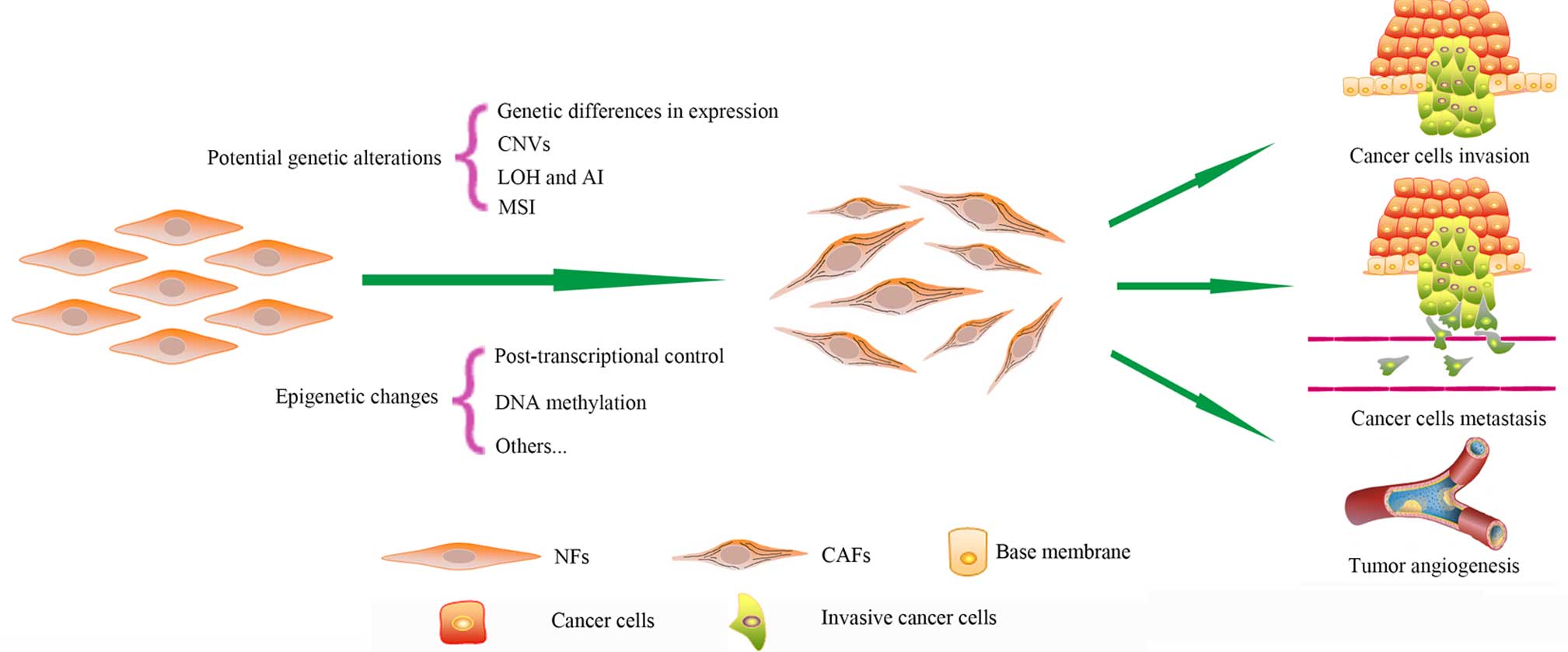
It is widely recognized that the accumulation of
various harmful genetic alterations in normal cells may induce
malignant cancer cells (1). Genetic
mutation is one genetic alteration, but not all genetic mutations
are harmful. Genetic mutation promotes biological evolution and
results in biodiversity (2,3). Genetic alterations include genetic
mutation, gene copy number variation (CNV), loss of heterozygosity
(LOH), allelic imbalance (AI) and microsatellite instability (MSI).
Epigenetic alterations, represented by post-transcriptional control
and DNA methylation, have been the focus of recent studies.
Cancer-associated fibroblasts (CAFs) are one major
type of component in the tumor microenvironment (4). CAFs differ phenotypically and
functionally from normal fibroblasts (NFs) (5). CAFs provide cancer cells with nutrition
and promote the proliferation, invasion and metastasis of cells
(6–9).
CAFs maintain their phenotype for numerous passages during culture
in vitro without exposure to cancer cells, while NFs cannot
be infinitely proliferous like cancer cells (10). Therefore, it has been demonstrated
that genetic or epigenetic alterations may be responsible for the
special features of CAFs (Fig. 1)
(10). Due to the critical role of
CAFs during cancer progression, the genetic characterization of
CAFs aids in the investigation of cancer therapy. The present
review summarizes the current knowledge regarding 7 possible
genetic and epigenetic alterations in CAFs.
CAFs are different from NFs, and their unique
phenotypes and functions are partly determined by differences in
gene expression. The differences between CAFs and NFs in gene
expression have been extensively compared; in one study, 31 genes
in breast CAFs, which were identified using Affymetrix Human Genome
U133 Plus 2.0 and an empirical Bayesian model, were different from
those in NFs (11). Of the 31 genes,
the 21 upregulated genes were primarily associated with cell
paracrine and intracellular signaling, transcription regulation and
cell adhesion and migration, and their transcriptional products
included transforming growth factor-β2 (TGF-β2), insulin-like
growth factor-binding protein 2 and transcriptional factor AP-2α/γ
(11). By contrast, the 10
downregulated genes were primarily associated with epithelial
membrane proteins (11). Genes mainly
involved in coding adhesion molecules and growth factors have also
been found to be upregulated in other types of CAFs, including
colon (12) and pancreatic (13). The prostaglandin-endoperoxide synthase
2 gene (PTGS2), which encodes cyclooxygenase-2, was found to
upregulate the expression of TGF-β2 (14). These results are consistent with
another study concerning the gene expression profiling of breast
CAFs, which were detected using a complementary DNA microarray
(15).
A study on the gene signature of CAFs in non-small
cell lung cancer (NSCLC) revealed similar results to those observed
in breast CAFs (16). In total, there
were 46 genes differently expressed in CAFs compared with NFs, and
the upregulated genes were enriched in cell signal transduction,
cell adhesion, cell response to stress and angiogenesis (16). In total, of 6/46 genes were also
associated with the TGF-β signaling pathway (16). TGF-β is a multifunctional signaling
factor (17–19), which is involved in the
epithelial-to-mesenchymal transition (EMT) (20,21). This
may be one origin of CAFs (5).
Cellular tumor antigen 53 (p53) signal transduction and genes
associated with cell apoptosis and death were downregulated in
colon CAFs (22). In addition, the
gene expression profiling of CAFs may be prognostically significant
for patients with NSCLC or colorectal cancer (CRC) (16,22).
The differences in gene expression between CAFs and
NFs are in keeping with the special function of CAFs in promoting
cancer progression. CAFs promote the growth of cancer cells by
providing nutrition to tumor cells (23,24), and
facilitate the invasion and metastasis of tumor cells by secreting
matrix metalloprotease to resolve the matrix surrounding cancer
cells (16,25–28).
Additionally, CAFs activate tumor angiogenesis (29–31).
CAFs have a gene expression signature different from
NFs, and upregulated genes are primarily associated with
angiogenesis, EMT, cell adhesion and cell interactions (32,33). The
gene expression profiles of CAFs are different among various
tumors, despite certain general features. In the 3 subtypes of
breast tumor, which are differentiated according to 3 receptors,
estrogen, human epidermal growth factor receptor 2 (HER2) and
progesterone, the gene expression profiles of CAFs are even
subtype-specific (34), which may be
used to distinguish the 3 subtypes (32). Cancer stage also affects the gene
expression profiling of CAFs in breast cancer (35).
