Introduction
Vascular endothelial growth factor (VEGF) and its
receptors play important roles in regulating angiogenesis during
physiological or pathological processes such as tumorigenesis
(1,2).
Angiogenesis has been shown to promote the growth of solid and
hematological tumors (1). VEGF, which
is a glycoprotein, has five isotypes, including VEGF-A, VEGF-B,
VEGF-C, VEGF-D and placental growth factor. VEGF-A is typically
referred to as VEGF (3). VEGF
receptors (VEGFRs) consist of three structurally similar tyrosine
kinase receptor proteins termed VEGFR1 [also called Fms-related
tyrosine kinase (FTL) 1], VEGFR2 (also called kinase insert domain
receptor) and VEGF3 (also called FLT4) (2). VEGF is the ligand of VEGFR1 and VEGFR2,
which, upon binding to VEGF, induce the proliferation of
endothelial cells via the Ras- or protein kinase C-activated
mitogen-activated protein kinase signaling pathway to promote
angiogenesis (2). The active form of
VEGF is a homodimer that is capable of binding to VEGFRs (1).
Acute myeloid leukemia (AML), which is a disease of
the hematopoietic progenitor cells characterized by heterogeneous
clonal proliferation, is the most common myeloid malignancy in
adults. The age of onset is late in life, and the median age is 70
years (4). Although several advances
in the molecular understanding of AML have been achieved,
additional studies are required in order to cure or conquer this
disease (4,5). Commonly, VEGF is expressed at higher
levels in AML cells, and autocrine regulation of VEGF affects the
clonal proliferation of AML cells (6). Compared with healthy individuals, 85% of
AML patients showed upregulation of VEGF in bone marrow biopsy
specimens (7). In addition, VEGFRs
have been shown to be recurrently upregulated in AML (8). AML cells may possess a VEGF autocrine
pathway, and its type I receptor (VEGFR1) may be involved in
cellular proliferation of the M3 subtype of AML (9,10).
MicroRNAs consist of a set of 19-25-nucleotide
single-stranded RNA molecules that are involved in the
transcription and translation of genes associated with growth,
development and pathological processes such as cancer (11). Increasingly, evidence has suggested
that there is a close association between AML and microRNAs
(12–15). MicroRNAs may have a role in the
regulation of the cellular biology and molecular genetics of AML,
since upregulation of miR-155, miR-10a, miR-10b and miR-191 has
commonly been observed in AML cells (15).
Although significant progress has been made, the
functions of VEGF and microRNAs in the development and progression
of AML, and the association between VEGF and microRNAs, are
unclear. Therefore, the present study aimed to determine the
association between VEGF overexpression and microRNA profiles in
AML cells and patients.
Materials and methods
Construction of lentiviral vectors and
stable cell lines
Polymerase chain reaction (PCR) was used to obtain
the VEGF gene expression sequence (NM_001171626), which was
inserted into a LV5 vector (Shanghai GenePharma, Co., Ltd.,
Shanghai, China) using the Not I/Nsi I restriction
enzymes to construct the LV-VEGF expression vector, as described
previously (15). LV-NC served as the
control vector. The VEGF-specific short hairpin (sh)RNA sequence,
TTC TCC GAA CGT GTC ACGT, was obtained from Shanghai GenePharma,
Co., Ltd. Following infection of the AML cell lines, K562, HL-60
and U937 (Type Culture Collection of the Chinese Academy of
Sciences, Shanghai, China) with the lentiviral vectors or
VEGF-specific shRNA, infected cells were selected using purimycin
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and
untransfected cells were removed 48 h later, as described
previously (16).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cells using TRIzol
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), as described
previously (17). Total RNA was
dissolved in diethylpyrocarbonate water and the optical density was
measured at 260 and 280 nm using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). The PrimeScript™ RT reagent kit
with gDNA Eraser (Perfect Real Time) (Takara Biotechnology Co.,
Ltd., Dalian, China) was used for cDNA production from RNA,
according to the manufacturer's protocol (18). The SYBR® Premix Ex Taq™ II
(Tli RNaseH Plus) (Takara Biotechnology Co., Ltd.) kit was used for
qPCR on a StepOnePlus™ Real-Time PCR instrument (Thermo Fisher
Scientific, Inc.). The cycling conditions were 95°C for 5 min,
followed by 40 cycles at 95°C for 10 sec and 60°C for 34 sec. The
primers were as follows: VEGF forward, 5′-CGCAGCTACTGCCATCCAAT-3′
and reverse, 5′-GTGAGGTTTGATCCGCATAATCT-3′; and GAPDH forward,
3′-ACCCAGAAGACTGTGGATGG-5′; and reverse,
3′-TCTAGACGGCAGGTCAGGTC-5′. The relative expression of the gene was
calculated using the 2−ΔΔCq method (19), with GAPDH as an internal control
gene.
