Introduction
Colorectal cancer, which is currently one of the
most common malignant diseases, is associated with a high annual
incidence of ~1 million cases (1).
Colorectal cancer is undoubtedly a major health threat to the
world's population. Despite advances in the screening and treatment
of colorectal cancer, which have improved the life expectancy of
patients, the prognosis of patients with colorectal cancer remains
poor (1). Therefore, understanding
the biological mechanisms underlying colorectal cancer progression
is important.
Increasingly, studies have shown that aberrant
microRNA (miRNA) expression participates in the development of
colorectal cancer (2,3). miRNAs are a family of small non-coding
RNAs that are able to post-transcriptionally regulate genes
involved in various biological processes (4–6).
microRNA-433 (miR-433) has been reported to be dysregulated in
several malignancies, including ovarian cancer (7), liver cancer (8) and hemopathy (9). Guo et al (10) reported that miR-433 was downregulated
in gastric cancer and functioned as a tumor suppressor. However,
the roles of miR-433 in colorectal cancer have yet to be
elucidated.
Metastasis associated in colon cancer-1 (MACC1) is
an oncogene, and its overexpression has been associated with the
development and progression of numerous tumors, including gastric
carcinoma (11), hepatocellular
carcinoma (12), lung adenocarcinoma
(13), esophageal cancer (14), glioma (15) and breast cancer (16). Zhen et al (17) demonstrated that MACC1 overexpression
resulted in the upregulation of Met and β-catenin, as well as its
downstream genes, including c-Myc, cyclin D1 and matrix
metallopeptidase 9, and the upstream gene phospho-glycogen synthase
kinase 3β (Ser9). In addition, MACC1 increased the expression of
vimentin and suppressed the expression of E-cadherin in colorectal
cancer (17).
In the present study, the expression of miR-433 and
MACC1 in colorectal cancer was evaluated in order to investigate
the role of miR-433 in the development and progression of
colorectal cancer. It was demonstrated that miR-433 was able to
reduce the viability and induce the apoptosis of colorectal cancer
cells by targeting MACC1.
Materials and methods
Human tissue specimens
A total of 79 patients with colorectal cancer who
had undergone routine surgery at Danyang Hospital Affiliated to
Nantong University (Zhenjiang, China) between July 2008 and April
2014 were enrolled in the present study. Colorectal cancer and the
corresponding adjacent tissues were collected from the 79 patients
during the routine surgery. All tissues were immediately frozen in
liquid nitrogen and stored at −80°C. The tumors were classified
according to the World Health Organization classification system
(18). This study was approved by the
Ethical Committee of The Affiliated Hospital of Nantong University,
and informed consent was obtained from all patients.
Cell culture
Five human colorectal cancer cell lines (SW480,
SW620, HT29, HCT116 and LoVo) were purchased from the American Type
Culture Collection (Manassas, VA, USA). The NCM460 normal human
colon mucosal epithelial cell line was purchased from INCELL
Corporation LLC (San Antonio, TX, USA). All cells were cultured in
RPMI-1640 medium or Dulbecco's modified Eagle's medium supplemented
with 10% fetal bovine serum (all Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100 µg/ml
streptomycin at 37°C in a humidified incubator containing 5%
CO2.
Isolation of total RNA and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cell lines
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA was quantified using a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA), after which miRNA
and mRNA were reverse transcribed into cDNA using the PrimeScript
RT Master Mix (Perfect Real Time) (Takara Biotechnology Co., Ltd.,
Dalian, China). The expression levels of miRNAs were assessed using
TaqMan miRNA assays with TaqMan® Universal Master Mix II
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to
standard protocol. U6 small nuclear RNA was used for normalization.
The primer sequences for miR-433 were as follows: RT primer,
GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACGAATAATG; forward primer,
5′-TGCGGTACGGTGAGCCTGTC-3′; and reverse primer,
5′-CCAGTGCAGGGTCCGAGGT-3′. One-tenth of the RT reaction was used
for qPCR using the TaqMan 2X Universal PCR Master Mix (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) on the ABI 7500 Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and with the following cycling conditions:
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 60 sec. Primer sequences for MACC1 were as follows: Forward,
5′-TTCTTTTGATTCCTCCGGTGA-3′ and reverse,
5′-ACTCTGATGGGCATGTGCTG-3′. Primer sequences for GAPDH were as
follows: Forward, 5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse,
5′-CAAAGTTGTCATGGATGHACC-3′. The relative mRNA expression levels of
MACC1 were normalized to GAPDH using the 2−∆∆Cq method
(19).