A karyotype, which is defined as the chromosomal
composition in an individual cell, reflects the number and
structure of chromosomes. Humans are diploid with 46 chromosomes;
however, cancer cells are usually polyploidy or aneuploidy with a
larger nuclei-cytoplasm ratio compared with normal cells.
Polyploidy helps cancer cells to resist adverse factors, and
tetraploid cell lines are more viable than diploid cell lines
(36). Whether or not CAFs have
various chromosome karyotypes in a way that is similar to cancer
cells remains controversial. No clear differences have been
identified in the chromosome karyotype of CAFs in oral carcinoma
(37). By contrast, CAFs from 2 types
of tumors, namely melanoma and prostate cancer (xenograft or
spontaneous), are characterized by aneuploid karyotypes, which are
caused by the attenuated activation of p53 in CAFs (38). Common reasons that account for the
conflicting results observed include the heterogeneity of CAFs and
the various anatomical locations. Furthermore, the largest
difference between the two aforementioned studies is the time in
cell culturing; the study that identified positive karyotype
alterations had cultured CAFs for ≥20 weeks prior to testing
(38). Since survival advantage in
the tumor microenvironment, caused by selection pressures, is the
reason for the alteration of karyotypes, a long-term culture is
more likely to obtain CAFs with altered karyotypes.
Senescence that is activated by signals in CAFs,
including growth-regulated oncogene-1 and the TGF-β/connective
tissue growth factor pathway, may fuel and promote the growth of
cancer cells (39,40). Polyploidy is one of the three steps
leading to the immortality of cancer cells, and cells with normal
karyotypes cannot avoid senescence (41). Consequently, whether the absence of
chromosome karyotype may cause CAF senescence and whether the
senescent CAFs then promote tumor growth remains unclear.
Numerous forms of genetic alterations, including
MSI, LOH, AI and CNV, have been observed in benign and malignant
epithelial neoplasms. The LOH-induced tumor suppressor gene (TSG)
mutation is common among cancer epithelial cells (48–50).
Whether or not LOH and AI are present in CAFs has been the focus of
studies in recent years.
Several types of cancer, including head and neck
squamous cell carcinoma (HNSCC), breast cancer, ovarian cancer,
bladder cancer and CRC, and their associated stromata have been
investigated since 2000. LOH or AI was revealed in all these cancer
stromata. The LOH of the p53 and BRCA1 genes in chromosome 17 was
identified in ulcerative colitis (UC)-associated carcinogenesis
(51) and CRC (52). The LOH in stromal cells is associated
with the colitis-associated carcinogenesis, and the frequency is
increased compared with cancer cells (51). In CAFs of HNSCC, LOH is identified in
the tumor protein 53 (TP53) and phosphatase and tensin homolog
(PTEN) genes (two important tumor-suppressing genes) and the
frequency is associated with tumor size and lymph node metastasis
(53). LOH in TP53 is also observed
in bladder cancer CAFs (54). The
frequency of LOH in chromosome 3p21.3, a region that harbors 3
hyaluronidase genes potentially encoding the hyaluronidase tumor
suppressor, in the stromal cells of ovarian cancer is similar to
the frequency in cancer cells (55).
However, LOH in these genes does not affect the accumulation of
hyaluronidase or tumor progress (55). Breast cancer is most commonly used in
the detection of LOH and AI, and numerous studies focus on TP53 and
PTEN (56). The LOHs of TP53 and PTEN
have been identified in hereditary and sporadic breast cancers:
Hereditary patients are more prone to TP53 mutation; however, it is
only in sporadic breast cancer that LOHs of TP53 and PTEN are
associated with cancer lymph node metastasis, tumor grade and type
of estrogen receptor (57,58). The frequency of LOH is high in
sporadic breast cancer, and LOH at certain loci is noted only in
stromal cells; these loci primarily code TSGs (51,53). LOH
and AI of certain genes occur in tumor and stromal cells, while
some occur only in tumor or stromal cells (53,56). The
occurrence of LOH in the cancer epithelium or stromal cells is not
completely random; certain unique loci may contain an increased
frequency of LOH compared with other loci, whilst several loci may
act as trigger points, at which a LOH may result in a LOH at
another locus (59,60). p53 mutations in CAFs also affect the
CAF in question. LOH or complete loss of p53 in CAFs leads to the
accumulation of stroma in cancer tissues, an increased
proliferation of mesenchymal cells and a mesenchymal response
(61). p53 status is involved in
clinical treatment, since a LOH of p53 in CAFs weakens the response
of cancer cells to radiotherapy and chemotherapy (38,62).
Recently, the acquired resistance to targeted chemotherapy of
pulmonary adenocarcinoma, including epidermal growth factor
receptor tyrosine kinase inhibitor (EGFR-TKI), has attracted more
attention, and EMT may be one major reason (63,64). Since
EMT may be one origin of CAFs (5),
the present study hypothesizes that the p53 status or LOH of p53 in
CAFs of pulmonary adenocarcinoma may play a role in the resistance
against EGFR-TKI. p53 status in cancer cells has been shown to be
associated with tumor angiogenesis (65–68).
Regarding the reason for the multifunctionality of CAFs, altered
p53 signal transduction in CAFs may also be associated with
angiogenesis.
Overall, the present review concludes that genetic
alterations in cancer and stromal cells may occur independently, at
least in breast cancer. In addition, interactions and cross-talk
exist between tumor epithelia and supportive stromal cells. Stromal
cells should be recognized as an equal and independent interactive
component, rather than a response or an accessory to the
carcinoma.
With the exception of MSI in the nuclei of CAFs, MSI
in DNA occurs in the cytoplasmic mitochondria (Mt) of CRC CAFs. The
Mt genome contains 16,569 nucleotides that encode 2 ribosomal RNAs
and 22 transfer RNAs, the production of which is critical for
oxidative phosphorylation (74).
MtMSI has already been reported in several types of cancer cells
(75–77). MtMSI in CAFs is primarily detected in
the D-loop of MtDNA, which contains transcription and replication
regulatory elements (78). Different
from nuclear MSI, MtMSI in CRC CAFs is not related to Duke's stage,
or to nuclear MSI of CAFs or cancer cells. MtMSI may work only in
the development, but not progression, of tumorigenesis (79).
An epigenetic alteration is a local and global gene
expression regulation, which is achieved primarily through DNA
modification and nucleosome rearrangement rather than changes in
DNA base pairs (80). The initiation
and progression of cancer may be accompanied by genetic alterations
and epigenetic regulations (81).
Epigenetic alterations are divided into several subtypes, including
post-transcriptional control, DNA (promoter) methylation,
long-range epigenetic regulation, local nucleosome remodeling,
deposition of histone proteins and covalent modification of
canonical core histones (82).
Certain subtypes associated with CAFs have already been
confirmed.
Post-transcriptional control is a key process in
gene expression, and its major tool is microRNA (miRNA). miRNAs are
small non-coding RNAs of 19–25 nucleotides. miRNA is involved in
numerous cellular progresses, including development,
differentiation, cell response to stress, cell proliferation and
apoptosis (83,84). miRNA acts as an oncogene and tumor
suppressor through various target genes (85). The functions of miRNA have been widely
investigated in various types of cancer cells (86).
