 | 0.58–2.50 | 0.624 |
| BM blast %, ≥median
vs. <median | 2.47 | 1.13–5.38 | 0.023 | 2.87 | 1.22–6.73 | 0.015 |
| BCR/ABL1, presence
vs. absence | 3.41 | 1.38–8.42 | 0.008 | 2.19 | 0.82–5.88 | 0.119 |
ERG expression in childhood B-ALL
patients
ERG expression levels were also analyzed in 91 B-ALL
patients. Among all the clinical features described above, there
was only a significant difference in WBC counts between the low ERG
group and the high ERG group (median in the low ERG group,
23.6×109/l vs. median in the high ERG group,
12.7×109/l; P=0.037) (Table
IV). Subsequently, the relevance between ERG expression levels
and outcomes in B-ALL patients was investigated. Patients with low
ERG expression had higher relapse rate (32 vs. 13%; P=0.029;
Table IV). No significant difference
was observed in CR rates (P=1.000) and prednisone response
(P=0.155) between the two groups based on ERG expression.
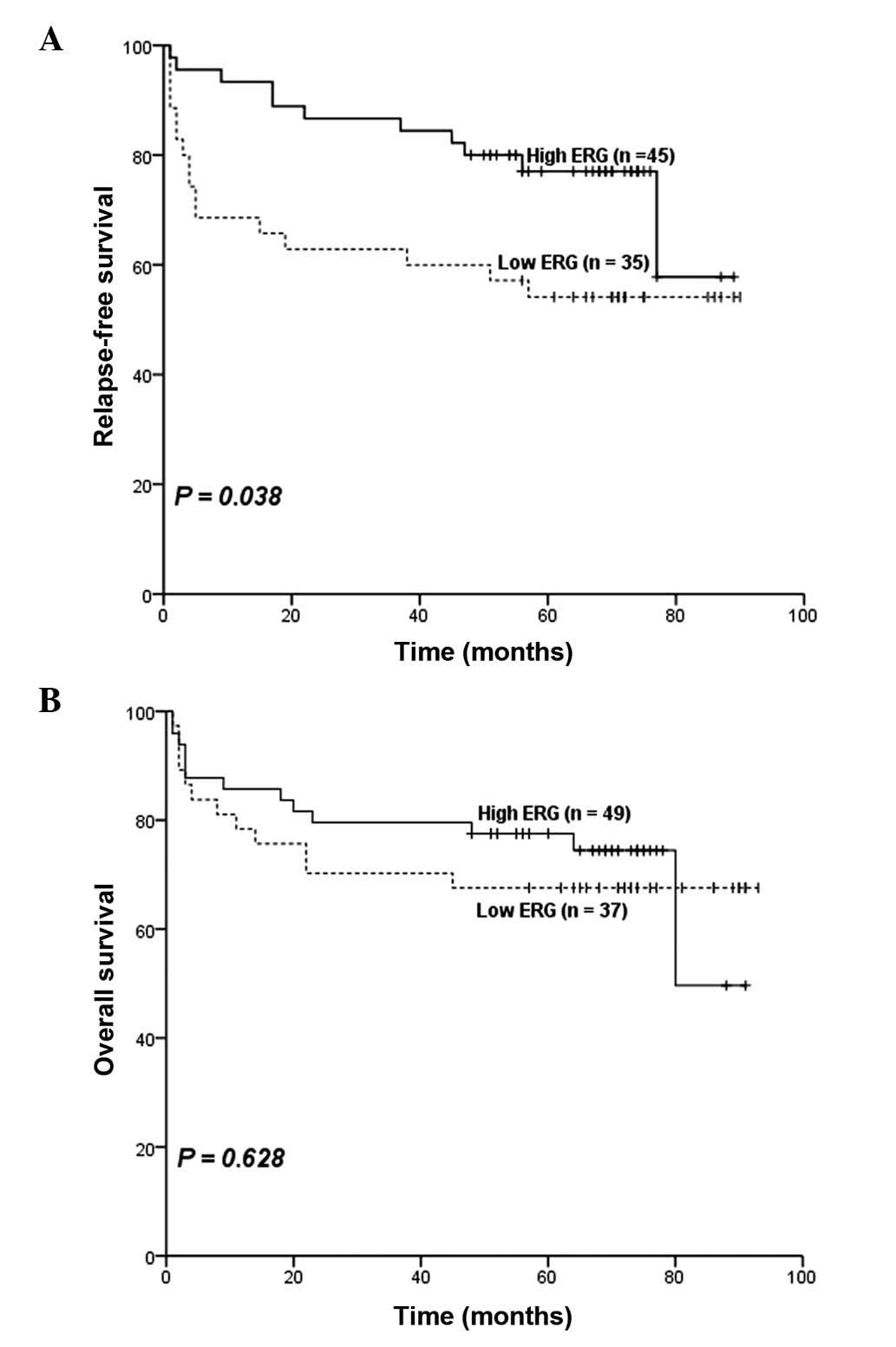
Furthermore, survival analyses suggested that patents with low ERG
expression had inferior RFS (P=0.038; estimated 5-year relapse-free
rate, 54 vs. 77%; Table IV and
Fig. 3A). However, no significant
difference was observed in OS (P=0.628; estimated 5-year survival
rate, 68% vs. 78%; Table IV and
Fig. 3B) was observed between
patients with high ERG expression and those with low ERG
expression. Results from multivariate analysis indicated that in
childhood B-ALL patients, ERG was not an independent prognostic
factor, for either OS, or for RFS. The independent prognostic
factors for OS were age (≤1 year, ≥10 years vs. 1–10 years) and WBC
counts (≥50×109/l vs. <50×109/l; Table V). In addition, age (≤1 year, ≥10
years vs. 1–10 years), WBC counts (≥50×109/l vs.
<50×109/l) and BCR/ABL1 (presence vs. absence) were
independent prognostic factors for RFS (Table V).
 | Table IV.ERG expression in childhood B-ALL