Transient transfection
Colorectal carcinoma and NCM460 cells were seeded
into 6-well plates at a density of 2×105 cells/ml per
well. Oligonucleotide hsa-miR-433 mimics (miR-433) and normal
control (NC; miR-control) oligonucleotides were purchased from
Shanghai GenePharma, Co., Ltd. (Shanghai, China). The cells were
transfected with hsa-miR-433 or miR-control at a final
concentration of 100 nM using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell viability assay
Cells were seeded into 96-well plates
(6.0×103 cells/well). MTT assays (Roche Diagnostics
GmbH, Penzberg, Germany) were performed to detect cell viability.
Cells were incubated with MTT (5 mg/ml per well) for 4 h. Dimethyl
sulfoxide was used to dissolve the formazan crystals. PBS was used
as a control. Absorbance at 490 nm was measured using the Infinite
M200 PRO multimode microplate reader (Tecan Benelux BVBA, Mechelen,
Belgium). Three independent experiments were performed in
quintuplicate.
Cell cycle distribution and apoptosis
analysis
The cells were seeded into 6-well plates at a
density of 2×105 cells/ml per well and transfected with
miR-433 mimics or NC for 48 h. Flow cytometry was performed with an
Annexin V-fluorescein isothiocyanate/propidium iodide kit (Miltenyi
Biotec GmbH, Bergisch Gladbach, Germany) using the BD FACSCalibur™
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Three
independent experiments were performed in quintuplicate.
Western blot analysis
For western blotting, the cells were harvested and
total protein was extracted from the cells using
radioimmunoprecipitation assay lysis buffer containing
phenylmethylsulfonyl fluoride (Roche Diagnostics, Basel,
Switzerland). Total protein was quantified using the bicinchoninic
acid protein assay (Beyotime Institute of Biotechnology, Haimen,
China). Proteins were separated by 10% SDS-PAGE and blotted onto
polyvinylidene fluoride membranes (Merck Millipore, Darmstadt,
Germany). After blocking with 5% non-fat milk in 1% TBST, the
membranes were incubated overnight at 4°C with goat anti-human
MACC1 (1:1,000; polyclonal; sc-163595; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), goat anti-human caspase-3 (1:1,000;
polyclonal; sc-22140; Santa Cruz Biotechnology, Inc.), goat
anti-human caspase-9 (1:1,000; polyclonal; sc-8297; Santa Cruz
Biotechnology, Inc.) and goat anti-human GAPDH (1:5,000;
polyclonal; sc-20357; Santa Cruz Biotechnology, Inc.). Next, the
membranes were washed four times with PBS containing 0.1% Tween 20
(PBST; Sigma-Aldrich; Merck Millipore). The secondary antibody,
rabbit anti-goat (1:5,000; polyclonal; sc-2922; Santa Cruz
Biotechnology, Inc.), was then added in PBST for 1 h at 37°C. The
membranes were then washed three times for 15 min with PBST, and
the secondary antibodies were detected using the Pierce™ ECL
Substrate Western Blot Detection system (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Protein bands were visualized using the
Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The integrated optical densities of the bands
were quantified using ImageJ software version 1.48u (https://imagej.nih.gov/ij/; National Institutes of
Health, Bethesda, MD, USA).
Bioinformatics analysis
The bioinformatics software microRNA.org (http://www.microrna.org/microrna/) and TargetScan
(http://www.targetscan.org/) were used to
identify candidate target genes of miR-433.
Plasmid construction and cell
transfection
The 3′-untranslated region (UTR) of MACC1 or a
mutant (MUT) sequence with the predicted target sites was inserted
into the Kpn I and Sac I sites of the pGL3 promoter
vector (GenScript, Nanjing, China). Cells were plated onto 6-well
plates at a density of 2×105 cells/ml per well and
transfected with 100 ng pGL3-MACC1 or pGL3-MACC1-MUT and 50 nM
miR-433 mimics using Lipofectamine 2000. In addition, the MACC1
gene (synthesized by GenScript) was digested with the Mlu I
restriction enzyme and subcloned into the pLV-green fluorescent
protein (GFP) plasmid (kindly donated by the Central Laboratory of
Nanjing Medical University, Nanjing, China) to form pLV-GFP-MACC1.
For amplification of the expression vector, 293T cells (Shanghai
Bioleaf Biotech Co., Ltd., Shanghai, China) were transfected with
pLV-GFP-MACC1 using the calcium phosphate co-precipitation method.
Colorectal cancer cells were subsequently transfected with
pLV-GFP-MACC1 using polybrene (8 µg/ml; Sigma-Aldrich; Merck
Millipore).
Dual luciferase assays
Following incubation with pLV-GFP-MACC1 for 48 h,
the luciferase activity was analyzed using the Dual Luciferase
Reporter Assay system (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. The relative luciferase
activities were determined by normalizing to the activity of
Renilla luciferase.