Two other studies looking at CAFs in endometrial
cancer have identified 12 miRNAs with various expressions (87). Among 12 miRNAs analyzed, miR-31 is the
most downregulated (93), the target
of which is the SATB2 homeobox gene. Special AT-rich
sequence-binding protein 2 (SATB2) is a matrix attachment
region-binding protein that codes cell type-specific
transcriptional factor involved in regulating transcription in
large chromatin domains (93). The
upregulated expression of SATB2 in breast cancer is associated with
increased tumor grade (94). SATB2 is
highly expressed in breast cancer CAFs, which results in the
increased capacity for promoting the migration of cancer cells and
invasion of CAFs (93). The
introduction of SATB2 into NFs could stimulate the expression of
genes involved in the scattering, migration and invasion of cells
(95). miR-148a is another reduced
miRNA in breast cancer, and by acting with its target Wnt family
member 10B (WNT10B), miR-148a promotes the migration of cancer
cells without affecting growth rate (96). WNT10B may also be involved in the
activation of CAFs (97).
miRNAs are also detected in other types of CAFs. Of
the 4 miRNAs investigated in bladder cancer CAFs, 2 miRNAs (miR-16
and −320) are upregulated and 2 miRNAs are downregulated (98). Certain studies suggested that miR-16
may pro-apoptotically act as a tumor suppressor and miR-320 may be
associated with DNA promoter methylation in cancer cells (99,100). On
the contrary, the expression of miR-143 and miR-145 is reduced in
CAFs, and both are involved in tumor suppression (101). Ovarian CAFs contain 2 downregulated
miRNAs (miR-31 and −214) and 1 upregulated miRNA (miR-155) compared
with NFs. Mimicking the miRNA status using miRNA suppressors and
transforming miRNAs could induce functional conversions from NFs to
CAFs, while the reverse experiment can result in the conversion
from CAFs to NFs (102). Notably,
the upregulation of miR-214 is directly associated with chemokine
(C-C motif) ligand 5, a chemokine that is abundant in the serum and
promotes the invasion and migration of cancer stem cells, and these
actions are similar to CAFs in ovarian cancer (103). However, miR-143 expression is
upregulated in the CAFs of scirrhous gastric cancer rather than in
the non-scirrhous types 40 (104).
miR-143 enhances the expression of collagen type III through
activating the TGF-β/SMAD pathway 40 (104). The increased expression of miR-143
is associated with poor cancer-specific mortality, thus miR-143 may
act as an independent prognostic factor (104). Two miRNAs (miR-15 and −16) with
lower expressions have been indicated in the CAFs of prostate
cancer, and the downregulation of each reduces the
post-transcriptional repression of fibroblast growth
factor-2/fibroblast growth factor receptor-1, which in turn
promotes the growth and progression of cancer cells (105) Therefore, the decreased expressions
of miR-15 and −16 also promote the growth of cancer cells.
Oncogenes, such as B cell lymphoma 2 and Wnt, and
angiogenesis-related genes such as vascular endothelial growth
factor and IL-6 (29), are involved
in the target genes of these two miRNAs (105).
miR-186 in CAFs is involved in glycolysis, and
combined with the 3′-untranslated region of its target gene glucose
transporter 1 (Glut 1), it is active in regulating the uptake of
glucose and production of lactate (106). The roles of CAFs in cancer glucose
and energy metabolism are recognized using a novel method named the
‘reverse Warburg effect’ (23,107,108).
Caveolin-1 is a key pathway in the CAF metabolism conversion
(109,110). Whether or not miR-186 and Glut 1 are
associated with the caveolin-1 pathway requires further
research.
DNA methylation is one epigenetic change and is
mainly divided into two subtypes, hypermethylation and
hypomethylation (80). DNA promoter
hypermethylation could result in the silence of TSGs, while
hypomethylation of oncogenes causes its activation or
overexpression (111,112). Thus, the two subtypes are critical
for tumorigenesis. DNA methylation in CAFs has been widely
investigated.
Tumor stroma is important for cancer progression. In
CRC, the components rich in connective tissues were found to have
an accumulation of chondroitin sulphate proteoglycan. The relevant
gene coding this proteoglycan contains a 3-fold decrease of
hypomethylation and this change only occurs in the CAFs of tumor
stroma, rather than in cancer cells (113,114).
The same hypomethylation is absent in the stroma of UC, indicating
that chronic inflammation is not powerful enough to change cytosine
methylation (115).
In prostate cancer, promoter hypermethylation of
cyclin-dependent kinase inhibitor 2A, hypermethylated in cancer 1
protein, tumor suppressor candidate 3 and glutathione S-transferase
P1 (GSTP1) is extremely abundant in cancer epithelial cells and
CAFs (116). The GSTP1 promoter is
methylated in >90% of prostate cancers. Though GSTP1 may not
suppress cancer cell growth and is not considered to be a TSG, it
may be a caretaker gene and its aberrant silencing in CAFs may
create a tumorigenic microenvironment (117). Promoter hypermethylation could also
be responsible for the inactivation of another candidate TSG, the
opioid binding protein/cell adhesion molecule-like gene, in
invasive cervical cancer CAFs (118). In addition to hypermethylation, DNA
promoter hypomethylation may also exist in CAFs. Activation of long
interspersed nucleotide element-1 in tumorigenesis-related CRC is
due to its hypomethylated promoter (119). Gene methylation profiles are
associated with the status of receptors. In HER2/neu-positive
breast cancer CAFs, 3 corresponding genes, including progesterone
receptor, type IV 17-β-hydroxysteroid dehydrogenase and H-cadherin,
are downregulated (120). These
genes are methylated in breast cancer, but are only slightly or not
methylated in non-neoplastic breast cancers (120). Gene methylation is a dynamic process
in esophageal squamous cancer and cervical cancer; thus, the level
of methylation changes with the progression of cancer (121,122).
Apart from locally methylated DNA, global gene hypomethylation was
confirmed in CAFs of gastric cancer (123). Most importantly, global methylation
of CAFs occurs as early as the dysplastic stage, which could
potentially provide a novel strategy for early diagnosis.
Mechanistically, several possible reasons may
explain the aberrant methylation in CAFs. The field effect could be
a cause of methylation in certain CAFs, as this effect induces the
spread of methylation among various types of cells (124,125);
for example, Septin 9 gene hypermethylation in CAFs occurs later
than in CRC epithelial cells, which may reflect the spread of
Septin 9 hypermethylation from CRC epithelial cells to CAFs. CAF
methylation possibly occurs through field effects from cancer cells
(126). In addition, abnormal cell
proliferation, a local decrease of methyl donors under certain
premalignant conditions, or both of these factors may result in
hypomethylation (123,126). As for DNA promoter hypermethylation
in cancer cells, 3 possible mechanisms have been put forward,
including the infidelity of maintenance DNA
(cytosine-5)-methyltransferase (DNMT)1, the de novo methylating
enzymes, DNMT3a and DNMT3b, and the faulty repair mechanism of
aberrantly methylated DNA (127).
Whether or not these 3 mechanisms are accurate and suitable for
CAFs should be validated through additional studies.
Perspective on the clinical significance. Clinical
trials have already used miRNAs as drug targets or biomarkers for
the stratification of patients, prognosis and drug efficacy
(128). miR-21 is upregulated in the
CAFs of several cancer types and is selected as a diagnostic and
prognostic marker. mir-21 is associated with the Duke's stage of
CRC. Highly expressed stromal miR-21 predicts a poor
recurrence-free survival (129).
Combined with other miRNAs, plasma miR-21 could be a novel method
for early cancer diagnosis. However, miR-143 usually shows a
decreased expression and is considered as a cancer suppressor
(104,130,131).
