Introduction
Neuroblastoma is one of the most prevalent types of
cancer that develops in young children, and is characterized by
aggressive progression, maturation or spontaneous regression. At
diagnosis, neuroblastoma usually presents with metastasis and a
poor outcome (1,2). A key stage of neuroblastoma metastasis
is the invasion of the cells of the primary tumor, which have
advanced invasive and migratory abilities, into the surrounding
normal tissues (3). At present, a
number of microRNAs (miRNAs or miRs), including miR-10b, miR29a/b,
miR-335, miR-7 and miR-338-3p, have been identified as being
associated with tumor progression in neuroblastoma (4). However, the precise mechanisms that
underlie this process, particularly the crucial molecular pathways,
remain to be elucidated.
Similar to other types of cancer, neuroblastoma
development is a complex process with accumulation of epigenetic
and genetic changes. The modified expression of miRNAs, including
miR-148a, miR-21 and miR-200a, has been identified to modulate cell
growth, migration, invasion and apoptosis in neuroblastoma
(5,6).
Thus, additional investigation into the role of miRNAs is required
in order to establish their function in the development of
neuroblastoma (7,8).
Previously, a miRNA assay identified the novel miRNA
miR-506, which regulated neuroblastoma cell differentiation
(9); however, the role of miR-506 in
the development of the disease remains unknown. In the current
study, the biological function of miR-506 in neuroblastoma was
investigated. The results demonstrated that Rho-associated,
coiled-coil containing protein kinase 1 (ROCK1) is a target of
miR-506, and the expression of ROCK1 is positively associated with
metastasis in patients with neuroblastoma. In addition, in
vitro experiments demonstrated that ROCK1 is implicated in
neuroblastoma cell invasion and motility. These results suggest
that miR-506 functions as a tumor suppressor, inhibiting
neuroblastoma metastasis by directly targeting ROCK1.
Materials and methods
Clinical specimens
Primary and normal biopsy specimens from children
with neuroblastoma were obtained from the Second Hospital of
Shandong University (Jinan, China). The identification of the tumor
and normal tissues was histologically confirmed by hematoxylin and
eosin staining. Informed consent was obtained from each patient,
and the research protocols were approved by the Ethics Committee of
the Second Hospital of Shandong University.
Cell culture and antibodies
IMR-32, N2A, SK-N-SH and SH-SY5Y cells were
originally obtained from the American Type Culture Collection
(Manassas, VA, USA), and were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich, St. Louis, MO, USA). The cells were cultured at
37°C in a 5% CO2 humidified atmosphere. ROCK1 primary
monoclonal rabbit antibody (1:1,000 dilution) was purchased from
Cell Signaling Technology, Inc. (Danvers, MA, USA; catalog no.,
4035). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA).
Transfection
miRNA mimics and miRNA antagomiRs were designed and
synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The
miRNA antagomiRs were composed of nucleotides with a 2′-O-methyl
modification. Cells were transiently transfected with miRNA mimics
and miRNA antagomiRs using Invitrogen Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Following the manufacturer's protocol, total RNA was
isolated from the cells using Invitrogen TRIzol® Reagent
(Thermo Fisher Scientific, Inc.). A total of 2 µg RNA was treated
with DNase (Promega Corporation, Madison, WI, USA) to remove
contaminating DNA prior to RT of the RNA to cDNA, which was
completed using the One Step SYBR® PrimeScript™ RT-PCR
kit (Takara Bio, Inc., Otsu, Japan). RT was performed with specific
primers, with the reaction mixtures incubated at 16°C for 30 min,
42°C for 30 min and 85°C for 5 min. To measure messenger (m)RNA
expression, RT-qPCR was performed using the ABI PRISM 7900HT
Sequence Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primers were purchased from Invitrogen (Thermo
Fisher Scientific, Inc.). Primer sequences were as follows:
Forward, 5′-TAAGGCACCCTTCTGAGTAGA-3′ and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′ for miR-506; forward,
5′-AGAGCCTGTGGTGTCCG-3′ and reverse 5′-CATCTTCAAAGCACTTCCCT-3′ for
U6; forward, 5′-AACCATGTGACTGAGTGCCC-3′ and reverse,
5′-TCAGTGTGTTGTGCCAAAGC-3′ for ROCK1; and forward
5′-AATCCCATCACCATCTTCCA-3′ and reverse, 5′-TGGACTCCACGACGTACTCA-3′
for GAPDH. PCR cycling conditions were as follows: An initial
denaturation step at 95°C for 10 min, followed by 40 cycles of
denaturing at 95°C for 10 sec, annealing and synthesis at 60°C for
60 sec. The relative expression levels were calculated by comparing
Cq values of the samples with those of the reference
(2−∆∆Cq), and all data were normalized to the internal
control GAPDH (10).