Statistical analysis
All statistical analyses were performed using the
Stata 11 software (StataCorp LP, College Station, TX, USA), and the
results were presented with GraphPad Prism software version 4.0
(GraphPad Software, Inc., La Jolla, CA, USA). Continuous variables
are expressed as the mean ± standard error of the mean. Differences
between groups were calculated using Student's t-tests. P<0.05
was considered statistically significant.
Results
miR-433 is downregulated in human
colorectal cancer tissues and cell lines
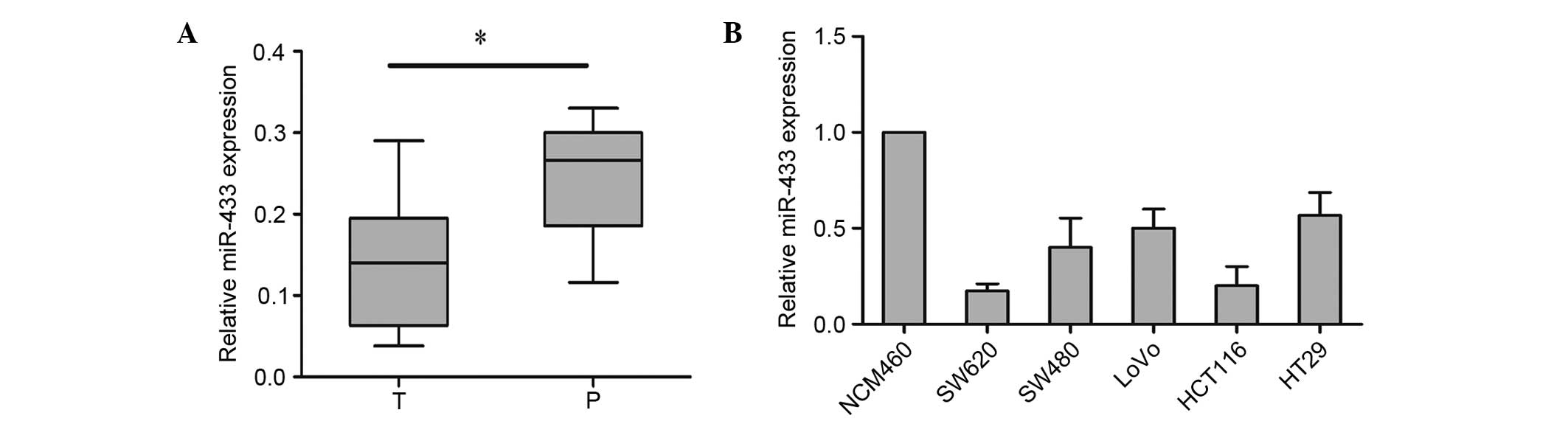
To explore the role of miR-433 in colorectal cancer,
the expression levels of miR-433 in human colorectal cancer samples
and their corresponding adjacent tissues were detected by RT-qPCR.
It was demonstrated that, as compared with the corresponding
adjacent tissues, the expression of miR-433 was significantly
decreased in colorectal cancer tissues (P=0.00085; Fig. 1A). The colorectal cancer tissues were
subsequently divided into either the miR-433 low-expression group
(n=40) or the miR-433 high-expression group (n=39) (Table I); the median expression level for all
samples (median=0.3022; Fig. 1A) was
regarded as the cut-off. From analyzing the clinicopathological
characteristics of all patients, it was determined that aberrant
expression of miR-433 was significantly associated with the tumor
size (P=0.008; Table I), suggesting
that miR-433 may have an important role the pathogenesis of
colorectal cancer. It was then demonstrated that the expression
level of miR-433 was markedly reduced in colorectal cancer cell
lines compared with the NCM460 normal human colon mucosal
epithelial cell line (Fig. 1B), which
was consistent with the result of colorectal cancer tissues. These
results suggest that miR-433 is downregulated in human colorectal
cancer tissues and cell lines.
 | Table I.Expression levels of miR-433 in
colorectal cancer and corresponding adjacent tissues. |
Table I.
Expression levels of miR-433 in
colorectal cancer and corresponding adjacent tissues.