Erb-b2 receptor tyrosine kinase 3 (ERBB3), one of the four members
of the ERBB family of receptor tyrosine kinases, is targeted by
miR-143, which in turn prevents breast cancer cells from
proliferation and invasion. EGFR-TKI drugs have a special effect in
targeting cancer cells, but patients have to endure drug-resistance
for several months after administration of the drugs. miR-143 could
potentially be a novel pathway for targeted drugs as it can inhibit
cancer cell invasion through an EGFR signalling pathway (132,133).
With regard to upregulated miR-221, a drug called
2′-O-methylphosphorothioate-modified anti-miRNA-221 has already
demonstrated an antitumor effect in mouse models (134). The in vivo administration of
tumor suppressor microRNA or inhibitors of tumor-promoting microRNA
that target CAFs may be an emerging tumor treatment; this may be
more effective compared with existing techniques, considering that
CAFs have a relatively steady phenotype compared with cancer cells
(134).
DNA methyltransferase inhibitors and DNA
methyltransferase maybe be useful for combating CAFs possessing the
corresponding hypermethylation and hypomethylation. The drugs,
5-azacytidine and 5-aza-2′-deoxycytidine, which have already been
used for treatment of acute and chronic myeloid leukemia and
myelodysplastic syndrome, have been investigated in the treatment
other tumors; however, they are less effective in the treatment of
solid tumors and have associated side effects (135).
Whether or not CAFs have somatic or genetic
alterations remains controversial, despite certain alterations that
have already been found. Studies that found a positive somatic
alteration in CAFs, without exception, have extracted DNA from
archival tissues. In other studies (10,47,136,137),
CAFs were isolated from fresh frozen tissues or flow cytometry; no
somatic genetic alterations were found, while CAFs from FFPE show a
high frequency of LOH and AI. This pattern indicates that FFPE
tissues could result in highly fragmented DNA and RNA molecules,
which are not suitable for large-scale genetic analysis. PCR
amplification then exacerbates the artificial false positive result
(138–143). Thus, certain studies refute genetic
alterations and instead attribute the results to epigenetic
alterations More reliable and accurate methods should be found to
solve these controversies.
CAFs retain diploid rather than polyploid cancer
cells, which may account for the senescence of CAFs. Since
telomerase is closely linked with cell senescence, particularly in
cancer cells, the activity and status of telomerase in CAFs may be
worth additional studies, though certain studies have already been
performed (144–146).
CAFs, which are phenotypically and functionally
different from NFs due to numerous potential genetic alterations
and epigenetic changes, are critical for tumor progression and have
attracted increasing numbers of studies in recent years (Fig. 1). With the potential genetic and
epigenetic alterations changes found in CAFs, the underlying
mechanisms regarding the features of CAFs are gradually revealed.
With the exception of genetic and epigenetic alterations, other
relevant changes may also explain the differential expressions of
genes in CAFs, and certain special genetic or epigenetic
alterations may be confirmed in the future. Further investigation
into the detailed genetic and epigenetic alterations of CAFs in the
tumor microenvironment increases the understanding of CAFs and
provides novel approaches for clinical application.
The present study was supported by the National
Natural Science Foundation of China, Beijing, China (grant no.
81071929), which awarded the grant to Professor Guowei Che.
|
1
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crispo E, Moore JS, Lee-Yaw JA, Gray SM
and Haller BC: Broken barriers: Human-induced changes to gene flow
and introgression in animals: An examination of the ways in which
humans increase genetic exchange among populations and species and
the consequences for biodiversity. Bioessays. 33:508–518. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Verde C, di Prisco G and Convey P:
Molecular and genetic advances to understanding evolution and
biodiversity in the polar regions. Mar Genomics. 8:1–2. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia CC, Wang TT, Liu W, Fu BS, Hua X, Wang
GY, Li TJ, Li X, Wu XY, Tai Y, et al: Cancer-associated fibroblasts
from hepatocellular carcinoma promote malignant cell proliferation
by HGF secretion. PLoS One. 8:e632432013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xing F, Saidou J and Watabe K: Cancer
associated fibroblasts (CAFs) in tumor microenvironment. Front
Biosci (Landmark Ed). 15:166–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franco OE, Shaw AK, Strand DW and Hayward
SW: Cancer associated fibroblasts in cancer pathogenesis. Semin
Cell Dev Biol. 21:33–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castello-Cros R, Bonnuccelli G, Molchansky
A, Capozza F, Witkiewicz AK, Birbe RC, Howell A, Pestell RG,
Whitaker-Menezes D, Sotgia F and Lisanti MP: Matrix remodeling
stimulates stromal autophagy, ‘fueling’ cancer cell mitochondrial
metabolism and metastasis. Cell Cycle. 10:2021–2034. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balliet RM, Capparelli C, Guido C, Pestell
TG, Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Chiavarina
B, Pestell RG, Howell A, et al: Mitochondrial oxidative stress in
cancer-associated fibroblasts drives lactate production, promoting
breast cancer tumor growth: Understanding the aging and cancer
connection. Cell Cycle. 10:4065–4073. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonuccelli G, Tsirigos A, Whitaker-Menezes
D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N,
Howell A, Martinez-Outschoorn UE, et al: Ketones and lactate ‘fuel’
tumor growth and metastasis: Evidence that epithelial cancer cells
use oxidative mitochondrial metabolism. Cell Cycle. 9:3506–3514.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qiu W, Hu M, Sridhar A, Opeskin K, Fox S,
Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, et
al: No evidence of clonal somatic genetic alterations in
cancer-associated fibroblasts from human breast and ovarian
carcinomas. Nat Genet. 40:650–655. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bauer M, Su G, Casper C, He R, Rehrauer W
and Friedl A: Heterogeneity of gene expression in stromal
fibroblasts of human breast carcinomas and normal breast. Oncogene.
29:1732–1740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakagawa H, Liyanarachchi S, Davuluri RV,
Auer H, Martin EW Jr, de la Chapelle A and Frankel WL: Role of
cancer-associated stromal fibroblasts in metastatic colon cancer to
the liver and their expression profiles. Oncogene. 23:7366–7377.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sato N, Maehara N and Goggins M: Gene
expression profiling of tumor-stromal interactions between
pancreatic cancer cells and stromal fibroblasts. Cancer Res.
64:6950–6956. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada C, Aikawa T, Okuno E, Miyagawa K,
Amano K, Takahata S, Kimata M, Okura M, Iida S and Kogo M: TGF-β in
jaw tumor fluids induces RANKL expression in stromal fibroblasts.
Int J Oncol. 49:499–508. 2016.PubMed/NCBI
|
|
15
|
Singer CF, Gschwantler-Kaulich D,
Fink-Retter A, Haas C, Hudelist G, Czerwenka K and Kubista E:
Differential gene expression profile in breast cancer-derived
stromal fibroblasts. Breast Cancer Res Treat. 110:273–281. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Navab R, Strumpf D, Bandarchi B, Zhu CQ,
Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L,
Barczyk M, et al: Prognostic gene-expression signature of
carcinoma-associated fibroblasts in non-small cell lung cancer.