patients. |
Table IV.
ERG expression in childhood B-ALL
patients.
| Characteristic | Low ERG (n=40) | High ERG
(n=51) | P-value |
|---|
| WBC count,
×109/l |
|
| 0.037 |
|
Median | 23.6 | 12.7 |
|
|
Range | 2.4–552.9 | 0.8–760 |
|
| Outcome |
|
|
|
| Response to
prednisone |
|
| 0.155 |
|
Sensitive, n (%) | 29 (79) | 43 (90) |
|
| CR |
|
| 1.000 |
| No, n
(%) | 2 (5.4) | 4 (7.2) |
|
| Relapse |
|
| 0.029 |
| Yes, n
(%) | 12 (32) | 6 (13) |
|
| Relapse free
survival |
|
| 0.038 |
|
Relapse-free at 5-year, % | 54 | 77 |
|
| 95%
CI | 38–71 | 64–89 |
|
| Overall
survival |
|
| 0.628 |
| Alive
at 5-year, % | 68 | 78 |
|
| 95%
CI | 53–83 | 66–89 |
|
 | Table V.Multivariate analyses for RFS and OS
in pediatric B-ALL patients. |
Table V.
Multivariate analyses for RFS and OS
in pediatric B-ALL patients.
|
| RFS | OS |
|---|
|
|
|
|
|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, ≤1 year, ≥10
years vs. 1–10 years | 2.41 | 1.1–5.26 | 0.028 | 2.79 | 1.25–6.20 | 0.012 |
| WBC,
≥50×109/l vs. <50×109/l | 2.77 | 1.25–6.17 | 0.013 | 2.87 | 1.29–6.37 | 0.010 |
| Risk, high vs. low
+ intermediate | 1.64 | 0.61–4.36 | 0.286 | 1.31 | 0.49–3.50 | 0.353 |
| ERG expression, low
vs. high | 1.85 | 0.83–4.16 | 0.153 | 1.21 | 0.55–2.68 | 0.632 |
| BM blast %, ≥median
vs. <median | 2.16 | 0.79–5.90 | 0.112 | 2.47 | 0.83–7.36 | 0.078 |
| BCR/ABL, presence
vs. absence | 4.60 | 1.76–12.02 | 0.002 | 1.98 | 0.67–5.80 | 0.098 |
Discussion
Treatment improvement in ALL requires an optimal
risk stratification of patients to guide subsequent risk-adapted
treatment. Thus, molecular study of ALL has always been one of the
most active areas of leukemia research. In the present study, the
ERG expression levels in a cohort of 119 childhood ALL patients and
its correlation to clinical features and outcome were investigated.