| Characteristics | All patients | miR-433 low
expression | miR-433 high
expression | P-value |
|---|
| Number of
patients | 79 | 40 | 39 |
|
| Age (years) |
|
|
| 0.930 |
|
<60 | 30 | 15 | 15 |
|
| ≥60 | 49 | 25 | 24 |
|
| Gender |
|
|
| 0.206 |
| Male | 45 | 20 | 25 |
|
|
Female | 34 | 20 | 14 |
|
| Histological
differentiation |
|
|
| 0.288 |
|
Well/moderate | 31 | 18 | 13 |
|
|
Poorly | 48 | 22 | 26 |
|
| Tumor size |
|
|
| 0.008a |
| T1,
T2 | 29 | 9 | 20 |
|
| T3,
T4 | 50 | 31 | 19 |
|
| Tumor stage |
|
|
| 0.002a |
| I or
II | 27 | 7 | 20 |
|
| III or
IV | 52 | 33 | 19 |
|
Upregulation of miR-433 promotes the
apoptosis and reduces the viability of colorectal cancer cells
without affecting the cell cycle
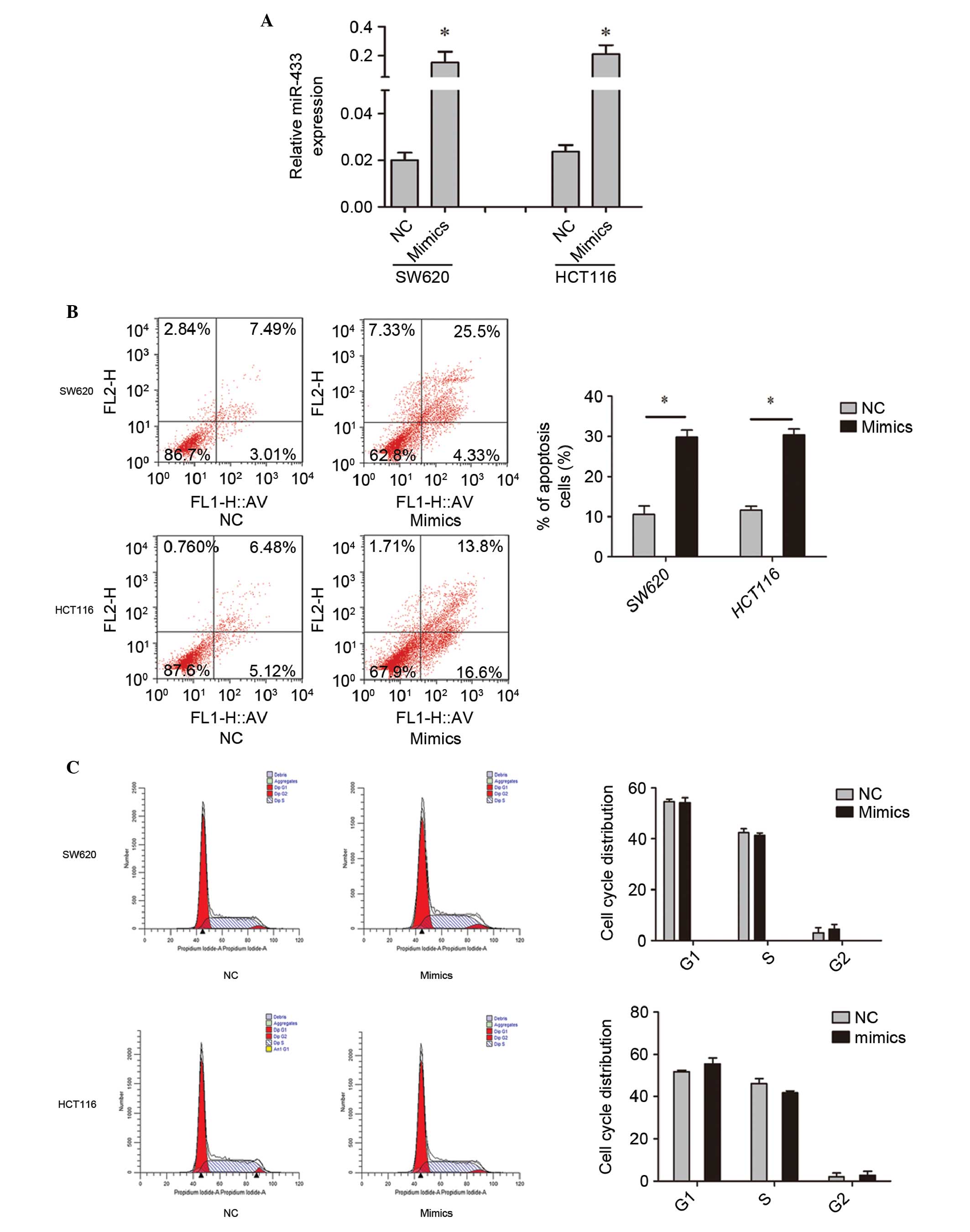
To investigate the functional roles of miR-433, cell
lines were transfected with miR-433 mimics or NC. RT-qPCR was used
to confirm the transfection efficiency (Fig. 2A). Subsequently, the effect of miR-433
on cell apoptosis, the cell cycle distribution and cell viability