Western blot analysis
After reaching 80–90% confluence, the cells in
culture plates were lysed using radioimmunoprecipitation assay
buffer (Beyotime Institute of Biotechnology, Haimen, China)
supplemented with 1 mM phenylmethylsulfonyl fluoride protease
inhibitor (Beyotime Institute of Biotechnology). The concentrations
of proteins were determined using the bicinchoninic acid assay kit
(Beyotime Institute of Biotechnology). The proteins were boiled
following addition of the sample loading buffer and the denatured
proteins were separated by 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes, which were blocked with 5%
bovine serum albumin (Beyotime Institute of Biotechnology), as
previously described (20).
Subsequently, the membranes were incubated with primary antibodies
against VEGF (cat. no. ab106041; 1:1,000 dilution; Abcam,
Cambridge, MA, USA) and β-actin (cat. no. ab106045; 1:5,000
dilution; Abcam), followed by incubation with the secondary
antibody (cat. no. ab6721; 1:5,000 dilution; Abcam). The bands of
proteins were visualized using enhanced chemiluminesence (EMD
Millipore, Billerica, MA, USA) and detected with the ChemiDoc MP
imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
microRNA array
Total RNA was extracted from cells showing
upregulation or downregulation of VEGF using the mirVana PARIS
miRNA isolation kit (Ambion; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. After quantitation of the
RNA using the NanoDrop 2000c spectrophotometer (Thermo Fisher
Scientific, Inc.), microRNA expression profiling was performed
using the Exiqon A/S Technology Platform (Exiqon A/S, Vedbaek,
Denmark). MicroRNAs were labeled using the miRCURY™ Hy3™/Hy5™ Power
kit and hybridized onto the miRCURY™ LNA (v.18.0) (Exiqon A/S).
After washing with the washing buffer in the kit, the slides were
scanned using the Axon GenePix 4000B Microarray Scanner, and the
data were analyzed using GenePix Pro 6.0 software (Molecular
Devices, LLC, Sunnyvale, CA, USA) (21,22).
Analysis of clinical samples and
miRNAs
The protocol of this study was approved by the
Ethics Committee of the First Affiliated Hospital of Zhejiang
University School of Medicine. Written informed consent was
obtained from all patients. In this study, 45 bone marrow samples
from patients with AML were collected at our hospital from
2013–2014, in strict accordance with the approved protocol. The
information of the patients in this experiment is summarized in
Table I. Total RNA, including small
RNAs, was isolated using the mirVana PARIS miRNA isolation kit from
blood mononuclear cells isolated using Ficoll-Paque PLUS (GE
Healthcare, Milwaukee, WI, USA), as described previously (23). RT-qPCR for the analysis of microRNAs
was performed using the miScript SYBR Green PCR kit (Qiagen GmbH,
Hilden, Germany). c-Abl served as the internal control gene. The
relative expression levels of microRNAs were calculated using the
2−ΔΔCq method (19).
Furthermore, the mRNA expression levels of VEGF in the bone marrow
samples were determined by RT-qPCR. The expression levels of VEGF
were subsequently ranked from maximum to minimum and then divided
into high and low groups.
 | Table I.Characteristics of acute myeloid
leukemia patients. |
Table I.
