Introduction
Taxanes, including paclitaxel and docetaxel, are
microtubule-stabilizing agents that are widely used to treat
various cancers. Taxane-resistant breast cancer is common;
therefore, the identification of resistance markers and a detailed
understanding of the mechanisms mediating paclitaxel resistance are
required to develop optimal treatment strategies and to identify
responsive patients (1). A previous
study described at least three potential mechanisms of paclitaxel
resistance (2). The first involves
decreased intracellular drug accumulation caused by the
overexpression of membrane-bound drug efflux proteins, such as
P-glycoprotein. However, clinical trials focusing on P-glycoprotein
inhibitors as chemosensitizing agents did not report promising
outcomes for patients with relapsing solid tumors and hematological
malignancies (3). The other two
mechanisms involve mutations in β-tubulin and overexpression of
β-tubulin isotypes (2). For example,
numerous studies reported that mutations in the paclitaxel-binding
sites of β-tubulin were associated with drug resistance, while
other studies were unable to detect these mutations in
paclitaxel-resistant breast cancers (4–7).
Overexpression of β-tubulin isotypes occurs in a restricted number
of patients with paclitaxel-resistant ovarian cancer and
occasionally in patients with breast cancer; however, knockdown of
β-tubulin expression by RNA interference had no effect on the
sensitivity of paclitaxel-resistant ovarian cancer cells (6,8,9). Therefore, the proposed mechanisms of
paclitaxel resistance remain controversial.
Global analysis of gene expression using cDNA
microarray is often used to determine the molecular mechanisms
underlying drug resistance. Since the correlation between mRNA
abundance and protein levels is poor, proteome analysis is
considered superior to cDNA microarrays for the analysis of cell
function. Furthermore, two-dimensional gel electrophoresis (2-DE)
analysis offers advantages because of its high resolution and
ability to detect posttranslational modifications (10,11).
Therefore, a proteomic approach using 2-DE in combination with drug
sensitivity studies may provide further insight into the mechanisms
of paclitaxel resistance.
In the present study, a proteomic analysis using
2-DE and matrix-assisted laser desorption/ionization (MALDI)-time
of flight (TOF) mass spectrometry was conducted to identify
proteins that play critical roles in paclitaxel resistance. The
proteomic analysis revealed 11 upregulated and 12 downregulated
proteins in paclitaxel-resistant MCF-7/PTX cells compared with the
paclitaxel-sensitive MCF-7 parental cells. Furthermore, it was
demonstrated that peptidyl-prolyl cis-trans isomerase A
(PPIA), which is also known as cyclophilin A, may have an important
role in the resistance of tumor cells to paclitaxel.
Materials and methods
Cell culture
The human breast cancer cell line MCF-7 and its
paclitaxel-resistant subclone MCF-7/PTX were obtained from Dr
Amadeo M. Parissenti (Tumor Biology Research Program, Sudbury
Regional Hospital, Sudbury, Canada) (12). Cells were cultured in Dulbecco's
modified Eagle's medium (Wako Pure Chemical Industries, Ltd.,
Osaka, Japan) supplemented with 10% fetal bovine serum (Tissue
Culture Biologicals, Long Beach, CA, USA) at 37°C in a humidified
atmosphere containing 5% CO2, and were harvested in
mid-log phase.
MTT assays for drug sensitivity
Cell viability was assessed 3 days later using MTT
assays, as described previously (13). The cells (5×103 per well)
were seeded into 96-well culture plates and pre-incubated for 24 h
at 37°C. Paclitaxel was added at various concentrations (0, 0.1,
0.3, 1, 3, 10, 30, 100, 300 and 1000 nM) and then incubated for 3
days at 37°C. MTT solution was added (final concentration, 0.45
µg/ml) and subsequently incubated for 2 h at 37°C. MTT formazan
crystals were dissolved in DMSO (Nacalai Tesque, Inc., Kyoto,
Japan). The absorbance of each well was read at 540 mn using a
SH-1000Lab microplate reader (Corona Electric Co., Ltd.,
Hitachinaka, Japan). Half maximal inhibitory concentration
(IC50) values were calculated from three independent
experiments performed in triplicate. The results are shown as a
percentage of the absorbance of the medium alone, and are expressed
as the mean ± standard error of the mean.
Protein preparation
Harvested cells were washed in ice-cold PBS prior to
lysing in chilled lysis buffer containing 7 M urea (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 2 M thiourea, 5%
(w/v) CHAPS, 5% (v/v) IPG buffer (pH 3–10 NL; GE Healthcare Life
Sciences, Chalfont, UK), 50 mM DTT and 25 µg/ml each of DNase I and
RNase A (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany).
Lysates were sonicated three times for 3 sec each (Handy Sonic
UR-20P; Tomy Seiko, Tokyo, Japan). Samples were centrifuged at
15,000 × g for 30 min, and the supernatants were subjected
to protein quantification using a 2-D quantification kit (GE
Healthcare Life Sciences) prior to use in the 2-DE experiments.