was assessed. Flow cytometry demonstrated that overexpression of
miR-433 promoted the apoptosis of the colorectal cancer cell lines
(Fig. 2B). In addition, it was shown
that miR-433 did not affect the cell cycle distribution compared
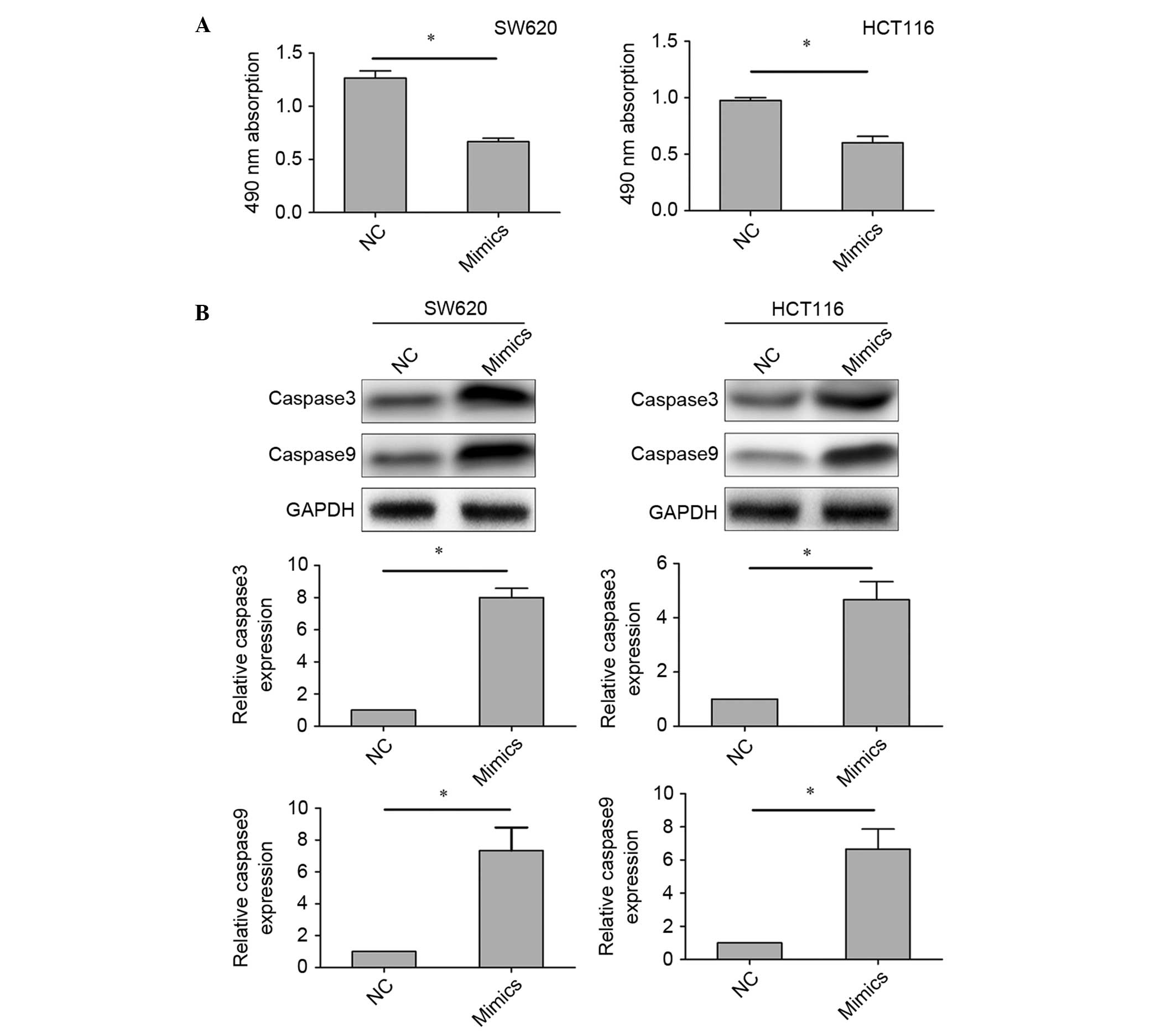
with the control (Fig. 2C). MTT
assays were performed to detect the effect of miR-433 on cell
viability. The results of MTT assays showed that miR-433 mimics
reduced the viability of the colorectal cancer cell lines (Fig. 3A). Furthermore, we investigated the
expression of caspase-3 and caspase-9 in cells transfected with
miR-433 mimics or NC, in order to further assess the effect of
miR-433 on cell apoptosis. Western blotting demonstrated that the
expression levels of caspase-3 and caspase-9 were increased in
cells transfected with miR-433 mimics compared with those
transfected with NC (Fig. 3B). These
results indicate that upregulation of miR-433 promotes cell
apoptosis and reduces cell viability, without affecting the cell
cycle of colorectal cancer cells.