Proc Natl Acad Sci USA. 108:7160–7165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wakefield LM and Roberts AB: TGF-beta
signaling: Positive and negative effects on tumorigenesis. Curr
Opin Genet Dev. 12:22–29. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herrera M, Islam AB, Herrera A, Martín P,
García V, Silva J, Garcia JM, Salas C, Casal I, de Herreros AG, et
al: Functional heterogeneity of cancer-associated fibroblasts from
human colon tumors shows specific prognostic gene expression
signature. Clin Cancer Res. 19:5914–5926. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Migneco G, Whitaker-Menezes D, Chiavarina
B, Castello-Cros R, Pavlides S, Pestell RG, Fatatis A, Flomenberg
N, Tsirigos A, Howell A, et al: Glycolytic cancer associated
fibroblasts promote breast cancer tumor growth, without a
measurable increase in angiogenesis: Evidence for
stromal-epithelial metabolic coupling. Cell Cycle. 9:2412–2422.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leiherer A, Geiger K, Muendlein A and
Drexel H: Hypoxia induces a HIF-1α dependent signalling cascade to
make a complex metabolic switch in SGBS-adipocytes. Mol Cell
Endocrinol. 383:21–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu C, Wang Z, Zhai L, Yang M, Shan L, Chai
C, Liu M and Wang L: Effects of cancer-associated fibroblasts on
the migration and invasion abilities of SGC-7901 gastric cancer
cells. Oncol Lett. 5:609–612. 2013.PubMed/NCBI
|
|
26
|
Kim SH, Choe C, Shin YS, Jeon MJ, Choi SJ,
Lee J, Bae GY, Cha HJ and Kim J: Human lung cancer-associated
fibroblasts enhance motility of non-small cell lung cancer cells in
co-culture. Anticancer Res. 33:2001–2009. 2013.PubMed/NCBI
|
|
27
|
Cao M, Seike M, Soeno C, Mizutani H,
Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L and Gemma A:
MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition
by targeting E-cadherin in lung cancer cells. Int J Oncol.
41:869–875. 2012.PubMed/NCBI
|
|
28
|
Schveigert D, Cicenas S, Bruzas S,
Samalavicius NE, Gudleviciene Z and Didziapetriene J: The value of
MMP-9 for breast and non-small cell lung cancer patients' survival.
Adv Med Sci. 58:73–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nagasaki T, Hara M, Nakanishi H, Takahashi
H, Sato M and Takeyama H: Interleukin-6 released by colon
cancer-associated fibroblasts is critical for tumour angiogenesis:
Anti-interleukin-6 receptor antibody suppressed angiogenesis and
inhibited tumour-stroma interaction. Br J Cancer. 110:469–478.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Ansari MM, Hendrayani SF, Tulbah A,
Al-Tweigeri T, Shehata AI and Aboussekhra A: P16INK4A represses
breast stromal fibroblasts migration/invasion and their
VEGF-A-dependent promotion of angiogenesis through Akt inhibition.
Neoplasia. 14:1269–1277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Orimo A, Gupta PB, Sgroi DC,
Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL
and Weinberg RA: Stromal fibroblasts present in invasive human
breast carcinomas promote tumor growth and angiogenesis through
elevated SDF-1/CXCL12 secretion. Cell. 121:335–348. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sadlonova A, Bowe DB, Novak Z, Mukherjee
S, Duncan VE, Page GP and Frost AR: Identification of molecular
distinctions between normal breast-associated fibroblasts and
breast cancer-associated fibroblasts. Cancer Microenviron. 2:9–21.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tchou J, Kossenkov AV, Chang L, Satija C,
Herlyn M, Showe LC and Puré E: Human breast cancer associated
fibroblasts exhibit subtype specific gene expression profiles. BMC
Med Genomics. 5:392012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park SU, Choi ES, Jang YS, Hong SH, Kim IH
and Chang DK: Effects of chromosomal polyploidy on survival of
colon cancer cells. Korean J Gastroenterol. 57:150–157. 2011.(In
Korean). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng XH, Liu Y, Zhou HM, Chen QM and Li
BQ: Analysis of chromosome karyotype of oral carcinoma-associated
Fibroblasts. Hua Xi Kou Qiang Yi Xue Za Zhi. 23:159–160. 2005.(In
Chinese). PubMed/NCBI
|
|
38
|
Dudley AC, Shih SC, Cliffe AR, Hida K and
Klagsbrun M: Attenuated p53 activation in tumour-associated stromal
cells accompanies decreased sensitivity to etoposide and
vincristine. Br J Cancer. 99:118–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang G, Rosen DG, Zhang Z, Bast RC Jr,
Mills GB, Colacino JA, Mercado-Uribe I and Liu J: The chemokine
growth-regulated oncogene 1 (Gro-1) links RAS signaling to the
senescence of stromal fibroblasts and ovarian tumorigenesis. Proc
Natl Acad Sci USA. 103:16472–16477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Capparelli C, Whitaker-Menezes D, Guido C,
Balliet R, Pestell TG, Howell A, Sneddon S, Pestell RG,
Martinez-Outschoorn U, Lisanti MP and Sotgia F: CTGF drives
autophagy, glycolysis and senescence in cancer-associated
fibroblasts via HIF1 activation, metabolically promoting tumor
growth. Cell Cycle. 11:2272–2284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Erenpreisa J and Cragg MS: Three steps to
the immortality of cancer cells: Senescence, polyploidy and
self-renewal. Cancer Cell Int. 13:922013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bowcock AM: Invited review DNA copy number
changes as diagnostic tools for lung cancer. Thorax. 69:495–496.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tuhkanen H, Anttila M, Kosma VM, Heinonen
S, Juhola M, Helisalmi S, Kataja V and Mannermaa A: Frequent gene
dosage alterations in stromal cells of epithelial ovarian
carcinomas. Int J Cancer. 119:1345–1353. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pelham RJ, Rodgers L, Hall I, Lucito R,
Nguyen KC, Navin N, Hicks J, Mu D, Powers S, Wigler M and Botstein
D: Identification of alterations in DNA copy number in host stromal
cells during tumor progression. Proc Natl Acad Sci USA.
103:19848–19853. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Carles-Kinch K, Kilpatrick KE, Stewart JC
and Kinch MS: Antibody targeting of the EphA2 tyrosine kinase
inhibits malignant cell behavior. Cancer Res. 62:2840–2847.
2002.PubMed/NCBI
|
|
46
|
Mao W, Luis E, Ross S, Silva J, Tan C,
Crowley C, Chui C, Franz G, Senter P, Koeppen H and Polakis P:
EphB2 as a therapeutic antibody drug target for the treatment of
colorectal cancer. Cancer Res. 64:781–788. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rummel S, Valente AL, Kane JL, Shriver CD
and Ellsworth RE: Genomic (in)stability of the breast tumor
microenvironment. Mol Cancer Res. 10:1526–1531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rohrbach H, Haas CJ, Baretton GB,
Hirschmann A, Diebold J, Behrendt RP and Löhrs U: Microsatellite
instability and loss of heterozygosity in prostatic carcinomas:
Comparison of primary tumors and of corresponding recurrences after
androgen-deprivation therapy and lymph-node metastases. Prostate.
40:20–27. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Smith HS, Lu Y, Deng G, Martinez O, Krams
S, Ljung BM, Thor A and Lagios M: Molecular aspects of early stages
of breast cancer progression. J Cell Biochem Suppl 17G. 144–152.
1993. View Article : Google Scholar
|
|
50
|
Agapova LS, Ivanov AV, Sablina AA, Kopnin
PB, Sokova OI, Chumakov PM and Kopnin BP: P53-dependent effects of
RAS oncogene on chromosome stability and cell cycle checkpoints.
Oncogene. 18:3135–3142. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Matsumoto N, Yoshida T and Okayasu I: High
epithelial and stromal genetic instability of chromosome 17 in
ulcerative colitis-associated carcinogenesis. Cancer Res.