Characteristics of acute myeloid
leukemia patients.
| Characteristic | Number of
patients |
|---|
| Total | 45 |
| Male | 25 |
| Female | 20 |
| FAB subtype |
|
| m0 | 1 |
| m2 | 20 |
| m3 | 12 |
|
m5b | 12 |
Statistical analysis
SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) was
used for statistical analysis. One-way analysis of variance was
performed for the comparisons of three groups and Student's t-test
for two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Construction of AML cells with stable
expression of VEGF and knockdown of VEGF
To confirm that cells with stable knockdown and
overexpression of VEGF were obtained, we extracted total protein
for western blotting and total RNA for RT-qPCR. The RT-qPCR
analysis showed that the mRNA expression levels of VEGF were
significantly higher in K562-VEGFa-O, HL-60-VEGFa-O and
U937-VEGFa-O cells compared with the control cells (K562-NC-LV,
HL-60-NC-LV and U937-NC-LV) (P<0.01), and that those of
K562-VEGF-shRNA, HL-60-VEGF-shRNA and U937-VEGF-shRNA cells were
significantly lower compared with the control cells (P<0.01;
Fig. 1A). In addition, the results of
western blotting confirmed that the VEGF protein had a pattern of
expression that resembled that of its mRNA (Fig. 1B).
VEGF changes the microRNA profile of
leukemia cells
For the detection of microRNA profiles, total RNA
was extracted from the cells for microarray analysis. Genes that
were screened using the microarray needed to meet the criteria of
≥2-fold increase or decrease in expression in the cells with