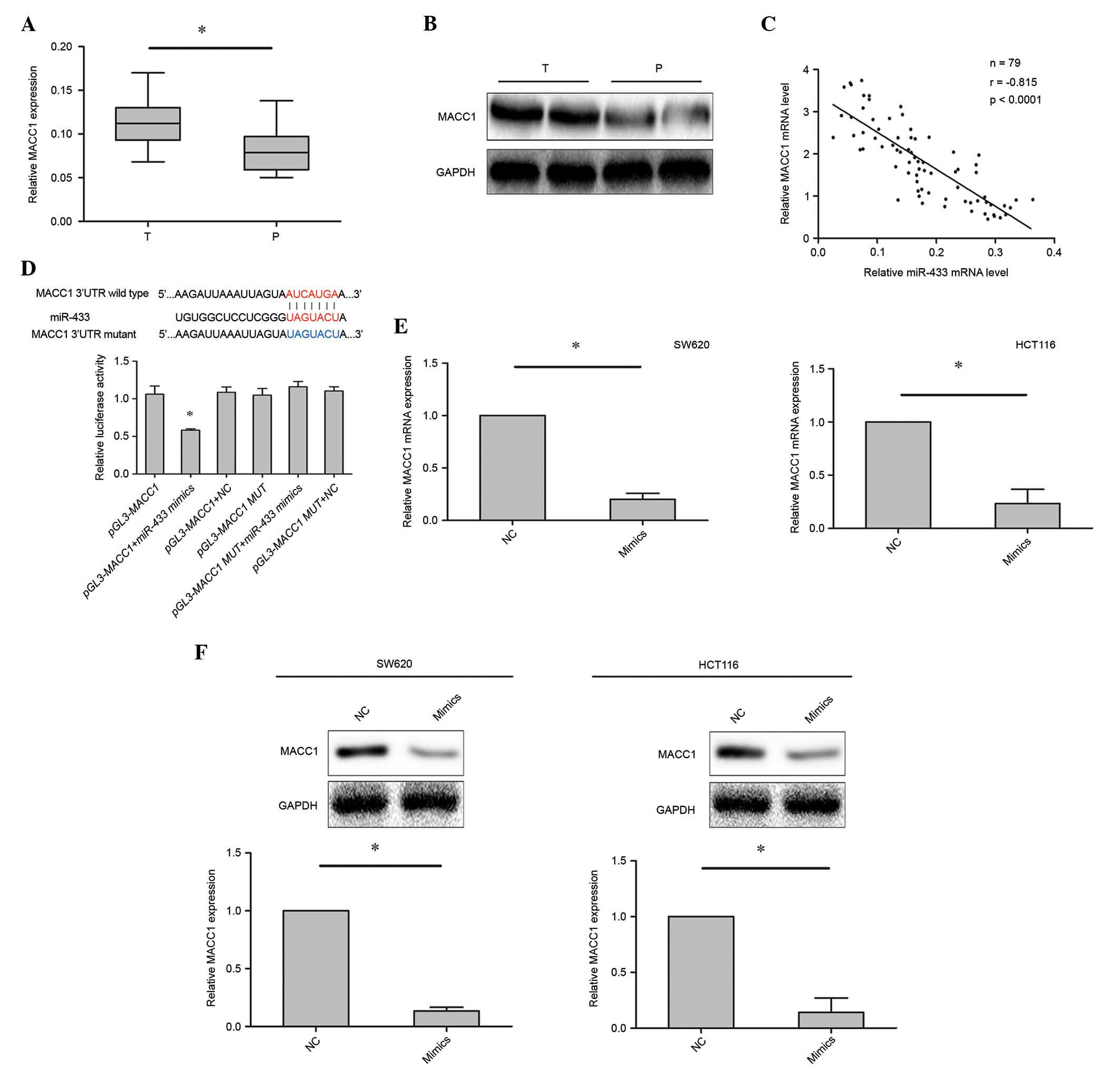
MACC1 is a target gene of miR-433
To investigate the potential role of miR-433 in
colorectal cancer cell apoptosis, a bioinformatics analysis, which
is commonly used to identify the candidate target genes of miRNAs
(20), was performed. The
bioinformatics analysis focused on candidate genes that have
previously been implicated in the regulation of cancer cell
apoptosis. microRNA.org and TargetScan were used to
select the MACC1 gene, which has previously been associated with
colorectal cancer cell proliferation, apoptosis and invasion
(17,21). The mRNA and protein expression levels
of MACC1 in 79 colorectal cancer samples were investigated using
RT-qPCR and western blotting. It was demonstrated that MACC1 was
upregulated in the colorectal cancer tissues compared with the
corresponding adjacent tissues (Fig. 4A
and B). In addition, an inverse correlation was observed
between the expression levels of miR-433 and MACC1 (r=−0.815;
P<0.0001; Fig. 4C) in colorectal
cancer tissues.