Methyl thiazolyl tetrazolium (MTT)
assay
An MTT assay was used to detect the growth of cells,
and the growth curve was delineated. Logarithmic phase cells were
collected, and the concentration of the cell suspension was
adjusted to 5,000 cells per well. The wells at the edge of the
plate were filled with aseptic phosphate-buffered saline. The cells
were incubated at 37°C with 5% CO2 and were cultured
once they had covered the bottom of the well in a flat-bottom
96-well plate (Corning Incorporated, Corning, NY, USA). A total of
20 µl MTT solution was added to each well (5 mg/ml; 0.5% MTT;
Sigma-Aldrich), and the cells were cultured for 4 h at 37°C.
Following incubation, the supernatant was discarded and 150 µl
dimethyl sulfoxide (Sigma-Aldrich) was added to each well, and the
culture plate was agitated at a low speed for 10 min until the
crystal had dissolved completely. An enzyme-linked immunosorbent
assay reader (MR-201 ELISA Microplate Reader) was used to measure
the absorbance at 570 nm.
Western blot analysis
Western blots were performed as previously described
(11). Cells were lysed in
radioimmunoprecipitation assay buffer (Sigma-Aldrich), containing
1X protease inhibitor cocktail (Sigma-Aldrich), and protein
concentrations were determined using the Quick Start™ Bradford
protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Proteins were separated by 12.5% sodium dodecyl sulfate
polyacrylamide gel electrophoresis and transferred to membranes
(EMD Millipore, Billerica, MA, USA) at 55 V for 4 h at 4°C.
Subsequent to blocking in 5% non-fat dry milk in Tris-buffered
saline (TBS; Sigma-Aldrich), the membranes were incubated with
primary antibodies in a 1:1,000 dilution in TBS overnight at 4°C.
The membranes were then washed 3 times with TBS-Tween 20
(Sigma-Aldrich), and incubated with secondary antibodies conjugated
with horseradish peroxidase in a 1:5,000 dilution in TBS for 1 h at
room temperature. The membranes were then washed 3 times in
TBS-Tween 20 at room temperature. The protein bands were visualized
on X-ray film using an enhanced chemiluminescence detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Migration assay
For the Transwell migration assay, 1×105
cells were plated into the top chamber of the non-coated membrane
(24-well insert; pore size, 8 µm; Corning Incorporated) and allowed
to migrate towards the serum-containing medium in the lower
chamber. Cells were fixed following 24 h of incubation with
methanol and stained with 0.1% crystal violet (2 mg/ml;
Sigma-Aldrich). A total of 3 random fields per well were selected,
and using a light microscope (x40 magnification; BX51; Olympus
Corporation, Tokyo, Japan), the number of cells invading through
the membrane was counted.
Invasion assay
For the invasion assay, 1×105
neuroblastoma cells were plated in the upper-chamber with a
Matrigel-coated membrane (24-well insert; pore size, 8 µm; BD
Biosciences, San Jose, CA, USA). Each well was freshly coated with
Matrigel (60 µg; BD Biosciences) prior to the invasion assay. The
cells were plated in medium without serum or growth factors, whilst
medium supplemented with serum was used as a chemoattractant in the
lower chamber. The cells were incubated for 48 h and cells that did
invade through the pores were removed by a cotton swab. Cells on
the lower surface of the membrane were fixed with methanol and
stained with crystal violet. A total of 3 random fields were
selected, and using a light microscope (x40 magnification; BX51),
the number of cells invading through the membrane was counted.
Statistical analysis
All experiments were performed in triplicate. The
data are expressed as the mean ± standard error of the mean and
were analyzed using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were performed using GraphPad Prism 5.01
software (GraphPad Software Inc., La Jolla, CA, USA).