overexpression or knockdown of VEGF compared with the controls.
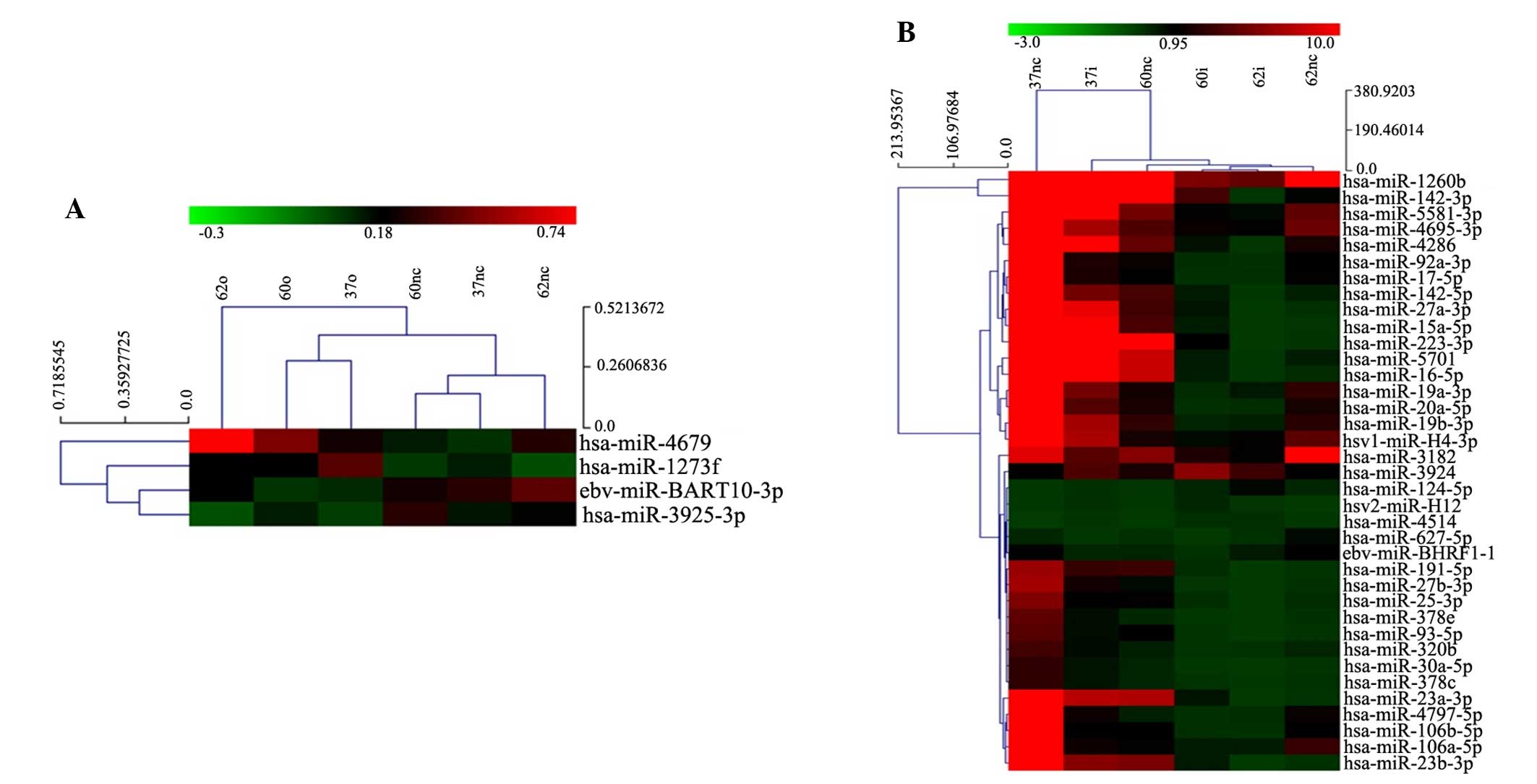
Overexpression of VEGF in leukemia cells (K562-VEGFa-O,
HL-60-VEGFa-O and U937-VEGFa-O), despite being upregulated by
3-fold, had little effect on microRNA expression; only four
microRNAs were identified as being altered in expression, including
has-miR-1273f and has-miR-4679, which were upregulated, and
eb-miR-BART10-3p and has-miR-3925-3p, which were downregulated
(Fig. 2A).
In K562-VEGF-shRNA, HL-60-VEGF-shRNA and
U937-VEGF-shRNA cells, microRNA profiling showed a significant
change compared with control cells (K562-NC-LV, HL-60-NC-LV and
U937-NC-LV). Among several microRNAs that showed a 5-fold change in
expression, hsv2-miR-H12, has-miR-124-5p, has-miR-3924 and
has-miR-4514 were upregulated, and 33 microRNAs, including
has-miR-20a, were downregulated (Fig.
2B).
Expression of microRNAs associated
with VEGF expression in human samples from AML patients
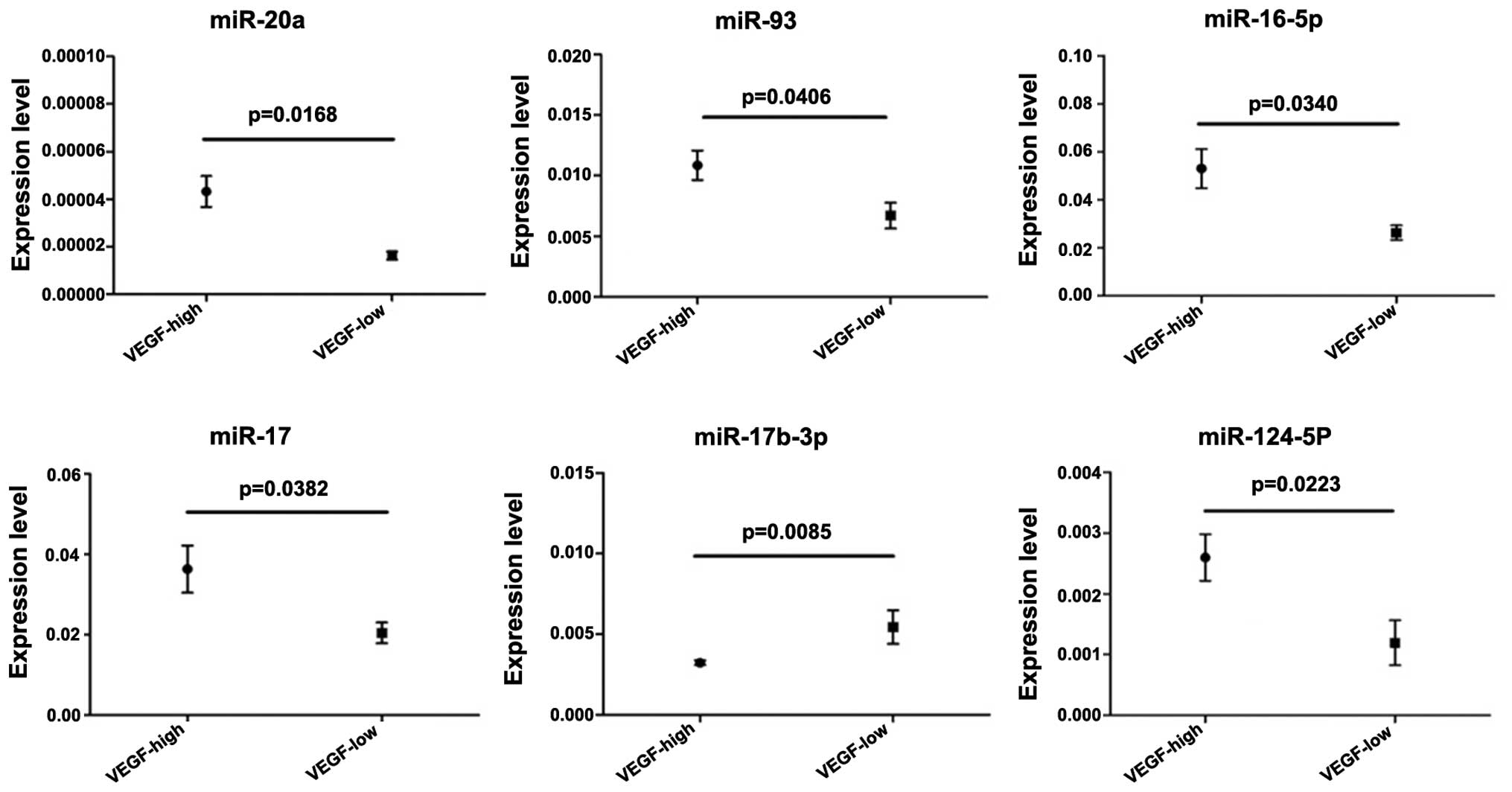
In the present study, several microRNAs showed
changes in their levels of expression following overexpression or
silencing of the expression of the VEGF gene. To confirm these
results, bone marrow samples from 45 patients with AML were used to
analyze the expression levels of microRNAs identified in the
microarray and VEGF. Subsequently, the expression levels of VEGF
were defined into low and high groups. The results indicated a
positive association between VEGF expression and the expression of
six microRNAs, including miR-20a, miR-93, miR-16-5p, miR-17-5p,