2-DE experiments
2-DE was performed as described previously (14). For isoelectric focusing (IEF), an
Immobiline DryStrip (pH 4–7 or 3–10, 7 cm; GE Healthcare Life
Sciences) was immersed in sample solution (~500 µg of protein) and
rehydration buffer (8 M urea, 0.5% [w/v] CHAPS, 20 mM DTT and 1.25%
[v/v] IPG buffer). IEF was performed using the NA-1410R7
electrophoresis apparatus (Nihon Eido, Co., Ltd., Tokyo, Japan) set
to 50 V for 6 h, 100 V for 6 h and 2,000 V for 7–9 h. Subsequently,
the IPG strips were equilibrated at room temperature for 15 min in
a solution containing 6 M urea, 1% SDS, 30% glycerol, 50 mM
Tris-HCl (pH 8.8), and 60 mM DTT. The strips were equilibrated for
an additional 15 min in the same solution, except that DTT was
replaced with 0.24 M iodoacetamide, prior to 2-D SDS-PAGE using
12.5% SDS gels. After electrophoresis, the gels were fixed in 10%
trichloroacetic acid solution for 1.5 h, washed with
double-distilled water three times for 5 min each, stained with
Coomassie brilliant blue (CBB) R-250, and then scanned using a
GS-800 Calibrated Densitometer (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). To verify the results, lysates from three
individual preparations were run on four pH 4–7 and pH 3–10 NL gels
each.
Image analysis
The 2-DE images were analyzed using Prodigy 2D
software (version 1; Nonlinear Dynamics, Ltd., Durham, NC, USA).
Spots were detected, matched automatically to a master gel and then
edited manually. The total intensity of valid spots was used for
normalization. Matched spots from triplicate gel sets with a
statistically significant difference in intensities (P<0.05) and
an average fold difference of >1.5 in spot volume were defined
as differentially expressed between MCF-7 and MCF-7/PTX cells.
In-gel digestion
A pipette tip was used to excise protein spots from
the 2-DE gels. Gel pieces were destained in a solution containing
30% acetonitrile and 25 mM ammonium bicarbonate for 10 min,
dehydrated in 100% acetonitrile for 10 min, and then dried using a
SpeedVac (Tomy Seiko). Dried gel pieces were rehydrated for 30 min
on ice in 5 µl of 50 mM ammonium bicarbonate containing 50 ng of
sequencing-grade modified trypsin (Promega Corporation, Madison,
WI, USA). After overnight incubation at 37°C, peptides were
extracted by vortexing for 30 min followed by sonicating for 3
min.
Protein identification by mass
spectrometry
The peptide extracts were desalted using C18 ZipTips
(Merck Millipore). Mass spectrometry was performed using an
Ultraflex II TOF/TOF mass spectrometer (Bruker Corporation,
Billerica, MA, USA) with an accelerating voltage of 20 kV, and
spectra were externally calibrated using the peptide calibration
standard II (Bruker Corporation). Proteins were identified by
matching the peptide mass fingerprinting and TOF/TOF results with
the Swiss-Prot database (http://www.uniprot.org/) using the MASCOT Search
engine v.2.2 (Matrix Science Ltd., London, UK). Database searches
were performed using the following parameters: Taxonomy, Homo
sapiens; and enzyme, trypsin. One missed cleavage was allowed.
Carbamidomethylation was selected as a fixed modification and
methionine oxidation was allowed as a variable. The peptide and
fragment mass tolerances were set to 50 or 100 ppm and 0.5 Da,
respectively.
Transfection of MCF-7/PTX cells with a
PPIA-specific small interfering (si)RNA
Cells were transfected with Silencer Select
Pre-designed siRNA specific for PPIA (s198123; Ambion; Thermo
Fisher Scientific, Inc.) at a final concentration of 5 nM.
Transfections were performed in six-well plates using Lipofectamine
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Silencer Select negative control siRNA
#1 (Invitrogen; Thermo Fisher Scientific, Inc.) was used as a
negative control. Gene silencing was assessed between 24 and 72 h
following transfection by western blotting.
Western blotting
Cells were lysed using Cell Lysis Buffer M
containing 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 0.05% Nonidet
P-40, and 2.5 mM MgCl2 (Wako Pure Chemical Industries,
Ltd.). Lysates were sonicated for 3 sec on ice and then centrifuged
at 15,000 × g for 15 min at 4°C. Protein concentration was
determined using the Quick Start Bradford Protein assay (Bio-Rad
Laboratories, Inc.) and protein samples (10 µg/lane) were separated
by 15% SDS-PAGE followed by semidry transfer to a polyvinylidene
difluoride (PVDF) membrane (GE Healthcare Life Sciences). The
membrane was blocked with 3% membrane blocking agent (GE Healthcare
Life Sciences) in PBS containing 0.1% Tween-20 (PBS-Tween),
incubated for 1 h at room temperature with an anti-PPIA antibody
(1:7,500 dilution; cat. no. 07-313; Merck Millipore) in 3% membrane
blocking agent in PBS-Tween for 2 h, and incubated further with a
horseradish peroxidase-conjugated species-specific donkey antibody
(1:50,000 dilution; cat. no. NA934V; GE Healthcare Life Sciences).
Immunoreactive bands were visualized using an ECL Prime detection
kit and Hyperfilm ECL (GE Healthcare Life Sciences). The films were
scanned with the GS-800 Calibrated Densitometer and analyzed by
Quantity One, version 4.5.0. software (Bio-Rad Laboratories, Inc.).