Results
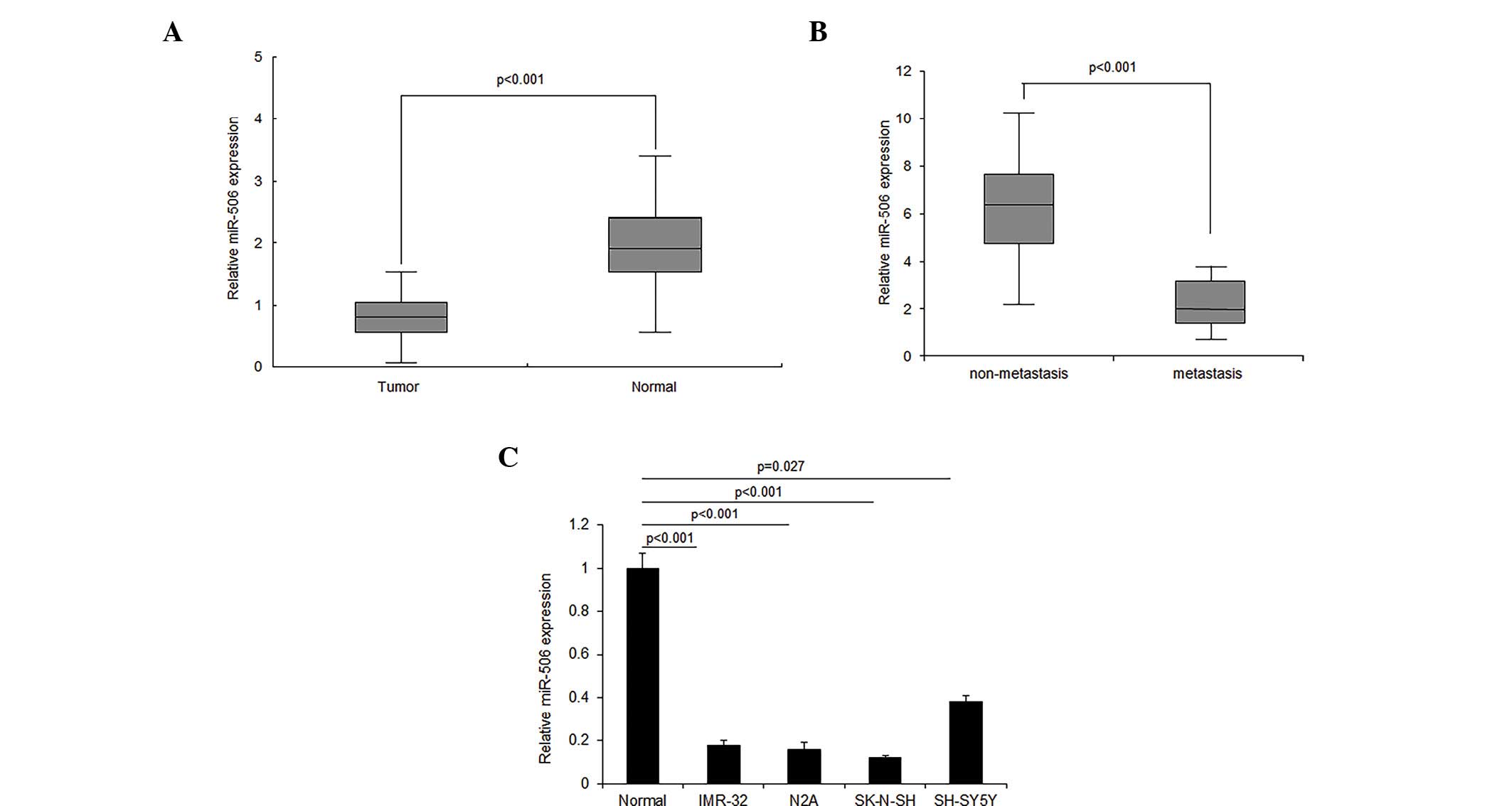
miR-506 expression is decreased in
neuroblastoma tissues and cell lines
The role of miR-506 in neuroblastoma remains
unknown. In order to investigate the role of miR-506, miR-506
expression was detected in neuroblastoma tissues using RT-qPCR. The
results demonstrated that miR-506 expression decreased in ~2/3 of
neuroblastoma tissues, compared with the normal tissues, and
increased in ~1/3 of tissues (Fig.