63:6158–6161. 2003.PubMed/NCBI
|
|
52
|
Wernert N, Löcherbach C, Wellmann A,
Behrens P and Hügel A: Presence of genetic alterations in
microdissected stroma of human colon and breast cancers. Anticancer
Res. 21:2259–2264. 2001.PubMed/NCBI
|
|
53
|
Moinfar F, Man YG, Arnould L, Bratthauer
GL, Ratschek M and Tavassoli FA: Concurrent and independent genetic
alterations in the stromal and epithelial cells of mammary
carcinoma: Implications for tumorigenesis. Cancer Res.
60:2562–2566. 2000.PubMed/NCBI
|
|
54
|
Paterson RF, Ulbright TM, MacLennan GT,
Zhang S, Pan CX, Sweeney CJ, Moore CR, Foster RS, Koch MO, Eble JN
and Cheng L: Molecular genetic alterations in the
laser-capture-microdissected stroma adjacent to bladder carcinoma.
Cancer. 98:1830–1836. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tuhkanen H, Anttila M, Kosma VM,
Ylä-Herttuala S, Heinonen S, Kuronen A, Juhola M, Tammi R, Tammi M
and Mannermaa A: Genetic alterations in the peritumoral stromal
cells of malignant and borderline epithelial ovarian tumors as
indicated by allelic imbalance on chromosome 3p. Int J Cancer.
109:247–252. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kurose K, Gilley K, Matsumoto S, Watson
PH, Zhou XP and Eng C: Frequent somatic mutations in PTEN and TP53
are mutually exclusive in the stroma of breast carcinomas. Nat
Genet. 32:355–357. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
57
|
Patocs A, Zhang L, Xu Y, Weber F, Caldes
T, Mutter GL, Platzer P and Eng C: Breast-cancer stromal cells with
TP53 mutations and nodal metastases. N Engl J Med. 357:2543–2551.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fukino K, Shen L, Patocs A, Mutter GL and
Eng C: Genomic instability within tumor stroma and
clinicopathological characteristics of sporadic primary invasive
breast carcinoma. JAMA. 297:2103–2111. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fukino K, Shen L, Matsumoto S, Morrison
CD, Mutter GL and Eng C: Combined total genome loss of
heterozygosity scan of breast cancer stroma and epithelium reveals
multiplicity of stromal targets. Cancer Res. 64:7231–7236. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kurose K, Hoshaw-Woodard S, Adeyinka A,
Lemeshow S, Watson PH and Eng C: Genetic model of multi-step breast
carcinogenesis involving the epithelium and stroma: Clues to
tumour-microenvironment interactions. Hum Mol Genet. 10:1907–1913.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hill R, Song Y, Cardiff RD and Van Dyke T:
Selective evolution of stromal mesenchyme with p53 loss in response
to epithelial tumorigenesis. Cell. 123:1001–1011. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hawsawi NM, Ghebeh H, Hendrayani SF,
Tulbah A, Al-Eid M, Al-Tweigeri T, Ajarim D, Alaiya A, Dermime S
and Aboussekhra A: Breast carcinoma-associated fibroblasts and
their counterparts display neoplastic-specific changes. Cancer Res.
68:2717–2725. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chung JH, Rho JK, Xu X, Lee JS, Yoon HI,
Lee CT, Choi YJ, Kim HR, Kim CH and Lee JC: Clinical and molecular
evidences of epithelial to mesenchymal transition in acquired
resistance to EGFR-TKIs. Lung Cancer. 73:176–182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shang Y, Cai X and Fan D: Roles of
epithelial-mesenchymal transition in cancer drug resistance. Curr
Cancer Drug Targets. 13:915–929. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schmid JO, Dong M, Haubeiss S, Friedel G,
Bode S, Grabner A, Ott G, Mürdter TE, Oren M, Aulitzky WE and van
der Kuip H: Cancer cells cue the p53 response of cancer-associated
fibroblasts to cisplatin. Cancer Res. 72:5824–5832. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Assadian S, El-Assaad W, Wang XQ, Gannon
PO, Barrès V, Latour M, Mes-Masson AM, Saad F, Sado Y, Dostie J and
Teodoro JG: P53 inhibits angiogenesis by inducing the production of
Arresten. Cancer Res. 72:1270–1279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ghahremani M Farhang, Goossens S, Nittner
D, Bisteau X, Bartunkova S, Zwolinska A, Hulpiau P, Haigh K,
Haenebalcke L, Drogat B, et al: P53 promotes VEGF expression and
angiogenesis in the absence of an intact p21-Rb pathway. Cell Death
Differ. 20:888–897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Teodoro JG, Parker AE, Zhu X and Green MR:
P53-mediated inhibition of angiogenesis through up-regulation of a
collagen prolyl hydroxylase. Science. 313:968–971. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Heinimann K: Toward a molecular
classification of colorectal cancer: The role of microsatellite
instability status. Front Oncol. 3:2722013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Matsumoto N, Yoshida T, Yamashita K,
Numata Y and Okayasu I: Possible alternative carcinogenesis pathway
featuring microsatellite instability in colorectal cancer stroma.
Br J Cancer. 89:707–712. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yagishita H, Yoshida T, Ishiguro K, Numata
Y and Okayasu I: Epithelial and stromal genetic instability linked
to tumor suppressor genes in ulcerative colitis-associated
tumorigenesis. Scand J Gastroenterol. 43:559–566. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu X, Goldblum JR, Zhao Z, Landau M,
Heald B, Pai R and Lin J: Distinct clinicohistologic features of
inflammatory bowel disease-associated colorectal adenocarcinoma: in
comparison with sporadic microsatellite-stable and Lynch
syndrome-related colorectal adenocarcinoma. Am J Surg Pathol.
36:1228–1233. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Shiraishi H, Mikami T, Yoshida T, Tanabe
S, Kobayashi N, Watanabe M and Okayasu I: Early genetic instability
of both epithelial and stromal cells in esophageal squamous cell
carcinomas, contrasted with Barrett's adenocarcinomas. J
Gastroenterol. 41:1186–1196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chomyn A and Attardi G: MtDNA mutations in
aging and apoptosis. Biochem Biophys Res Commun. 304:519–529. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liu VW, Shi HH, Cheung AN, Chiu PM, Leung
TW, Nagley P, Wong LC and Ngan HY: High incidence of somatic
mitochondrial DNA mutations in human ovarian carcinomas. Cancer
Res. 61:5998–6001. 2001.PubMed/NCBI
|
|
76
|
Habano W, Sugai T, Nakamura SI, Uesugi N,
Yoshida T and Sasou S: Microsatellite instability and mutation of
mitochondrial and nuclear DNA in gastric carcinoma.
Gastroenterology. 118:835–841. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Habano W, Nakamura S and Sugai T:
Microsatellite instability in the mitochondrial DNA of colorectal
carcinomas: Evidence for mismatch repair systems in mitochondrial
genome. Oncogene. 17:1931–1937. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Suzuki M, Toyooka S, Miyajima K, Iizasa T,
Fujisawa T, Bekele NB and Gazdar AF: Alterations in the
mitochondrial displacement loop in lung cancers. Clin Cancer Res.
9:5636–5641. 2003.PubMed/NCBI
|
|
79
|
Kim HS, Lim HS, Lee SH, Lee JW, Nam SW,
Park WS, Lee YS, Lee JY and Yoo NJ: Mitochondrial microsatellite
instability of colorectal cancer stroma. Int J Cancer.