In parallel, the blotted PVDF membranes were stained with 0.008%
Direct Blue 71 (Sigma-Aldrich; Merck Millipore) as described
previously (15), analyzed by the
GS-800 and Quantity One software version 4.5.0 (Bio-Rad
Laboratories, Inc.), and the total protein intensity of each lane
was used as the sample loading control, as described previously
(16).
Statistical analysis
Statistical analyses were performed using Excel 2010
(Microsoft Corporation, Redmond, WA, USA). The results are
expressed as the mean ± standard error of the mean, and P<0.05
was considered to indicate a statistically significant
difference.
Results
Paclitaxel resistance in human breast
cancer cell lines
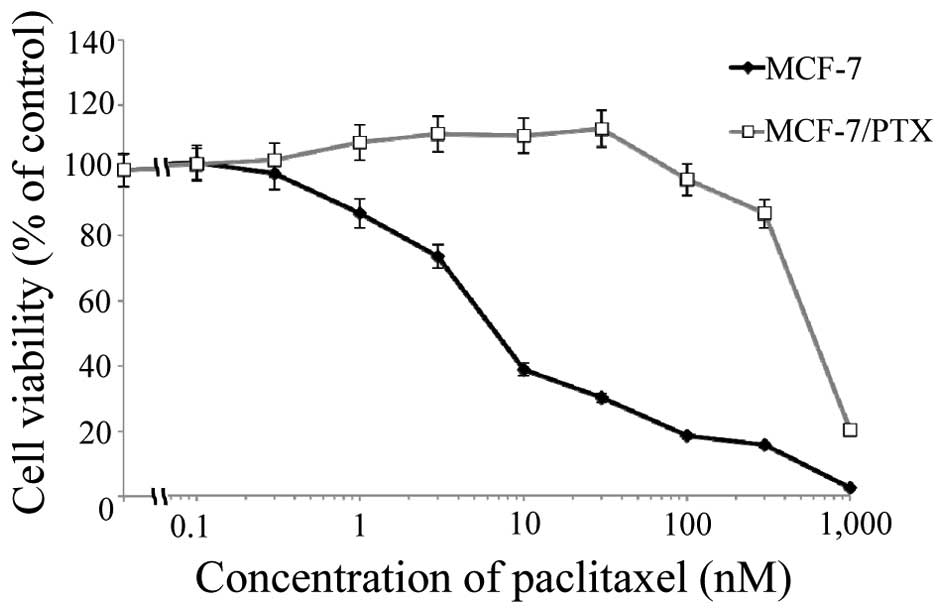
The cytotoxicity of paclitaxel on the human breast
cancer cell line MCF-7 and its paclitaxel-resistant subclone
MCF-7/PTX was compared using MTT assays (Fig. 1). The IC50 values for
paclitaxel were 7.7±1.5 and 580±50 nM for MCF-7 and MCF-7/PTX
cells, respectively (P<0.05). The cytotoxicity of paclitaxel on
MCF-7/PTX cells was 75-fold lower than its cytotoxicity on MCF-7
cells.
Identification of differentially
expressed proteins
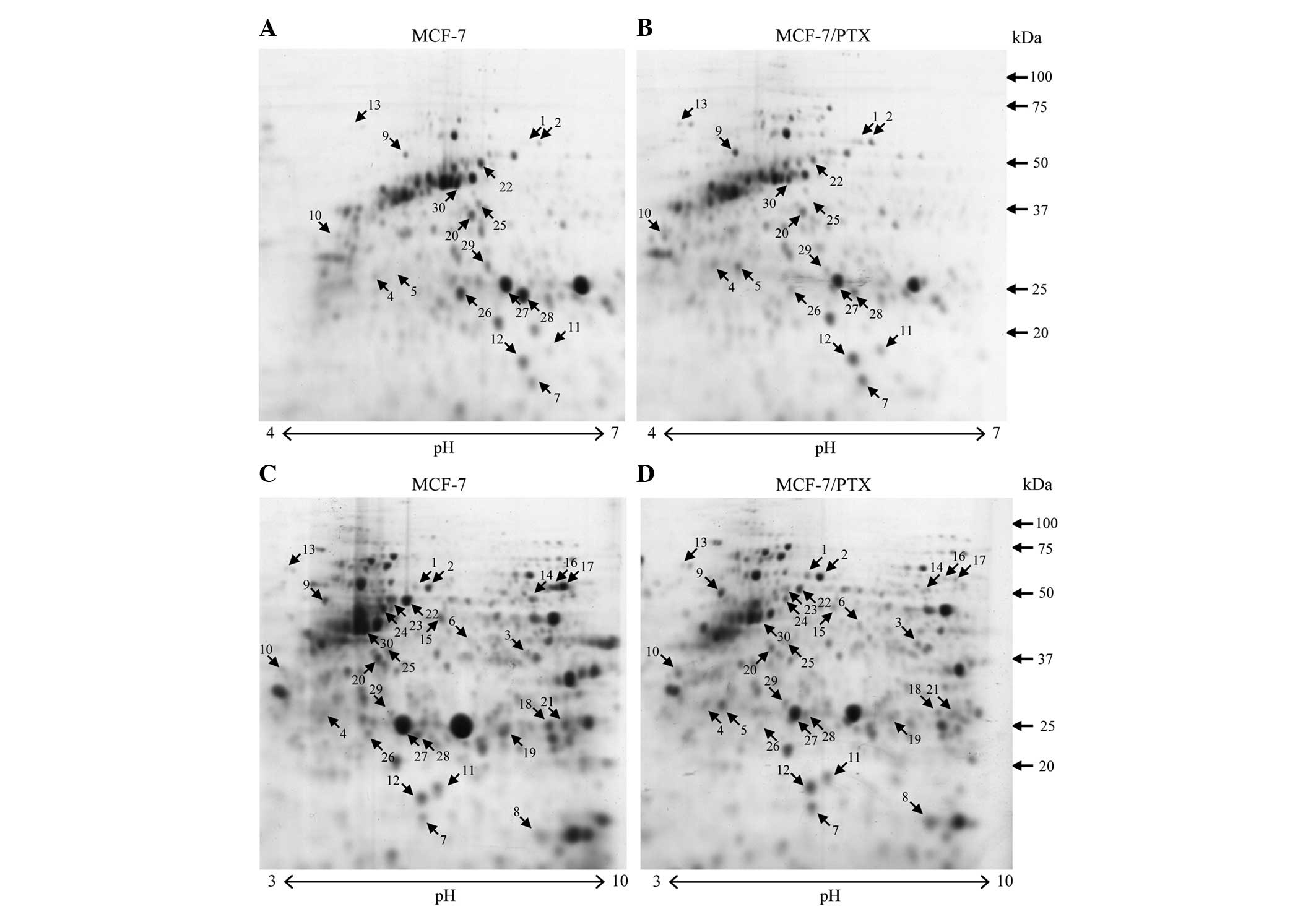
The protein expression profiles of the MCF-7 and
MCF-7/PTX cells were compared to identify proteins associated with
paclitaxel sensitivity. To confirm reproducibility, proteins were
extracted from each cell line three times and four 2-D gels were
prepared for each cell extract. Gel analysis showed a consistent
image with reproducible resolution of the 2-DE maps. CBB R-250
staining revealed >400 protein spots on each 7-cm gel (pH 3–10
NL) (Fig. 2C and D). Six and seven
images were analyzed for the pH 3–10 and pH 4–7 gels,
respectively.
Differences in protein expression that were
≥1.5-fold (P<0.05), as indicated by changes in the intensity of