1A). It was also observed that the loss of miR-506 expression
was associated with metastasis in neuroblastoma tissue (Fig. 1B). Similar results were confirmed by
RT-qPCR in the neuroblastoma cell lines (Fig. 1C). The downregulation of miR-506 in
neuroblastoma tissue and cells indicates that miR-506 may function
as a tumor suppressor gene.
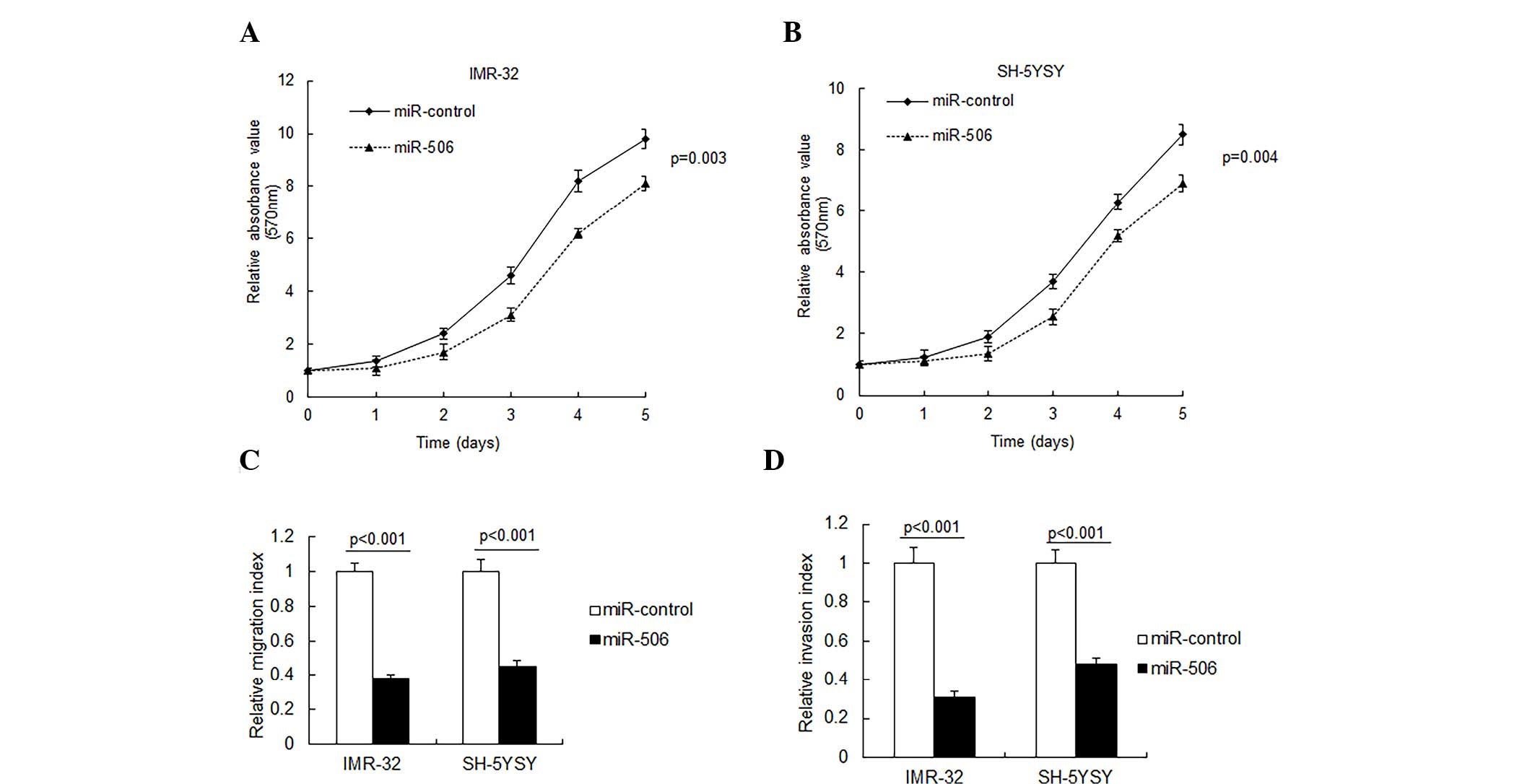
miR-506 inhibits neuroblastoma
proliferation and metastasis
To investigate the role of miR-506 in neuroblastoma
cells, IMR-32 and SH-SY5Y cells were transfected with miR-506 and
cell proliferation was observed by an MTT assay. As presented in
Fig. 2A and B, miR-506 overexpression
in the IMR-32 and SH-SY5Y cells resulted in a significant
inhibition of cell proliferation. Metastasis is an important
characteristic of neuroblastoma, thus cell migration and invasion
were analyzed by Transwell migration assay. It was demonstrated
that miR-506 significantly inhibited cell migration and invasion in
the IMR-32 and SH-5YSY cells (P<0.001; Fig. 2C and D).