miR-17b-3p and miR-124-5p. In addition, a negative association was
observed between VEGF expression and the expression of miR-17b-3p
(Fig. 3).
Discussion
Elevated expression of the VEGF gene is commonly
observed in major types of cancer, including AML, and is essential
for angiogenesis, which supplies nutrients, oxygen and other
factors for the rapid proliferation and growth of malignancies
(2–4,6,8). VEGF could play important roles in
various physical events of malignant cells using autocrine or
paracrine pathways (24–30). VEGF and its receptors are commonly
upregulated in bone marrow precursor cells and leukemia progenitor
cells and influence the secretion of various factors from bone
marrow cells (31). Furthermore, the
autocrine pathway of VEGF may have effects on the survival of
hematopoietic stem cells, and VEGFR-1 and VEGFR-2 have also been
shown to contribute to this process (31). Therefore, VEGF performs essential
functions in the initiation and development of solid and
hematological tumors.
In the present study, a microRNA microarray was used
to detect the influence of VEGF on hundreds of microRNAs in
leukemia cells. It was observed that overexpression of VEGF had a
limited impact on leukemia cells, but that silencing of VEGF
significantly altered the microRNA profiles of leukemia cells. At
least seven microRNAs (miR-20a, miR-93, miR-16-5p, miR-17,
miR-124-5p and miR-17b-3p) were identified in the peripheral blood
samples of AML patients, of which six were positively associated,
and one (miR-17b-3p) was negatively associated, with VEGF
expression.