the spots on the gels, were defined as statistically significant
(Fig. 2). In total, 30 differentially
expressed protein spots were detected, including 13 upregulated and
17 downregulated spots. Using this procedure, 11 proteins were
identified as upregulated, while 12 proteins were determined to be
downregulated (Tables I and II). The functions of identified proteins
were assigned using information from the Swiss-Prot database
(www.uniprot.org/uniprot/) and the
protein function databases Pfam (pfam.xfam.org/).
 | Table I.Identification of upregulated
proteins in MCF-7/PTX cells compared with MCF-7 cells. |
Table I.
Identification of upregulated
proteins in MCF-7/PTX cells compared with MCF-7 cells.
| No. | Protein name | Locus name | Mr
(Da)a | pIb | Mascot
scorec | Fold change |
|---|
| 1 | Stress-70 protein,
mitochondrial | GRP75 | 73,920 | 5.87 | 233 | 1.9 |
| 2 | Stress-70 protein,
mitochondrial | GRP75 | 73,920 | 5.87 | 240 | 1.7 |
| 3 | Heterogeneous
nuclear ribonucleoprotein H3 | HNRH3 | 36,960 | 6.37 | 172 | 1.7 |
| 4 | Heat shock cognate
71 kDa protein | HSP7C | 71,082 | 5.37 | 124 | 2.4 |
| 5 | Heat shock cognate
71 kDa protein | HSP7C | 71,082 | 5.37 | 126 | 2.5 |
| 6 | Pyruvate kinase
M1/M2 | KPYM | 58,470 | 7.96 | 148 | 1.9 |
| 7 | Stathmin | STMN1 | 17,292 | 5.76 | 127 | 1.5 |
| 8 | Peptidyl-prolyl
cis-trans isomerase A | PPIA | 18,229 | 7.68 | 124 | 1.5 |
| 9 | ATP synthase
β-subunit | ATPB | 56,525 | 5.26 | 216 | 1.7 |
| 10 | Tropomyosin α-1
chain | TPM1 | 32,746 | 4.69 | 146 | 1.8 |
| 11 | Nucleoside
diphosphate kinase A | NDKA | 17,309 | 5.83 | 122 | 1.7 |
| 12 | Superoxide
dismutase [Cu-Zn] | SODC | 16,154 | 5.7 | 168 | 1.5 |
| 13 | 78 kD
glucose-regulated protein | GRP78 | 72,402 | 5.07 | 161 | 2.4 |
 | Table II.Identification of downregulated
proteins in MCF-7/PTX cells compared with MCF-7 cells. |
Table II.