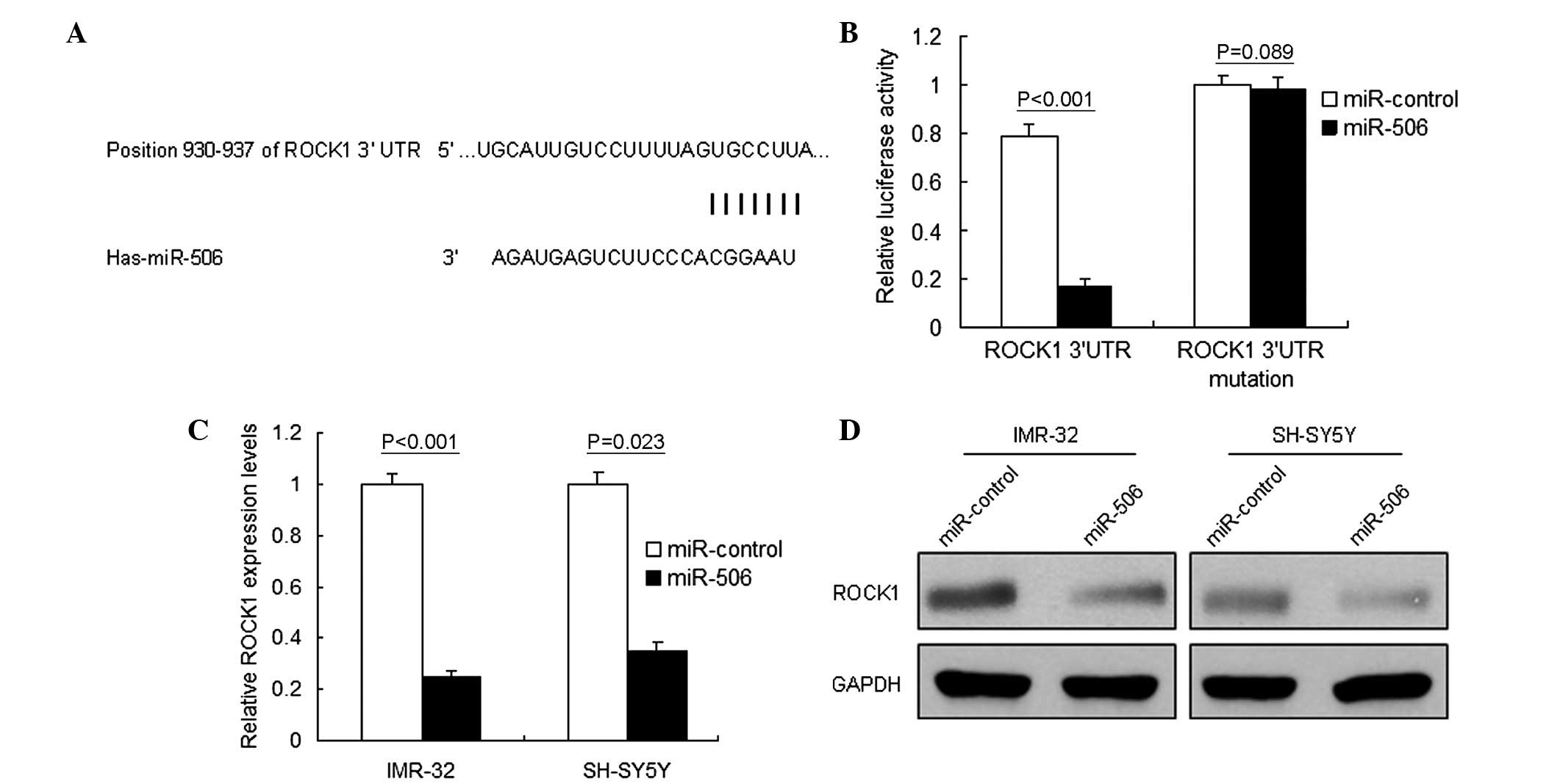
ROCK1 is a target gene of miR-506 in
neuroblastoma
A single miRNA, including miR-506, may regulate the
expression of numerous target genes. Using Targetscan (http://www.targetscan.org) and miRBase (www.mirbase.org), multiple genes were predicted to be
potential target genes of miR-506. The target genes with higher
scores were selected for RT-qPCR analysis, and ROCK1 was noted as
one of the downregulated genes (Fig.