Among the microRNAs identified, miR-17 and miR-20a
belong to the miR-17-92 cluster, which includes six members
(miR-17, miR-18a, miR-19a, miR-20a, miR-19b and miR-92a), all of
which are spliced from a single pre-RNA (32). This cluster of miRNAs is overexpressed
in a variety of cancers, including hematopoietic and solid tumors
(33,34). miR-93 is encoded by the polycistronic
miR-106b-25 cluster, which contains miR-106, miR-93 and miR-25
(33). The miR-106b-25 cluster is a
paralog of the miR-17-92 cluster and both exert similar functions
in development and tumorigenesis (35,36). In
previous studies, miR-93 could promote angiogenesis, causing
endothelial cells to spread and promoting tube formation by
suppressing the expression of integrin-β8 in brain and breast
cancers (37,38). Unlike miR-17, miR-20 and miR-93,
miR-124-5p may predominantly have an anticancer effect (39,40). These
reports are not completely consistent with the results of the
present study, which showed that a lower level of VEGF was
associated with downregulation of miR-124-5p. miR-17-3p, whose
precursor is microRNA-17, is a passenger strand microRNA that is
able to target and repress the expression of TIMP metallopeptidase
inhibitor 3, phosphatase and tensin homolog deleted on chromosome
10, GalNT7 and vimentin, all of which are known to promote the
proliferation and metastasis of cancer cells (41,42). These
reports are in agreement with the results of the present study.
Previous studies have determined that AML and other
subtypes of leukemia show elevated expression of VEGF and its
receptors (4,9,43–45). The results of the present study
suggested that knockdown of VEGF could induce large changes in the
expression of microRNAs, likely because VEGF signaling has an
important role in the development and progression of AML cells. The
differences in the results of the microarray and clinical samples
may be due to the differences between the immortal cell lines and
AML cells in patients, which emphasizes the requirement for further
clinical examinations.
In conclusion, the present study demonstrated that
autocrine or paracrine expression of VEGF by AML cells directly or
indirectly affected AML cells themselves or neighboring cells. The
changes in the microRNA expression profile of leukemia cells
following downregulation of VEGF suggests that several targets for
anti-AML therapy may exist in the VEGF-microRNA axis. Additional
related studies are required in the future to address this.
Acknowledgements
The authors would like to thank Dr Guowei Qu for his
critical suggestions on the processing of data and design of
experiments. This study was supported by grants from the Science
and Technology Department of Zhejiang Province (no. 2011C23015),
Zhejiang Provincial Department of Education (no. Y201120523) and
Zhejiang Provincial Administration of Traditional Chinese Medicine
(no. Y201120523).
References
|
1
|
Kerbel RS: Tumor angiogenesis. N Engl J
Med. 358:2039–2049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Olsson AK, Dimberg A, Kreuger J and
Claesson-Welsh L: VEGF receptor signalling-in control of vascular
function. Nat Rev Mol Cell Biol. 7:359–371. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song G, Li Y and Jiang G: Role of
VEGF/VEGFR in the pathogenesis of leukemias and as treatment
targets (Review). Oncol Rep. 28:1935–1944. 2012.PubMed/NCBI
|
|
4
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Donnell MR, Abboud CN, Altman J,
Appelbaum FR, Arber DA, Attar E, Borate U, Coutre SE, Damon LE,
Goorha S, et al: Acute myeloid leukemia. J Natl Compr Canc Netw.
10:984–1021. 2012.PubMed/NCBI
|
|
6
|
Hou HA, Chou WC, Lin LI, Tang JL, Tseng
MH, Huang CF, Yao M, Chen CY, Tsay W and Tien HF: Expression of
angiopoietins and vascular endothelial growth factors and their
clinical significance in acute myeloid leukemia. Leuk Res.