To further evaluate whether the inhibitory effect of
MACC1 induced by miR-433 mimics was dependent on the binding of
miR-433 to the 3′-UTR of MACC1, the 3′-UTR fragment of MACC1
containing the predicted binding site was cloned into the pGL3
luciferase reporter vector (pGL3-MACC1). The 3′-UTR fragment
containing the MUT sequence was also cloned into the pGL3
luciferase reporter vector as a control group (pGL3-MACC1-MUT).
Dual luciferase reporter assays showed that the luciferase activity
was decreased in cells transfected with miR-433 mimics and
pGL3-MACC1 vectors, as compared with the control- and
pGL3-MACC1-MUT-transfected cells (Fig.
4D). Furthermore, the mRNA and protein expression levels of
MACC1 in cells transfected with miR-433 mimics or control were
investigated using RT-qPCR and western blotting. Marked inhibition
of MACC1 expression was observed in cells transfected with miR-433
mimics compared with control, suggesting that miR-433 negatively
regulates MACC1 in colorectal cancer cell lines (Fig. 4E and F). These findings suggest that
the MACC1 gene is a target of miR-433.
Discussion
Colorectal cancer is one of the most common causes
of cancer-associated mortality worldwide (1). miRNAs have been reported to have
important regulatory roles in the pathogenesis of colorectal cancer
(2,3).
miR-378 suppresses the proliferation and induces the apoptosis of
colorectal cancer cells by targeting BRAF (22). Regulation of ubiquitin-like PHD and
RING finger domain-containing protein 1 by miR-9 modulates
colorectal cancer cell proliferation and apoptosis (23). The present study aimed to provide
evidence for miR-433 targeting MACC1 in the regulation of the
viability and apoptosis of colorectal cancer. It was demonstrated
that the expression level of miR-433 was downregulated in
colorectal cancer tissues compared with the corresponding adjacent
tissues. Furthermore, the upregulation of miR-433 in colorectal
cancer cells using miR-433 mimics was shown to reduce the viability
and promote the apoptosis of colorectal cancer cells. In addition,
an inverse correlation between the expression levels of miR-433 and
MACC1 was detected in 79 colorectal cancer tissues.
MACC1 is recurrently upregulated in colorectal
cancer, and numerous studies have reported that MACC1 significantly
contributes to lymph node metastases, an advanced TNM stage and
tumor size (20,21); thus suggesting that MACC1 has an
important role in the development of colorectal cancer. MACC1 is a
newly identified key regulator of hepatocyte growth factor-Met
signaling and has been associated with the progression and
prognosis of various carcinomas (15). In a previous study, high MACC1
expression was an independent prognostic indicator for reduced
overall survival in patients with colorectal cancer (24). Furthermore, overexpression of MACC1
has been reported to promote the metastasis and recurrence of
colorectal cancer (24), and it was
associated with peritoneal dissemination and a higher TNM stage
(25). The results of the present
study suggested that miR-433 regulates the function of MACC1 by
binding to its 3-UTR.
In conclusion, the present study demonstrated that
downregulation of miR-433 in colorectal cancer was markedly
associated with cancer development. Upregulation of miR-433 in
colorectal cancer cell lines promoted cell apoptosis and reduced
the cell viability. Furthermore, miR-433 was shown to induce the
apoptosis of colorectal cancer cells by regulating the expression
of MACC1, which has a crucial role in the development of colorectal
cancer. Because of the limited number of colorectal cancer samples
and cell types in the present study, further studies investigating
the potential role of miR-433 in the development of colorectal
cancer are required. In addition, future studies should investigate
whether the miR-433-MACC1 pathway might be exploited in a
therapeutic approach for the treatment of colorectal cancer.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
3′-UTR
|
3′-untranslated region
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
MACC1
|
metastasis associated in colon
cancer-1
|
References
|
1
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang YX, Chen YR, Liu SS, Ye YP, Jiao HL,
Wang SY, Xiao ZY, Wei WT, Qiu JF, Liang L, Liao WT and Ding YQ:
MiR-384 inhibits human colorectal cancer metastasis by targeting
KRAS and CDC42. Oncotarget. doi: 10.18632/oncotarget.12704, 2016
(Epub ahead of print).
|
|
3
|
Wei W, Yang Y, Cai J, Cui K, Li RX, Wang
H, Shang X and Wei D: mir-30a-5p suppresses tumor metastasis of
human colorectal cancer by targeting ITGB3. Cell Physiol Biochem.
39:1165–1176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shao Y, Zhang SQ, Quan F, Zhang PF and Wu
SL: MicroRNA-145 inhibits the proliferation, migration and invasion
of the human TCA8113 oral cancer line. Oncol Lett. 6:1636–1640.