3A). The present study focused on ROCK1 in particular due to
its important role in various types of cancer, including
neuroblastoma. Luciferase activity assay identified ROCK1 as the
target gene of miR-506 in IMR-32 cells (P<0.001; Fig. 3B). RT-qPCR and western blot analysis
were performed to analyze ROCK1 mRNA and protein levels in
neuroblastoma cells. It was demonstrated that ROCK1 mRNA
(P<0.001; Fig. 3C) and protein
(Fig. 3D) levels decreased in the
IMR-32 cells treated with miR-506. These results verified that
ROCK1 was a direct target gene of miR-506.
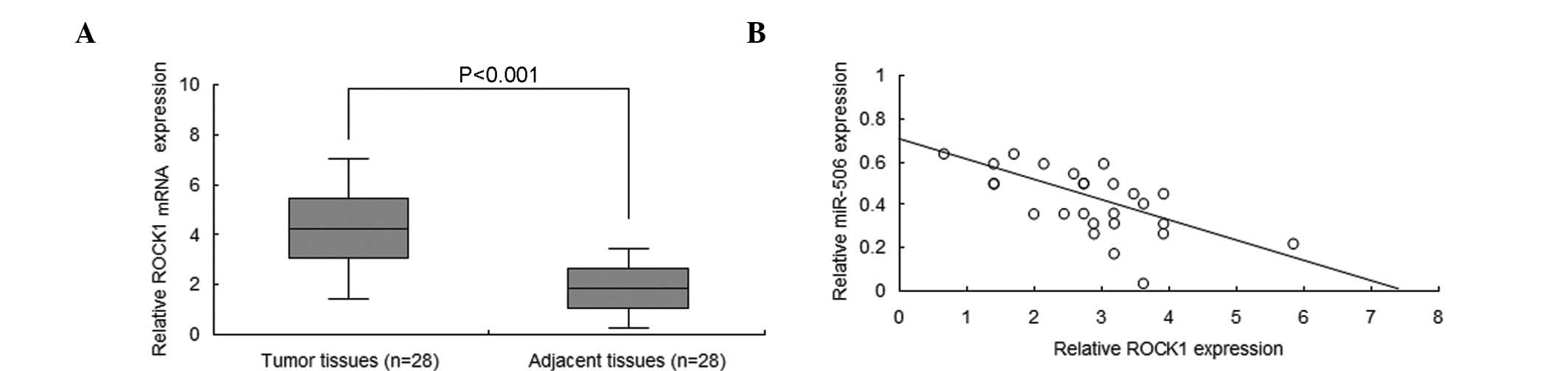
miR-506 expression is negatively
associated with ROCK1 in neuroblastoma tissues
To increase the understanding of the association
between miR-506a and ROCK1 in neuroblastoma, ROCK1 mRNA levels were
analyzed by RT-qPCR. The results demonstrated that the ROCK1 mRNA
is significantly overexpressed in neuroblastoma tissues when
compared with normal tissues (P<0.001; Fig. 4A). In addition, miR-506 was negatively
associated with ROCK1 in the neuroblastoma tissue (Fig. 4B).