32:904–912. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kampen KR, Ter Elst A and de Bont ES:
Vascular endothelial growth factor signaling in acute myeloid
leukemia. Cell Mol Life Sci. 70:1307–1317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Padró T, Bieker R, Ruiz S, Steins M,
Retzlaff S, Bürger H, Büchner T, Kessler T, Herrera F, Kienast J,
et al: Overexpression of vascular endothelial growth factor (VEGF)
and its cellular receptor KDR (VEGFR-2) in the bone marrow of
patients with acute myeloid leukemia. Leukemia. 16:1302–1310. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hiramatsu A, Miwa H, Shikami M, Ikai T,
Tajima E, Yamamoto H, Imai N, Hattori A, Kyo T, Watarai M, et al:
Disease-specific expression of VEGF and its receptors in AML cells:
Possible autocrine pathway of VEGF/type1 receptor of VEGF in t(15;
17) AML and VEGF/type2 receptor of VEGF in t(8; 21) AML. Leuk
Lymphoma. 47:89–95. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santos SC and Dias S: Internal and
external autocrine VEGF/KDR loops regulate survival of subsets of
acute leukemia through distinct signaling pathways. Blood.
103:3883–3889. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garzon R, Garofalo M, Martelli MP,
Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG,
Schnittger S, Haferlach T, et al: Distinctive microRNA signature of
acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin.
Proc Natl Acad Sci USA. 105:3945–3950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garzon R, Volinia S, Liu CG,
Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K,
Alder H, Nakamura T, et al: MicroRNA signatures associated with
cytogenetics and prognosis in acute myeloid leukemia. Blood.
111:3183–3189. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie WJ, Flamant S and Rasko J:
MicroRNA in acute myeloid leukemia. N Engl J Med. 359:653author
reply 653–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Havelange V, Stauffer N, Heaphy CC,
Volinia S, Andreeff M, Marcucci G, Croce CM and Garzon R:
Functional implications of microRNAs in acute myeloid leukemia by
integrating microRNA and messenger RNA expression profiling.
Cancer. 117:4696–4706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei J, Zhao ZX, Li Y, Zhou ZQ and You TG:
Cortactin expression confers a more malignant phenotype to gastric
cancer SGC-7901 cells. World J Gastroenterol. 20:3287–3300. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li S, Moffett HF, Lu J, Werner L, Zhang H,
Ritz J, Neuberg D, Wucherpfennig KW, Brown JR and Novina CD:
MicroRNA expression profiling identifies activated B cell status in
chronic lymphocytic leukemia cells. PloS One. 6:e169562011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei R, Yuan D, Wang T, Zhou C, Lin F, Chen
H, Wu H, Yang S, Wang Y, Liu J, et al: Characterization, tissue
distribution and regulation of agouti-related protein (AgRP) in a
cyprinid fish (Schizothorax prenanti). Gene. 527:193–200. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ward IM and Chen J: Histone H2AX is
phosphorylated in an ATR-dependent manner in response to
replicational stress. J Biol Chem. 276:47759–47762. 2001.PubMed/NCBI
|
|
22
|
Gao W, Yu Y, Cao H, Shen H, Li X, Pan S
and Shu Y: Deregulated expression of miR-21, miR-143 and miR-181a
in non small cell lung cancer is related to clinicopathologic
characteristics or patient prognosis. Biomed Pharmacother.
64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lutherborrow M, Bryant A, Jayaswal V,
Agapiou D, Palma C, Yang YH and Ma DD: Expression profiling of
cytogenetically normal acute myeloid leukemia identifies microRNAs
that target genes involved in monocytic differentiation. Am J
Hematol. 86:2–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kirschner MB, Kao SC, Edelman JJ,
Armstrong NJ, Vallely MP, van Zandwijk N and Reid G: Haemolysis
during sample preparation alters microRNA content of plasma. PloS
One. 6:e241452011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koch KR, Refaian N, Hos D, Schlereth SL,
Bosch JJ, Cursiefen C and Heindl LM: Autocrine impact of VEGF-A on
uveal melanoma cells. Invest Ophthalmol Vis Sci. 55:2697–2704.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Larsen AK, Poindessous V, Sabbah M, de
Gramont A and Mesange P: Autocrine signaling plays a major role in
the response of colorectal cancer cells to VEGF blockage.
International Journal of Molecular Medicine. 34:S22. 2014.
|
|
27
|
Barr MP, Gray G, Gately KA, Harns E,
Fallon PG, Davies AM, Richard DJ, Pidgeon GP, Cuffe S, Finn S and
O'Byrne KJ: VEGF autocrine survival signalling is mediated via
neuropilin 1 receptor in NSCLC cells. Lung Cancer. 83:S5. 2014.