Identification of downregulated
proteins in MCF-7/PTX cells compared with MCF-7 cells.
| No. | Protein name | Locus name | Mr
(Da)a | pIb | Mascot
scorec | Fold change |
|---|
| 14 | Glucose-6-phosphate
1-dehydrogenase | G6PD | 59,675 | 6.39 | 187 | 0.67 |
| 15 | 26S protease
regulatory subunit 7 | PRS7 | 49,002 | 5.71 | 181 | 0.67 |
| 16 | UDP-glucose 6
dehydrogenase | UGDH | 55,674 | 6.73 | 258 | 0.33 |
| 17 | UDP-glucose 6
dehydrogenase | UGDH | 55,674 | 6.73 | 233 | 0.34 |
| 18 | Phosphoglycerate
mutase 1 | PGAM1 | 28,900 | 6.67 | 116 | 0.67 |
| 19 |
Peroxiredoxin-6 | PRDX6 | 27,838 | 6.28 | 116 | 0.63 |
| 20 | Tubulin β-4B
chain | TBB4B | 50,255 | 4.79 | 132 | 0.59 |
| 21 |
Peroxiredoxin-1 | PRDX1 | 22,324 | 8.27 | 185 | 0.67 |
| 22 | Cytokeratin 8 | K2C8 | 53,671 | 5.52 | 307 | 0.67 |
| 23 | Cytokeratin 8 | K2C8 | 53,671 | 5.52 | 323 | 0.56 |
| 24 | Cytokeratin 8 | K2C8 | 53,671 | 5.52 | 199 | 0.36 |
| 25 | Tubulin α-1A
chain | TBA1A | 50,788 | 4.94 | 140 | 0.67 |
| 26 | Heat shock protein
β-1 | HSPB1 | 22,826 | 5.98 | 153 | 0.43 |
| 27 | Heat shock protein
β-1 | HSPB1 | 22,826 | 5.98 | 123 | 0.71 |
| 28 | Heat shock protein
β-1 | HSPB1 | 22,826 | 5.98 | 104 | 0.42 |
| 29 | Actin, cytoplasmic
1 | ACTB | 42,052 | 5.29 | 81 | 0.53 |
| 30 | Cytokeratin 18 | K1C18 | 48,629 | 5.34 | 208 | 0.67 |
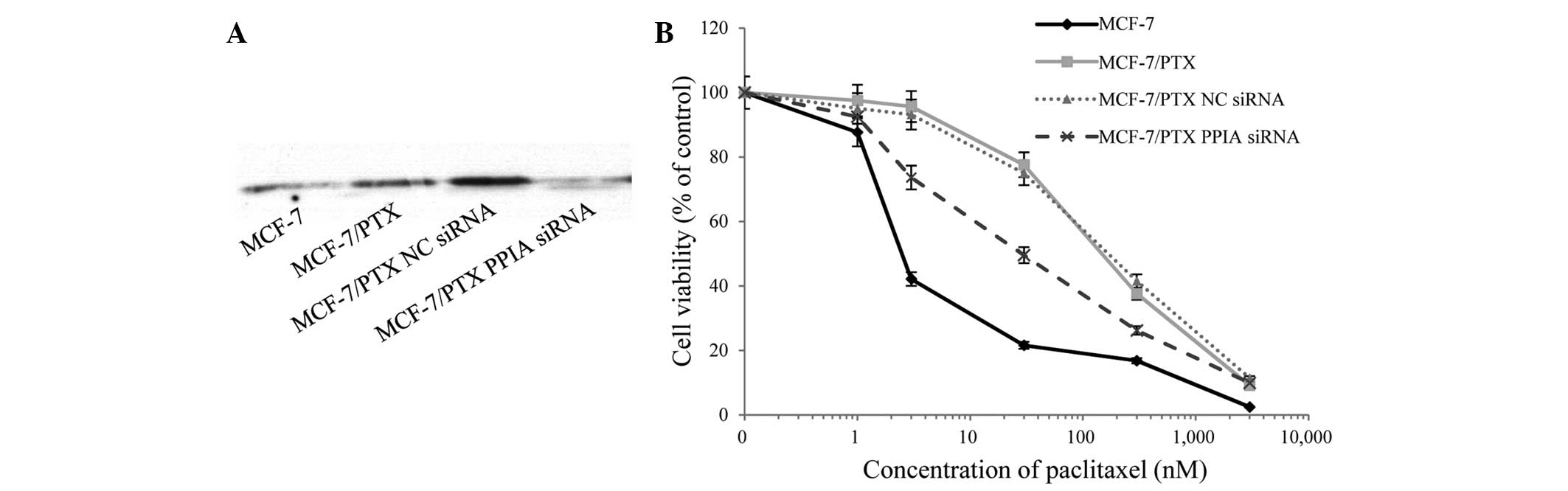
Knockdown of PPIA expression in
MCF-7/PTX cells
PPIA expression was significantly higher in
MCF-7/PTX cells compared with MCF-7 cells (Fig. 2C and D, spot no. 8). To investigate
the role of PPIA in paclitaxel resistance, siRNA-mediated knockdown
of PPIA was performed and paclitaxel-induced cell death was
assessed by viability assays (Fig.
3A). Notably, paclitaxel sensitivity was significantly
increased in siRNA-treated cells compared with untransfected
MCF-7/PTX cells and negative control siRNA-treated counterparts
(Fig. 3B). The IC50 values
of paclitaxel were 3.2±1.1, 160±57, 175±100 and 14±9 nM in MCF-7
cells, MCF-7/PTX cells, negative control siRNA transfected
MCF-7/PTX cells and PPIA-siRNA-transfected MCF-7/PTX cells,
respectively. These results indicate a close association between
PPIA and paclitaxel resistance in MCF-7 cells, and suggest that
PPIA levels may predict MCF-7 resistance to paclitaxel.