The results of the current study support that ERG expression has an
association with the outcome of childhood ALL patients.
Together with its >30 ETS family members, ERG
acts as downstream nuclear targets of signal transduction pathways
regulating and promoting cell differentiation, proliferation, and
tissue invasion (5,7,23). ERG
rearrangements with EWS RNA binding protein 1 and FUS RNA binding
protein have been respectively identified in Ewing sarcomas
harboring t(21;22) (q22;q12) and in AML with t(16;21) (p11;q22)
(8,24), and are likely to increase the
oncogenic activity of the resulting chimeric transcription factors
by redirecting them to specific targets (10).
Furthermore, in hematopoietic tissues, ERG
expression is restricted to progenitor cells (25). High expression of ERG is significantly
correlated with the immature subtypes of AML (10). The present study compared BM samples
from childhood ALL patients with those of controls, and ERG
overexpression was observed in pediatric ALL patients with more
immature leukemia cells in their bone marrow. The above results
suggest that ERG may be associated with the outcomes of pediatric
ALL patients.
BCR/ABL1 positivity is a particularly poor
prognostic marker for ALL (26–28). In
addition, dominant-negative Ikaros isoform, notably Ik6, has been
reported to be highly expressed in ALL, and correlated with
clinical outcome (29). The relevance
between ERG expression and the presence of BCR/ABL1 or IK6
mutations in pediatric ALL required further elucidation. The
present study analyzed ERG expression values in patients according
to BCR/ABL1 and IK6 mutation status. The results demonstrated that
there was no significant difference in ERG expression levels
between patients with BCR/ABL1 or IK6 and those without these
genetic abnormities.
The current study demonstrated that patients in the
low ERG group demonstrated a significantly inferior outcome
compared with the high ERG group. Low ERG expression identified a
group of patients with higher WBC counts and higher percentages of
T-ALL, which have been treated as clinically poor prognostic
factors in childhood ALL. In addition, low ERG expression conferred
an impact on relapse rate and a significant influence on RFS was
suggested. In multivariate analysis, subsequent to adjusting for
the impact of other factors, ERG remained an independent prognostic
factor for RFS. Furthermore, in childhood B-ALL patients, low ERG
expression was associated with higher WBC counts, higher relapse
rate and poor RFS. The conclusions on how ERG expression levels
affect patient outcome and survival between the pediatric group in
the present study and other adult groups are inconsistent. As there
is no previous study available regarding ERG expression on
pediatric ALL, the possible explanations for this discrepancy are
as follows: i) Compared with other studies focused on adult
leukemia and childhood AML, the current study included patients
from a childhood ALL population with marked differences in biology,
etiology, treatment protocol and outcome in their diseases; and ii)
ERG expression level has been reported to correlate with other
genes, such as meningioma (disrupted in balanced translocation) 1
and brain and acute leukemia, cytoplasmic (14), which may also influence the prognostic
role of ERG (30), thus, there may be
other genes cooperating with or acting against ERG that directly
affect its influence on survival in childhood ALL patients; iii) in
addition, the inconsistent conclusions may be a result of
unadjusted study design confounder from different research centers,
including different diseases, ethnic background, medical resources
and treatment protocols.
To the best of our knowledge, this is the first
study to focus on ERG expression in pediatric ALL patients.
However, certain shortcomings remain, including that a relatively
small sample size may not be enough to draw a definitive
conclusion, and the retrospective nature of the present study may
present a certain bias.
In conclusion, the present study suggests that ERG
expression level may be useful as a prognostic factor for pediatric
ALL stratification, however, further large-scale and well-designed
prospective studies are required.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 30971283,
81170502, 81470304, 81200386 and 81300400), the Zhejiang Provincial
Natural Science Foundation of China (grant nos. LZ12H08001 and
Q12H080008), the Department of Health of Zhejiang Province of China
(grant no. 2013KYA107) and the Leukemia Research Innovative Team of
Zhejiang Province (grant no. 2011R50015).
References
|
1
|
Harvey RC, Mullighan CG, Wang X, Dobbin
KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et
al: Identification of novel cluster groups in pediatric high-risk
B-precursor acute lymphoblastic leukemia with gene expression
profiling: Correlation with genome-wide DNA copy number
alterations, clinical characteristics, and outcome. Blood.