119:2607–2611. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dey P: Epigenetic changes in tumor
microenvironment. Indian J Cancer. 48:507–512. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ting AH, McGarvey KM and Baylin SB: The
cancer epigenome-components and functional correlates. Genes Dev.
20:3215–3231. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lund AH and van Lohuizen M: Epigenetics
and cancer. Genes Dev. 18:2315–2335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo
H, Tang X, Zeng Z and Liu M: MiRNA expression analysis of
cancer-associated fibroblasts and normal fibroblasts in breast
cancer. Int J Biochem Cell Biol. 44:2051–2059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Bronisz A, Godlewski J, Wallace JA,
Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ,
Martin CK, Li F, et al: Reprogramming of the tumour
microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol.
14:159–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Rask L, Balslev E, Jørgensen S, Eriksen J,
Flyger H, Møller S, Høgdall E, Litman T and Nielsen BS: High
expression of miR-21 in tumor stroma correlates with increased
cancer cell proliferation in human breast cancer. APMIS.
119:663–673. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Pathmanathan N and Balleine RL: Ki67 and
proliferation in breast cancer. J Clin Pathol. 66:512–516. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yamamichi N, Shimomura R, Inada K, Sakurai
K, Haraguchi T, Ozaki Y, Fujita S, Mizutani T, Furukawa C,
Fujishiro M, et al: Locked nucleic acid in situ hybridization
analysis of miR-21 expression during colorectal cancer development.
Clin Cancer Res. 15:4009–4016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Nouraee N, Roosbroeck K, Vasei M, Semnani
S, Samaei NM, Naghshvar F, Omidi AA, Calin GA and Mowla SJ:
Expression, tissue distribution and function of miR-21 in
esophageal squamous cell carcinoma. PLoS One. 8:e730092013.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dobreva G, Dambacher J and Grosschedl R:
SUMO modification of a novel MAR-binding protein, SATB2, modulates
immunoglobulin mu gene expression. Genes Dev. 17:3048–3061. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Patani N, Jiang W, Mansel R, Newbold R and
Mokbel K: The mRNA expression of SATB1 and SATB2 in human breast
cancer. Cancer Cell Int. 9:182009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Aprelikova O, Yu X, Palla J, Wei BR, John
S, Yi M, Stephens R, Simpson RM, Risinger JI, Jazaeri A and
Niederhuber J: The role of miR-31 and its target gene SATB2 in
cancer-associated fibroblasts. Cell Cycle. 9:4387–4398. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Aprelikova O, Palla J, Hibler B, Yu X,
Greer YE, Yi M, Stephens R, Maxwell GL, Jazaeri A, Risinger JI, et
al: Silencing of miR-148a in cancer-associated fibroblasts results
in WNT10B-mediated stimulation of tumor cell motility. Oncogene.
32:3246–3253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wei J, Melichian D, Komura K, Hinchcliff
M, Lam AP, Lafyatis R, Gottardi CJ, MacDougald OA and Varga J:
Canonical Wnt signaling induces skin fibrosis and subcutaneous
lipoatrophy: A novel mouse model for scleroderma? Arthritis Rheum.
63:1707–1717. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Enkelmann A, Heinzelmann J, von Eggeling
F, Walter M, Berndt A, Wunderlich H and Junker K: Specific protein
and miRNA patterns characterise tumour-associated fibroblasts in
bladder cancer. J Cancer Res Clin Oncol. 137:751–759. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Schepeler T, Reinert JT, Ostenfeld MS,
Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ,
Kruhøffer M, Laurberg S, et al: Diagnostic and prognostic microRNAs
in stage II colon cancer. Cancer Res. 68:6416–6424. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lee KH, Lotterman C, Karikari C, Omura N,
Feldmann G, Habbe N, Goggins MG, Mendell JT and Maitra A:
Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent
kinase 6 expression in pancreatic cancer. Pancreatology. 9:293–301.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Mitra AK, Zillhardt M, Hua Y, Tiwari P,
Murmann AE, Peter ME and Lengyel E: MicroRNAs reprogram normal
fibroblasts into cancer-associated fibroblasts in ovarian cancer.
Cancer Discov. 2:1100–1108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Long H, Xie R, Xiang T, Zhao Z, Lin S,
Liang Z, Chen Z and Zhu B: Autocrine CCL5 signaling promotes
invasion and migration of CD133+ovarian cancer stem-like cells via
NF-κB-mediated MMP-9 upregulation. Stem Cells. 30:2309–2319. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Naito Y, Sakamoto N, Oue N, Yashiro M,
Sentani K, Yanagihara K, Hirakawa K and Yasui W: MicroRNA-143
regulates collagen type III expression in stromal fibroblasts of
scirrhous type gastric cancer. Cancer Sci. 105:228–235. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Musumeci M, Coppola V, Addario A, Patrizii
M, Maugeri-Saccà M, Memeo L, Colarossi C, Francescangeli F, Biffoni
M, Collura D, et al: Control of tumor and microenvironment
cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene.
30:4231–4242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sun P, Hu JW, Xiong WJ and Mi J: MiR-186
regulates glycolysis through Glut1 during the formation of
cancer-associated fibroblasts. Asian Pac J Cancer Prev.
15:4245–4250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Pavlides S, Whitaker-Menezes D,
Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro
MC, Wang C, Fortina P, Addya S, et al: The reverse Warburg effect:
Aerobic glycolysis in cancer associated fibroblasts and the tumor
stroma. Cell Cycle. 8:3984–4001. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Sotgia F, Martinez-Outschoorn UE, Pavlides
S, Howell A, Pestell RG and Lisanti MP: Understanding the Warburg
effect and the prognostic value of stromal caveolin-1 as a marker
of a lethal tumor microenvironment. Breast Cancer Res. 13:2132011.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Sotgia F, Martinez-Outschoorn UE, Howell
A, Pestell RG, Pavlides S and Lisanti MP: Caveolin-1 and cancer
metabolism in the tumor microenvironment: Markers, models, and
mechanisms. Annu Rev Pathol. 7:423–467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Razani B, Zhang XL, Bitzer M, von
Gersdorff G, Böttinger EP and Lisanti MP: Caveolin-1 regulates
transforming growth factor (TGF)-beta/SMAD signaling through an
interaction with the TGF-beta type I receptor. J Biol Chem.
276:6727–6738. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
112
|
Wilson AS, Power BE and Molloy PL: DNA
hypomethylation and human diseases. Biochim Biophys Acta.
1775:138–162. 2007.PubMed/NCBI
|
|
113
|
Adany R, Heimer R, Caterson B, Sorrell JM
and Iozzo RV: Altered expression of chondroitin sulfate
proteoglycan in the stroma of human colon carcinoma.
Hypomethylation of PG-40 gene correlates with increased PG-40
content and mRNA levels. J Biol Chem. 265:11389–11396.
1990.PubMed/NCBI
|
|
114
|
Adany R and Iozzo RV: Altered methylation
of versican proteoglycan gene in human colon carcinoma. Biochem
Biophys Res Commun. 171:1402–1413. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Adany R and Iozzo RV: Hypomethylation of
the decorin proteoglycan gene in human colon cancer. Biochem J.