Discussion
The purpose of this study was to investigate the
mechanisms underlying paclitaxel resistance in breast cancer cells
and to identify markers of drug resistance. To this end, a
proteomic analysis using 2-DE coupled with MALDI-TOF/TOF mass
spectrometry was conducted, and 23 differentially expressed
proteins between paclitaxel-resistant MCF-7/PTX cells and parental
MCF-7 cells were identified. These proteins were classified into
several functional groups, including roles in the stress response,
metabolism, cytoskeleton and apoptosis. Differences between the
experimental and theoretical molecular mass/isoelectric point
values were observed for stress-70 protein (GRP75), heat shock
cognate 71-kDa protein (HSP7C), UDP-glucose 6 dehydrogenase (UGDH),
cytokeratin 8 (CK8) and heat shock protein β-1 (HSPB1), which may
be attributed to posttranslational modifications such as cleavage
and/or phosphorylation.
Notably, it was observed that the stress-response
chaperones, GRP75, HSP7C, 78-kD glucose-regulated protein (GRP78),
PPIA and superoxide dismutase [Cu-Zn] (SODC) were upregulated in
MCF-7/PTX cells, whereas 26S protease regulatory subunit 7,
peroxiredoxin-1, peroxiredoxin-6 and HSPB1 were downregulated in
MCF-7/PTX cells. GRP75, HSP7C and GRP78 are members of the heat
shock protein 70 (HSP70) superfamily, which perform essential roles
in facilitating proper protein folding and in preventing the
aggregation of denatured proteins in order to maintain protein
homeostasis (17). These proteins are
upregulated by heat, hypoxia, oxidative stress and toxic chemicals,
and subsequently enhance cell survival (17). HSP70 family members are highly
expressed by cancer cells, wherein they promote cell growth and
survival via multiple anti-apoptotic functions (17). For instance, GRP75 overexpression has
been associated with increased cancer cell malignancy, implicating
its use as a biomarker of metastatic cancer (18,19).
Furthermore, knockdown of endogenous GRP78 expression by siRNA
sensitized human breast cancer cells to estrogen starvation-induced
apoptosis (20), while its
upregulation is associated with resistance to chemotherapeutic
drugs such as doxorubicin, 5-fluorouracil (5-FU) and vincristine
(21). In addition, SODC eliminates
reactive oxygen species and promotes cisplatin resistance in
ovarian cancer cells (22). The
authors of the present study hypothesize that these proteins may
also function as anti-apoptotic factors in MCF-7/PTX cells and may
play a role in paclitaxel resistance.
The present study demonstrated that PPIA was
upregulated in paclitaxel-resistant breast cancer cells, and that
its siRNA-mediated knockdown restored paclitaxel sensitivity to
MCF-7/PTX cells. PPIA is a peptidyl-prolyl cis-trans
isomerase that catalyzes the cis-trans isomerization of
proline imidic peptide bonds, promotes protein folding and binds to
the immunosuppressive drug cyclosporin A (23). A previous study reported that PPIA is
overexpressed in many cancers and is involved in the various stages
of tumorigenesis (24). Furthermore,
PPIA upregulation was shown to prevent cisplatin-induced apoptosis
by limiting the subsequent accumulation of reactive oxygen species,
while PPIA knockdown increased the rate of cell death (25). Therefore, we hypothesized that PPIA
functions in the regulation of cell death and paclitaxel resistance
in MCF-7/PTX cells. In future studies, we plan to investigate
whether PPIA is a prognostic biomarker for paclitaxel resistance
using clinical tumor specimens.
The present study demonstrated that three metabolic
proteins, pyruvate kinase M2 (PKM2), ATP synthase β and nucleoside
diphosphate kinase A, were upregulated, whereas glucose-6-phosphate
1-dehydrogenase, UGDH and phosphoglycerate mutase 1 were
downregulated, in MCF-7/PTX cells. Notably, the differential
expression of PKM2 (26) and ATP
synthase β (27) has been shown to
have important roles in multi-drug resistance and apoptosis.
Furthermore, heterogeneous nuclear ribonucleoprotein (hnRNP) H3
(HNRH3), which is a member of the hnRNP protein family that
includes numerous nucleic acid binding proteins and spliceosome
components and which function in the splicing of selected target
mRNAs (28), has been shown to
prevent the apoptosis of cancer cells by regulating the alternative
splicing of transcripts that have important roles in apoptosis
(29). Therefore, the upregulation of
certain anti-apoptotic proteins in MCF-7/PTX cells may also prevent
apoptosis induced by paclitaxel.
The present study also demonstrated the upregulation
of the cytoskeletal proteins, stathmin and tropomyosin α-1, and the
downregulation of tubulin α-1A, tubulin β-4B, β-actin, CK8 and
CK18, in MCF-7/PTX cells. In our previous study, it was
demonstrated that CK8 undergoes differential phosphorylation and/or
cleavage in 5-FU-resistant colon cancer cell lines (14). Similarly, in the present study, full
length 53-kDa CK8 (Fig. 2, spots no.