View Article : Google Scholar
|
|
28
|
Ji Y, Chen S, Li K, Xiao X, Xu T and Zheng
S: Upregulated autocrine vascular endothelial growth factor
(VEGF)/VEGF receptor-2 loop prevents apoptosis in
haemangioma-derived endothelial cells. Brit J Dermatol. 170:78–86.
2014. View Article : Google Scholar
|
|
29
|
Barr MP, Gray SG, Gately KA and O'Byrne K:
Vegf is an autocrine survival factor in non-small cell lung cancer,
mediating its effects via the neuropilin-1 receptor. J Thorac
Oncol. 8:S426. 2013.
|
|
30
|
Lee S, Chen TT, Barber CL, Jordan MC,
Murdock J, Desai S, Ferrara N, Nagy A, Roos KP and Iruela-Arispe
ML: Autocrine VEGF signaling is required for vascular homeostasis.
Cell. 130:691–703. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reinmuth N, Liu W, Jung YD, Ahmad SA,
Shaheen RM, Fan F, Bucana CD, McMahon G, Gallick GE and Ellis LM:
Induction of VEGF in perivascular cells defines a potential
paracrine mechanism for endothelial cell survival. FASEB J.
15:1239–1241. 2001.PubMed/NCBI
|
|
32
|
Bellamy WT, Richter L, Sirjani D, Roxas C,
Glinsmann-Gibson B, Frutiger Y, Grogan TM and List AF: Vascular
endothelial cell growth factor is an autocrine promoter of abnormal
localized immature myeloid precursors and leukemia progenitor
formation in myelodysplastic syndromes. Blood. 97:1427–1434. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van Haaften G and Agami R: Tumorigenicity
of the miR-17-92 cluster distilled. Genes Dev. 24:1–4. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et
al: Identification of the miR-106b~25 microRNA cluster as a
proto-oncogenic PTEN-targeting intron that cooperates with its host
gene MCM7 in transformation. Sci Signal. 3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiao C, Srinivasan L, Calado DP, Patterson
HC, Zhang B, Wang J, Henderson JM, Kutok JL and Rajewsky K:
Lymphoproliferative disease and autoimmunity in mice with increased
miR-17-92 expression in lymphocytes. Nat Immunol. 9:405–414. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17- through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW and Yang BB: MicroRNA
miR-93 promotes tumor growth and angiogenesis by targeting
integrin-β8. Oncogene. 30:806–821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fang L, Du WW, Yang W, Rutnam ZJ, Peng C,
Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al: MiR-93
enhances angiogenesis and metastasis by targeting LATS2. Cell
Cycle. 11:4352–4365. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Q, Lu G, Cai Y, Li Y, Xu R, Ke Y and
Zhang S: MiR-124-5p inhibits the growth of high-grade gliomas
through posttranscriptional regulation of LAMB1. Neuro Oncol.
16:637–651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang X, Du WW, Li H, Liu F, Khorshidi A,
Rutnam ZJ and Yang BB: Both mature miR-17-5p and passenger strand
miR-17-3p target TIMP3 and induce prostate tumor growth and
invasion. Nucleic Acids Res. 41:9688–9704. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shan SW, Fang L, Shatseva T, Rutnam ZJ,
Yang X, Du W, Lu W, Xuan JW, Deng Z and Yang BB: Mature miR-17-5p
and passenger miR-17-3p induce hepatocellular carcinoma by
targeting PTEN, GalNT7 and vimentin in different signal pathways. J
Cell Sci. 126:1517–1530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruan GR, Liu YR, Chen SS, Fu JY, Chang Y,
Qin YZ, Li JL, Yu H and Wang H: Effect of antisense VEGF cDNA
transfection on the growth of chronic myeloid leukemia K562 cells
in vitro and in nude mice. Leuk Res. 28:763–769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bamba H, Ota S, Kato A, Kawamoto C and
Fujiwara K: Prostaglandins up-regulate vascular endothelial growth
factor production through distinct pathways in differentiated U937
cells. Biochem Biophys Res Commun. 273:485–491. 2000. View Article : Google Scholar : PubMed/NCBI
|