2013.PubMed/NCBI
|
|
5
|
Yang L, Wang YL, Liu S, Zhang PP, Chen Z,
Liu M and Tang H: miR-181b promotes cell proliferation and reduces
apoptosis by repressing the expression of adenylyl cyclase 9 (AC9)
in cervical cancer cells. FEBS Lett. 588:124–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Billeter AT, Barnett RE, Druen D, Polk HC
Jr and van Berkel VH: MicroRNA as a new factor in lung and
esophageal cancer. Semin Thorac Cardiovasc Surg. 24:155–165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weiner-Gorzel K, Dempsey E, Milewska M,
McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A,
Sharpe D, et al: Overexpression of the microRNA miR-433 promotes
resistance to paclitaxel through the induction of cellular
senescence in ovarian cancer cells. Cancer Med. 4:745–758. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Z, Tsuchiya H, Zhang Y, Hartnett ME
and Wang L: MicroRNA-433 inhibits liver cancer cell migration by
repressing the protein expression and function of cAMP response
element-binding protein. J Biol Chem. 288:28893–28899. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin X, Rice KL, Buzzai M, Hexner E, Costa
FF, Kilpivaara O, Mullally A, Soares MB, Ebert BL, Levine R and
Licht JD: miR-433 is aberrantly expressed in myeloproliferative
neoplasms and suppresses hematopoietic cell growth and
differentiation. Leukemia. 27:344–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ge SH, Wu XJ, Wang XH, Xing XF, Zhang LH,
Zhu YB, Du H, Dong B, Hu Y and Ji JF: Over-expression of
metastasis-associated in colon cancer-1 (MACC1) associates with
better prognosis of gastric cancer patients. Chin J Cancer Res.
23:153–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Wu J, Yun J, Lai J, Yuan Y, Gao Z,
Li M, Li J and Song L: MACC1 as a prognostic biomarker for
early-stage and AFP-normal hepatocellular carcinoma. PLoS One.
8:e642352013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Li Z, Wu C, Wang Y, Xia Y, Chen L,
Zhu Q and Chen Y: MACC1 overexpression predicts a poor prognosis
for non-small cell lung cancer. Med Oncol. 31:7902014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu M, Xu Y, Mao X, Gao Y, Shao L and Yan
F: Overexpression of metastasis-associated in colon cancer-1
associated with poor prognosis in patients with esophageal cancer.
Pathol Oncol Res. 19:749–753. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang T, Kong B, Kuang YQ, Cheng L, Gu JW,
Zhang JH, Shu HF, Yu SX, He WQ, Xing XM and Huang HD:
Overexpression of MACC1 protein and its clinical implications in
patients with glioma. Tumour Biol. 35:815–819. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Y, Zhang H, Cai J, Fang L, Wu J, Ye
C, Zhu X and Li M: Overexpression of MACC1 and Its significance in
human breast cancer progression. Cell Biosci. 3:162013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhen T, Dai S, Li H, Yang Y, Kang L, Shi
H, Zhang F, Yang D, Cai S, He Y, et al: MACC1 promotes
carcinogenesis of colorectal cancer via β-catenin signaling
pathway. Oncotarget. 5:3756–3769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quirke P, Williams GT, Ectors N, Ensari A,
Piard F and Nagtegaal I: The future of the TNM staging system in
colorectal cancer: Time for a debate? Lancet Oncol. 8:651–657.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia Y, Zhu Y, Ma T, Pan C, Wang J, He Z,
Li Z, Qia X and Chen Y: miR-204 functions as a tumor suppressor by
regulating SIX1 in NSCLC. FEBS Letters. 588:3703–3712. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kokoszyńska K, Kryński J, Rychlewski L and
Wyrwicz LS: Unexpected domain composition of MACC1 links MET
signaling and apoptosis. Acta Biochim Pol. 56:317–323.
2009.PubMed/NCBI
|
|
22
|
Wang Z, Ma B, Ji X, Deng Y, Zhang T, Zhang
X, Gao H, Sun H, Wu H, Chen X and Zhao R: MicroRNA-378-5p
suppresses cell proliferation and induces apoptosis in colorectal
cancer cells by targeting BRAF. Cancer Cell Int. 15:402015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu M, Xu Y, Ge M, Gui Z and Yan F:
Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell
proliferation and apoptosis. Cancer Sci. 106:833–839. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boardman LA: Overexpression of MACC1 leads
to downstream activation of HGF/MET and potentiates metastasis and
recurrence of colorectal cancer. Genome Med. 1:362009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shirahata A, Shinmura K, Kitamura Y,
Sakuraba K, Yokomizo K, Goto T, Mizukami H, Saito M, Ishibashi K,
Kigawa G, et al: MACC1 as a marker for advanced colorectal
carcinoma. Anticancer Res. 30:2689–2692. 2010.PubMed/NCBI
|