22 and 23) and its 50-kDa cleavage product (Fig. 2, spot no. 24) were downregulated in
paclitaxel-resistant cells. CK8 and CK18 function in the
cytoskeleton as intermediate filament components, and are
phosphorylated in response to cellular stress (30). Furthermore, cleaved, soluble fragments
of CK released from apoptotic cancer cells can be detected in
bodily fluids, including serum (31).
Therefore, we plan to analyze clinical samples in future studies to
determine whether the phosphorylated and/or cleaved forms of CK8
and CK18 are potential prognostic biomarkers for paclitaxel and
5-FU resistance.
Stathmin is a microtubule-destabilizing protein and
plays an important role in the regulation of mitosis (32). Previous studies have determined that
stathmin expression is associated with paclitaxel resistance in
breast cancer cell lines (33) and
with clinical responses to taxanes in patients receiving standard
treatment (34). Notably, stathmin
and the stress-response chaperones, GRP75 and GRP78, bind to
tubulin (35), which is the target of
paclitaxel. Among the 14 identified tubulin heterodimer-associated
proteins, GRP75, GRP78 and stathmin were all upregulated in
MCF-7/PTX cells. This may be the result of direct or indirect
effects on paclitaxel resistance and/or stress-response chaperones,
which could convey protection against paclitaxel-induced stress and
apoptosis.
In conclusion, the present study identified 23
proteins that were differentially expressed in paclitaxel-resistant
MCF-7/PTX cells compared with the paclitaxel-sensitive parental
MCF-7 cell line using proteomic techniques. Among these proteins,
PPIA levels were upregulated in MCF-7/PTX cells, the knockdown of
which restored paclitaxel resistance. These results suggested that
PPIA plays an important role in paclitaxel resistance in MCF-7/PTX
cells, likely by inhibiting apoptosis through various means.
Acknowledgements
This study was supported by JSPS KAKENHI (grant nos.
23510267 and 25870924). This manuscript was prepared with the
assistance of a scientific editing service.
Glossary
Abbreviations
Abbreviations:
|
5-FU
|
5-fluorouracil
|
|
GRP78
|
78-kD glucose-regulated protein
|
|
CK
|
cytokeratin
|
|
HSP7C
|
heat shock cognate 71-kDa protein
|
|
HSPB1
|
heat shock protein β-1
|
|
hnRNP
|
heterogeneous nuclear
ribonucleoprotein
|
|
IEF
|
isoelectric focusing
|
|
MALDI
|
matrix-assisted laser
desorption/ionization
|
|
PPIA
|
peptidyl-prolyl cis-trans
isomerase A
|
|
PKM2
|
pyruvate kinase M2
|
|
GRP75
|
stress-70 protein
|
|
SODC
|
superoxide dismutase [Cu-Zn]
|
|
2-DE
|
two-dimensional gel
electrophoresis
|
|
TOF
|
time of flight
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
References
|
1
|
Tan M and Yu D: Molecular mechanisms of
erbB2-mediated breast cancer chemoresistance. Adv Exp Med Biol.
608:119–129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kavallaris M: Microtubules and resistance
to tubulin-binding agents. Nat Rev Cancer. 10:194–204. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pusztai L, Wagner P, Ibrahim N, Rivera E,
Theriault R, Booser D, Symmans FW, Wong F, Blumenschein G, Fleming
DR, et al: Phase II study of tariquidar, a selective P-glycoprotein
inhibitor, in patients with chemotherapy-resistant, advanced breast
carcinoma. Cancer. 104:682–691. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katsetos CD, Herman MM and Mörk SJ: Class
III beta-tubulin in human development and cancer. Cell Motil
Cytoskeleton. 55:77–96. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee KM, Cao D, Itami A, Pour PM, Hruban
RH, Maitra A and Ouellette MM: Class III beta-tubulin, a marker of
resistance to paclitaxel, is overexpressed in pancreatic ductal
adenocarcinoma and intraepithelial neoplasia. Histopathology.
51:539–546. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sève P and Dumontet C: Is class III
beta-tubulin a predictive factor in patients receiving
tubulin-binding agents? Lancet Oncol. 9:168–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katsetos CD, Dráberová E, Legido A,
Dumontet C and Dráber P: Tubulin targets in the pathobiology and
therapy of glioblastoma multiforme. I. Class III beta-tubulin. J
Cell Physiol. 221:505–513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kavallaris M, Kuo DY, Burkhart CA, Regl
DL, Norris MD, Haber M and Horwitz SB: Taxol-resistant epithelial
ovarian tumors are associated with altered expression of specific
beta-tubulin isotypes. J Clin Invest. 100:1282–1293. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gan PP, Pasquier E and Kavallaris M: Class
III beta-tubulin mediates sensitivity to chemotherapeutic drugs in
non small cell lung cancer. Cancer Res. 67:9356–9363. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wilhelm M, Schlegl J, Hahne H, Gholami AM,
Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx
H, et al: Mass-spectrometry-based draft of the human proteome.