116:4874–4884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bailey LC, Lange BJ, Rheingold SR and
Bunin NJ: Bone-marrow relapse in paediatric acute lymphoblastic
leukaemia. Lancet Oncol. 9:873–883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moorman AV, Ensor HM, Richards SM, Chilton
L, Schwab C, Kinsey SE, Vora A, Mitchell CD and Harrison CJ:
Prognostic effect of chromosomal abnormalities in childhood B-cell
precursor acute lymphoblastic leukaemia: Results from the UK
Medical Research Council ALL97/99 randomised trial. Lancet Oncol.
11:429–438. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moorman AV: The clinical relevance of
chromosomal and genomic abnormalities in B-cell precursor acute
lymphoblastic leukaemia. Blood Rev. 26:123–135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oikawa T: ETS transcription factors:
possible targets for cancer therapy. Cancer Sci. 95:626–633. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anderson MK, Hernandez-Hoyos G, Diamond RA
and Rothenberg EV: Precise developmental regulation of Ets family
transcription factors during specification and commitment to the T
cell lineage. Development. 126:3131–3148. 1999.PubMed/NCBI
|
|
7
|
Hart AH, Corrick CM, Tymms MJ, Hertzog PJ
and Kola I: Human ERG is a proto-oncogene with mitogenic and
transforming activity. Oncogene. 10:1423–1430. 1995.PubMed/NCBI
|
|
8
|
Sorensen PH, Lessnick SL, Lopez-Terrada D,
Liu XF, Triche TJ and Denny CT: A second Ewing's sarcoma
translocation, t(21;22), fuses the EWS gene to another ETS-family
transcription factor, ERG. Nat Genet. 6:146–151. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marcucci G, Baldus CD, Ruppert AS,
Radmacher MD, Mrózek K, Whitman SP, Kolitz JE, Edwards CG, Vardiman
JW, Powell BL, et al: Overexpression of the ETS-related gene, ERG,
predicts a worse outcome in acute myeloid leukemia with normal
karyotype: A Cancer and Leukemia Group B study. J Clin Oncol.
23:9234–9242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marcucci G, Maharry K, Whitman SP,
Vukosavljevic T, Paschka P, Langer C, Mrózek K, Baldus CD, Carroll
AJ, Powell BL, et al: High expression levels of the ETS-related
gene, ERG, predict adverse outcome and improve molecular risk-based
classification of cytogenetically normal acute myeloid leukemia: A
Cancer and Leukemia Group B Study. J Clin Oncol. 25:3337–3343.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eid MA, Attia M, Abdou S, El-Shazly SF,
Elahwal L, Farrag W and Mahmoud L: BAALC and ERG expression in
acute myeloid leukemia with normal karyotype: Impact on prognosis.
Int J Lab Hematol. 32:197–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Metzeler KH, Dufour A, Benthaus T, Hummel
M, Sauerland MC, Heinecke A, Berdel WE, Büchner T, Wörmann B,
Mansmann U, et al: ERG expression is an independent prognostic
factor and allows refined risk stratification in cytogenetically
normal acute myeloid leukemia: A comprehensive analysis of ERG,
MN1, and BAALC transcript levels using oligonucleotide microarrays.
J Clin Oncol. 27:5031–5038. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schwind S, Marcucci G, Maharry K,
Radmacher MD, Mrózek K, Holland KB, Margeson D, Becker H, Whitman
SP, Wu YZ, et al: BAALC and ERG expression levels are associated
with outcome and distinct gene and microRNA expression profiles in
older patients with de novo cytogenetically normal acute myeloid
leukemia: A Cancer and Leukemia Group B study. Blood.