Discussion
miRNAs perform important functions in the regulation
of gene expression (12). The
majority of miRNAs are involved in regulating the biological
functions of cancer, including tumorigenesis, invasion and
recurrence (13,14). The deletion or overexpression of
miRNAs may induce the initiation and development of human cancer,
including neuroblastoma (14). Due to
the multiple mechanisms that may lead to the resistance of
neuroblastoma to therapy, targeting single elements of a signalling
pathway may not be a sufficient treatment. Therefore, it is
desirable to identify molecules, including miRNAs, that are able to
regulate multiple cellular processes. This may minimise the risk of
resistance to therapy and improve the overall clinical response to
treatment. miRNAs are upstream regulators that are able to
simultaneously target large numbers of protein-coding genes and
multiple cancer signaling pathways. Emerging cumulative data reveal
fucntional roles of miRNAs in the origin and progression of
neuroblastoma (14). In the present
study, it was demonstrated that miR-506 functions as a tumor
suppressor in neuroblastoma by directly targeting ROCK1.
Previous studies have reported that the expression
of miR-506 is reduced in numerous types of cancer, including
epithelial ovarian cancer (15–18),
hepatocellular carcinoma (19),
cervical cancer (20), lung cancer
(21), breast cancer (22) and colon cancer (23), and is involved in cancer progression,
notably cell proliferation, metastasis and apoptosis. In the
current study, it was observed that miR-506 was decreased in
neuroblastoma tissues and cell lines when compared to normal
tissues, supporting the notion that miR-506 functions as a
potential tumor suppressor. Additionally, the results demonstrated
that miR-506 inhibited neuroblastoma cell proliferation and
metastasis. The previously reported target genes of miR-506 include
cyclin-dependent kinase4/6-forkhead box protein M1 (18), yes-associated protein (19), GLI family zinc finger 3 (20), nuclear factor-κB p65 (21) and others. In the present study, it was
verified that ROCK1 was a novel target gene for miR-506 in
neuroblastoma.
The Rho-associated serine/threonine kinase family,
of which ROCK1 is a member, operates as a key regulator of actin
cytoskeleton dynamics and organization (24). ROCK1 has various different roles,
including involvement in invasion, metastasis and migration during
tumorigenesis (25). Several miRNAs
target ROCK1 in numerous types of cancer, including miRNA-148a in
nasopharyngeal carcinoma (26) and
miRNA-340 in pediatric osteosarcoma (27). ROCK1 has been positively correlated
with lymph node metastasis and the tumor-node-metastasis stage, and
is implicated in miRNA-148a-induced inhibition of gastric cancer
cell invasion and migration (28).
The present study demonstrated that ROCK1 is a predicted target
gene of miR-506 in neuroblastoma cells, and acts as an oncogene.
Furthermore, it was observed that ROCK1 expression was negatively
associated with miR-506 in neuroblastoma tissue.
In conclusion, the present study indicates that
miR-506 functions as a tumor suppressor in neuroblastoma and
negatively regulates cell proliferation and metastasis.
Furthermore, the current study verified that ROCK1 is direct target
gene of miR-506 in neuroblastoma. The expression of miR-506 is
downregulated in neuroblastoma tissues and cell lines and may have
significant functions in neuroblastoma suppression by
downregulation of ROCK1. ROCK1 may be a novel therapeutic target
molecule for the treatment of neuroblastoma.
References
|
1
|
Park JR, Bagatell R, London WB, Maris JM,
Cohn SL, Mattay KK and Hogarty M: COG Neuroblastoma Committee:
Children's Oncology Group's 2013 blueprint for research:
Neuroblastoma. Pediatr Blood Cancer. 60:985–993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Irwin MS and Park JR: Neuroblastoma:
Paradigm for precision medicine. Pediatr Clin North Am. 62:225–256.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM,
Xiao SD and Ran ZH: MicroRNA 506 regulates expression of PPAR alpha
in hydroxycamptothecin-resistant human colon cancer cells. FEBS
Lett. 585:3560–3568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shalaby T, Fiaschetti G, Baumgartner M and
Grotzer MA: Significance and therapeutic value of miRNAs in
embryonal neural tumors. Molecules. 19:5821–5862. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhi F, Wang R, Wang Q, Xue L, Deng D, Wang
S and Yang Y: MicroRNAs in neuroblastoma: Small-sized players with
a large impact. Neurochem Res. 39:613–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mei H, Lin ZY and Tong QS: The roles of
microRNAs in neuroblastoma. World J Pediatr. 10:10–16. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Domingo-Fernandez R, Watters K, Piskareva
O, Stallings RL and Bray I: The role of genetic and epigenetic
alterations in neuroblastoma disease pathogenesis. Pediatr Surg
Int. 29:101–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Verissimo CS, Molenaar JJ, Fitzsimons CP
and Vreugdenhil E: Neuroblastoma therapy: What is in the pipeline?