Nature. 509:582–587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sá-Correia I and Teixeira MC: 2D
electrophoresis-based expression proteomics: A microbiologist's
perspective. Expert Rev Proteomics. 7:943–953. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villeneuve DJ, Hembruff SL, Veitch Z,
Cecchetto M, Dew WA and Parissenti AM: cDNA microarray analysis of
isogenic paclitaxel- and doxorubicin-resistant breast tumor cell
lines reveals distinct drug-specific genetic signatures of
resistance. Breast Cancer Res Treat. 96:17–39. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carmichael J, DeGraff WG, Gazdar AF, Minna
JD and Mitchell JB: Evaluation of a tetrazolium-based semiautomated
colorimetric assay: Assessment of chemosensitivity testing. Cancer
Res. 47:936–942. 1987.PubMed/NCBI
|
|
14
|
Sakai A, Otani M, Miyamoto A, Yoshida H,
Furuya E and Tanigawa N: Identification of phosphorylated serine-15
and −82 residues of HSPB1 in 5-fluorouracil-resistant colorectal
cancer cells by proteomics. J Proteomics. 75:806–818. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aldridge GM, Podrebarac DM, Greenough WT
and Weiler IJ: The use of total protein stains as loading controls:
an alternative to high-abundance single-protein controls in
semi-quantitative immunoblotting. J Neurosci Methods. 172:250–254.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong HY, Yoo GS and Choi JK: Direct Blue
71 staining of proteins bound to blotting membranes.
Electrophoresis. 21:841–845. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Murphy ME: The HSP70 family and cancer.
Carcinogenesis. 34:1181–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yi X, Luk JM, Lee NP, Peng J, Leng X, Guan
XY, Lau GK, Beretta L and Fan ST: Association of mortalin (HSPA9)
with liver cancer metastasis and prediction for early tumor
recurrence. Mol Cell Proteomics. 7:315–325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rozenberg P, Kocsis J, Saar M, Prohászka
Z, Füst G and Fishelson Z: Elevated levels of mitochondrial
mortalin and cytosolic HSP70 in blood as risk factors in patients
with colorectal cancer. Int J Cancer. 133:514–518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu Y, Li J and Lee AS: GRP78/BiP inhibits
endoplasmic reticulum BIK and protects human breast cancer cells
against estrogen starvation-induced apoptosis. Cancer Res.
67:3734–3740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roller C and Maddalo D: The molecular
chaperone GRP78/BiP in the development of chemoresistance:
Mechanism and possible treatment. Front Pharmacol. 4:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim JW, Sahm H, You J and Wang M:
Knock-down of superoxide dismutase 1 sensitizes cisplatin-resistant
human ovarian cancer cells. Anticancer Res. 30:2577–2581.
2010.PubMed/NCBI
|
|
23
|
Obchoei S, Wongkhan S, Wongkham C, Li M,
Yao Q and Chen C: Cyclophilin A: Potential functions and
therapeutic target for human cancer. Med Sci Monit. 15:RA221–RA232.
2009.PubMed/NCBI
|
|
24
|
Nigro P, Pompilio G and Capogrossi MC:
Cyclophilin A: A key player for human disease. Cell Death Dis.
4:e8882013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J,
Choe W and Kim SS: Overexpressed cyclophilin A in cancer cells
renders resistance to hypoxia- and cisplatin-induced cell death.
Cancer Res. 67:3654–3662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pandita A, Kumar B, Manvati S, Vaishnavi
S, Singh SK and Bamezai RN: Synergistic combination of gemcitabine
and dietary molecule induces apoptosis in pancreatic cancer cells
and down regulates PKM2 expression. PLoS One. 9:e1071542014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao X, Yang J, Li R, Liu S, Xu Y, Zheng
W, Yi Y, Luo Y, Gong F, Peng H, et al: Deregulation of
mitochondrial ATPsyn-β in acute myeloid leukemia cells and with
increased drug resistance. PLoS One. 8:e836102013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dreyfuss G, Kim VN and Kataoka N:
Messenger-RNA-binding proteins and the messages they carry. Nat Rev
Mol Cell Biol. 3:195–205. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rauch J, O'Neill E, Mack B, Matthias C,
Munz M, Kolch W and Gires O: Heterogeneous nuclear
ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells
by regulating A-Raf transcription. Cancer Res. 70:1679–1688. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao J, Ku NO and Omary MB: Stress,
apoptosis, and mitosis induce phosphorylation of human keratin 8 at
Ser-73 in tissues and cultured cells. J Biol Chem. 272:17565–17573.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barak V, Goike H, Panaretakis KW and
Einarsson R: Clinical utility of cytokeratins as tumor markers.
Clin Biochem. 37:529–540. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rubin CI and Atweh GF: The role of
stathmin in the regulation of the cell cycle. J Cell Biochem.
93:242–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alli E, Yang JM, Ford JM and Hait WN:
Reversal of stathmin-mediated resistance to paclitaxel and
vinblastine in human breast carcinoma cells. Mol Pharmacol.
71:1233–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Werner HM, Trovik J, Halle MK, Wik E,
Akslen LA, Birkeland E, Bredholt T, Tangen IL, Krakstad C and
Salvesen HB: Stathmin protein level, a potential predictive marker
for taxane treatment response in endometrial cancer. PLoS One.
9:e901412014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gache V, Louwagie M, Garin J, Caudron N,
Lafanechere L and Valiron O: Identification of proteins binding the
native tubulin dimer. Biochem Biophys Res Commun. 327:35–42. 2005.
View Article : Google Scholar : PubMed/NCBI
|