116:5660–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baldus CD, Burmeister T, Martus P,
Schwartz S, Gökbuget N, Bloomfield CD, Hoelzer D, Thiel E and
Hofmann WK: High expression of the ETS transcription factor ERG
predicts adverse outcome in acute T-lymphoblastic leukemia in
adults. J Clin Oncol. 24:4714–4720. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldus CD, Martus P, Burmeister T,
Schwartz S, Gökbuget N, Bloomfield CD, Hoelzer D, Thiel E and
Hofmann WK: Low ERG and BAALC expression identifies a new subgroup
of adult acute T-lymphoblastic leukemia with a highly favorable
outcome. J Clin Oncol. 25:3739–3745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pigazzi M, Masetti R, Martinolli F, Manara
E, Beghin A, Rondelli R, Locatelli F, Fagioli F, Pession A and
Basso G: Presence of high-ERG expression is an independent
unfavorable prognostic marker in MLL-rearranged childhood myeloid
leukemia. Blood. 119:1086–1087; author reply 1087–1088. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hermkens MC, van den Heuvel-Eibrink MM,
Arentsen-Peters ST, Baruchel A, Stary J, Reinhardt D, Zimmerman M,
de Haas V, Pieters R and Zwaan CM: The clinical relevance of BAALC
and ERG expression levels in pediatric AML. Leukemia. 27:735–737.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subspecialty Group of Hematology Diseases,
The Society of Pediatrics, Chinese Medical Association; Editorial
Board, Chinese Journal of Pediatrics, . Recommendations for
diagnosis and treatment of acute lymphoblastic leukemia in
childhood (3rd revised version). Zhonghua Er Ke Za Zhi. 44:392–395.
2006.(In Chinese). PubMed/NCBI
|
|
20
|
Xu XJ, Tang YM, Shen HQ, Song H, Yang SL,
Shi SW and Xu WQ: Day 22 of induction therapy is important for
minimal residual disease assessment by flow cytometry in childhood
acute lymphoblastic leukemia. Leuk Res. 36:1022–1027. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mu Q, Wang Y, Chen B, Qian W, Meng H, Tong
H, Chen F, Ma Q, Ni W, Chen S and Jin J: High expression of
Musashi-2 indicates poor prognosis in adult B-cell acute
lymphoblastic leukemia. Leuk Res. 37:922–927. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mullighan CG, Miller CB, Radtke I,
Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui
CH, et al: BCR-ABL1 lymphoblastic leukaemia is characterized by the
deletion of Ikaros. Nature. 453:110–114. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oikawa T and Yamada T: Molecular biology
of the Ets family of transcription factors. Gene. 303:11–34. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ichikawa H, Shimizu K, Hayashi Y and Ohki
M: An RNA-binding protein gene, TLS/FUS, is fused to ERG in human
myeloid leukemia with t(16;21) chromosomal translocation. Cancer
Res. 54:2865–2868. 1994.PubMed/NCBI
|
|
25
|
Rainis L, Toki T, Pimanda JE, Rosenthal E,
Machol K, Strehl S, Göttgens B, Ito E and Izraeli S: The
proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res.
65:7596–7602. 2005.PubMed/NCBI
|
|
26
|
Dombret H, Gabert J, Boiron JM,
Rigal-Huguet F, Blaise D, Thomas X, Delannoy A, Buzyn A,
Bilhou-Nabera C, Cayuela JM, et al: Outcome of treatment in adults
with Philadelphia chromosome-positive acute lymphoblastic
leukemia-results of the prospective multicenter LALA-94 trial.
Blood. 100:2357–2366. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gleissner B, Gökbuget N, Bartram CR,
Janssen B, Rieder H, Janssen JW, Fonatsch C, Heyll A, Voliotis D,
Beck J, et al: Leading prognostic relevance of the BCR-ABL
translocation in adult acute B-lineage lymphoblastic leukemia: A
prospective study of the German Multicenter Trial Group and
confirmed polymerase chain reaction analysis. Blood. 99:1536–1543.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mancini M, Scappaticci D, Cimino G, Nanni
M, Derme V, Elia L, Tafuri A, Vignetti M, Vitale A, Cuneo A, et al:
A comprehensive genetic classification of adult acute lymphoblastic
leukemia (ALL): Analysis of the GIMEMA 0496 protocol. Blood.
105:3434–3441. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu P, Lin Z, Qian S, Qiao C, Qiu H, Wu Y,
Li J and Ge Z: Expression of dominant-negative Ikaros isoforms and
associated genetic alterations in Chinese adult patients with
leukemia. Ann Hematol. 91:1039–1049. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Staffas A, Kanduri M, Hovland R,
Rosenquist R, Ommen HB, Abrahamsson J, Forestier E, Jahnukainen K,
Jónsson ÓG, Zeller B, et al: Presence of FLT3-ITD and high BAALC
expression are independent prognostic markers in childhood acute
myeloid leukemia. Blood. 118:5905–5913. 2011. View Article : Google Scholar : PubMed/NCBI
|