276:301–306. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Kekeeva TV, Popova OP, Shegaĭ PV, Alekseev
BIa, Adnreeva IuIu, Zaletaev DV and Nemtsova MV: Abberant
methylation of p16, HIC1, N33 and GSTP1 genes in tumor epitelium
and tumor-associated stromal cells of prostate cancer. Mol Biol
(Mosk). 41:79–85. 2007.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Rodriguez-Canales J, Hanson JC, Tangrea
MA, Erickson HS, Albert PS, Wallis BS, Richardson AM, Pinto PA,
Linehan WM, Gillespie JW, et al: Identification of a unique
epigenetic sub-microenvironment in prostate cancer. J Pathol.
211:410–419. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ye F, Zhang SF, Xie X and Lu WG: OPCML
gene promoter methylation and gene expression in tumor and stroma
cells of invasive cervical carcinoma. Cancer Invest. 26:569–574.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Matsunoki A, Kawakami K, Kotake M, Kaneko
M, Kitamura H, Ooi A, Watanabe G and Minamoto T: LINE-1 methylation
shows little intra-patient heterogeneity in primary and synchronous
metastatic colorectal cancer. BMC Cancer. 12:5742012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Fiegl H, Millinger S, Goebel G,
Müller-Holzner E, Marth C, Laird PW and Widschwendter M: Breast
cancer DNA methylation profiles in cancer cells and tumor stroma:
Association with HER-2/neu status in primary breast cancer. Cancer
Res. 66:29–33. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Dawsey SP, Roth MJ, Adams L, Hu N, Wang
QH, Taylor PR and Woodson K: COX-2 (PTGS2) gene methylation in
epithelial, subepithelial lymphocyte and stromal tissue
compartments in a spectrum of esophageal squamous neoplasia. Cancer
Detect Prev. 32:135–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhuang J, Jones A, Lee SH, Ng E, Fiegl H,
Zikan M, Cibula D, Sargent A, Salvesen HB, Jacobs IJ, et al: The
dynamics and prognostic potential of DNA methylation changes at
stem cell gene loci in women's cancer. PLoS Genet. 8:e10025172012.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Jiang L, Gonda TA, Gamble MV, Salas M,
Seshan V, Tu S, Twaddell WS, Hegyi P, Lazar G, Steele I, et al:
Global hypomethylation of genomic DNA in cancer-associated
myofibroblasts. Cancer Res. 68:9900–9908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Wasserkort R, Kalmar A, Valcz G, Spisak S,
Krispin M, Toth K, Tulassay Z, Sledziewski AZ and Molnar B:
Aberrant septin 9 DNA methylation in colorectal cancer is
restricted to a single CpG island. BMC Cancer. 13:3982013.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Hanson JA, Gillespie JW, Grover A, Tangrea
MA, Chuaqui RF, Emmert-Buck MR, Tangrea JA, Libutti SK, Linehan WM
and Woodson KG: Gene promoter methylation in prostate
tumor-associated stromal cells. J Natl Cancer Inst. 98:255–261.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Kim YI, Fawaz K, Knox T, Lee YM, Norton R,
Arora S, Paiva L and Mason JB: Colonic mucosal concentrations of
folate correlate well with blood measurements of folate status in
persons with colorectal polyps. Am J Clin Nutr. 68:866–872.
1998.PubMed/NCBI
|
|
127
|
Momparler RL: Cancer epigenetics.
Oncogene. 22:6479–6483. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Nielsen BS, Jørgensen S, Fog JU, Søkilde
R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S and
Nielsen HJ: High levels of microRNA-21 in the stroma of colorectal
cancers predict short disease-free survival in stage II colon
cancer patients. Clin Exp Metastasis. 28:27–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Dou L, Zheng D, Li J, Li Y, Gao L, Wang L
and Yu L: Methylation-mediated repression of microRNA-143 enhances
MLL-AF4 oncogene expression. Oncogene. 31:507–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Liu R, Liao J, Yang M, Sheng J, Yang H,
Wang Y, Pan E, Guo W, Pu Y, Kim SJ and Yin L: The cluster of
miR-143 and miR-145 affects the risk for esophageal squamous cell
carcinoma through co-regulating fascin homolog 1. PLoS One.
7:e339872013. View Article : Google Scholar
|
|
132
|
Zhu H, Dougherty U, Robinson V, Mustafi R,
Pekow J, Kupfer S, Li YC, Hart J, Goss K, Fichera A, et al: EGFR
signals downregulate tumor suppressors miR-143 and miR-145 in
Western diet-promoted murine colon cancer: role of G1 regulators.
Mol Cancer Res. 9:960–975. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wang Q, Cai J, Wang J, Xiong C and Zhao J:
MiR-143 inhibits EGFR-signaling-dependent osteosarcoma invasion.
Tumour Biol. 35:12743–12748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wang X, Baumgartner C, Shields DC, Deng HW
and Beckmann JS: Application of Clinical Bioinformatics. Springer;
Netherlands: pp. 1262016
|
|
135
|
Anestopoulos I, Voulgaridou GP,
Georgakilas AG, Franco R, Pappa A and Panayiotidis MI: Epigenetic
therapy as a novel approach in hepatocellular carcinoma. Pharmacol
Ther. 145:103–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Corver WE, Ter Haar NT, Fleuren GJ and
Oosting J: Cervical carcinoma-associated fibroblasts are DNA
diploid and do not show evidence for somatic genetic alterations.
Cell Oncol (Dordr). 34:553–563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Walter K, Omura N, Hong SM, Griffith M and
Goggins M: Pancreatic cancer associated fibroblasts display normal
allelotypes. Cancer Biol Ther. 7:882–888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Erez N, Truitt M, Olson P, Arron ST and
Hanahan D: Cancer-associated fibroblasts are activated in incipient
neoplasia to orchestrate tumor-promoting inflammation in an
NF-kappaB-dependent manner. Cancer Cell. 17:135–147. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Martinez-Outschoorn UE, Whitaker-Menezes
D, Lin Z, Flomenberg N, Howell A, Pestell RG, Lisanti MP and Sotgia
F: Cytokine production and inflammation drive autophagy in the
tumor microenvironment: Role of stromal caveolin-1 as a key
regulator. Cell Cycle. 10:1784–1793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Servais C and Erez N: From sentinel cells
to inflammatory culprits: Cancer-associated fibroblasts in
tumour-related inflammation. J Pathol. 229:198–207. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Shiels MS, Engels EA, Shi J, Landi MT,
Albanes D, Chatterjee N, Chanock SJ, Caporaso NE and Chaturvedi AK:
Genetic variation in innate immunity and inflammation pathways
associated with lung cancer risk. Cancer. 118:5630–5636. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Pavlides S, Tsirigos A, Vera I, Flomenberg
N, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, Pestell RG,
et al: Loss of stromal caveolin-1 leads to oxidative stress, mimics
hypoxia and drives inflammation in the tumor microenvironment,
conferring the ‘reverse Warburg effect’ a transcriptional
informatics analysis with validation. Cell Cycle. 9:2201–2219.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Lehmann U and Kreipe H: Real-time PCR
analysis of DNA and RNA extracted from formalin-fixed and
paraffin-embedded biopsies. Methods. 25:409–418. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Ale-Agha N, Dyballa-Rukes N, Jakob S,
Altschmied J and Haendeler J: Cellular functions of the
dual-targeted catalytic subunit of telomerase, telomerase reverse
transcriptase-potential role in senescence and aging. Exp Gerontol.
56:189–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Urquidi V, Tarin D and Goodison S: Role of
telomerase in cell senescence and oncogenesis. Annu Rev Med.
51:65–79. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Shawi M and Autexier C: Telomerase,
senescence and ageing. Mech Ageing Dev. 129:3–10. 2008. View Article : Google Scholar : PubMed/NCBI
|