Endocr Relat Cancer. 18:R213–R231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stallings RL, Foley NH, Bray IM, Das S and
Buckley PG: MicroRNA and DNA methylation alterations mediating
retinoic acid induced neuroblastoma cell differentiation. Semin
Cancer Biol. 21:283–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R,
Zhang W, Lv Z, Zheng S and Zhou L: NDRG1 as a biomarker for
metastasis, recurrence and of poor prognosis in hepatocellular
carcinoma. Cancer Lett. 310:35–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Almeida MI, Reis RM and Calin GA:
MicroRNAs and metastases - the neuroblastoma link. Cancer Biol
Ther. 9:453–454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schulte JH, Horn S, Schlierf S, Schramm A,
Heukamp LC, Christiansen H, Buettner R, Berwanger B and Eggert A:
MicroRNAs in the pathogenesis of neuroblastoma. Cancer Lett.
274:10–15. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stallings RL: MicroRNA involvement in the
pathogenesis of neuroblastoma: Potential for microRNA mediated
therapeutics. Curr Pharm Des. 15:456–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y,
Rupaimoole R, Rodriguez-Aguayo C, Lopez-Berestein G, Ji P, Chen K,
et al: MiR-506 inhibits multiple targets in the
epithelial-to-mesenchymal transition network and is associated with
good prognosis in epithelial ovarian cancer. J Pathol. 235:25–36.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koutsaki M, Spandidos DA and Zaravinos A:
Epithelial-mesenchymal transition-associated miRNAs in ovarian
carcinoma, with highlight on the miR-200 family: Prognostic value
and prospective role in ovarian cancer therapeutics. Cancer Lett.
351:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot
CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al: Integrated
analyses identify a master microRNA regulatory network for the
mesenchymal subtype in serous ovarian cancer. Cancer Cell.
23:186–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang
D, Kerrigan BC Parker, Shmulevich I, Chen K, Sood AK, et al:
MiR-506 suppresses proliferation and induces senescence by directly
targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol.
233:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Cui M, Sun BD, Liu FB, Zhang XD
and Ye LH: MiR-506 suppresses proliferation of hepatoma cells
through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin. 35:1207–1214.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao Z, Ma X, Hsiao TH, Lin G, Kosti A, Yu
X, Suresh U, Chen Y, Tomlinson GE, Pertsemlidis A and Du L: A
high-content morphological screen identifies novel microRNAs that
regulate neuroblastoma cell differentiation. Oncotarget.
5:2499–2512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin M, Ren X, Zhang X, Luo Y, Wang G,
Huang K, Feng S, Bao X, Huang K, He X, et al: Selective killing of
lung cancer cells by miRNA-506 molecule through inhibiting NF-κB
p65 to evoke reactive oxygen species generation and p53 activation.
Oncogene. 34:691–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nunes KP, Rigsby CS and Webb RC:
RhoA/Rho-kinase and vascular diseases: What is the link? Cell Mol
Life Sci. 67:3823–3836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schofield AV and Bernard O: Rho-associated
coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem
Mol Biol. 48:301–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li HP, Huang HY, Lai YR, Huang JX, Chang
KP, Hsueh C and Chang YS: Silencing of miRNA-148a by
hypermethylation activates the integrin-mediated signaling pathway
in nasopharyngeal carcinoma. Oncotarget. 5:7610–7624. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cai H, Lin L, Cai H, Tang M and Wang Z:
Combined microRNA-340 and ROCK1 mRNA profiling predicts tumor
progression and prognosis in pediatric osteosarcoma. Int J Mol Sci.
15:560–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC,
Lee JH, Lee SS, Ashktorab H, Smoot DT, Ryu KW, et al: MicroRNA 135a
suppresses lymph node metastasis through down-regulation of ROCK1
in early gastric cancer. PLoS One. 9:e852052014. View Article : Google Scholar : PubMed/NCBI
|