Introduction
Cancer/testis (CT) antigens are a heterogeneous
group of >200 proteins with an eponymous expression pattern of
being restricted to tumor cells of different histological origins,
and to germ cells in the testes and placenta (1–4). CT
antigens are classified as those that are encoded on the ×
chromosome, known as CT-X antigens, and those that are not, which
are named non-X CT antigens (5). In
total, >50% of all the identified CT antigens to date are CT-X
antigens, which are frequently multicopy genes (5). The genes encoding non-X CT antigens,
however, are distributed throughout the genome and are typically
single-copy genes (5,6). Despite >200 CT antigens have been
categorized, the function of the majority of CT antigens in
gametogenesis and carcinogenesis remains unclear (2,7,8).
The CT antigen NY-SAR-35, also referred to as CT
antigen 37 or fragile × mental retardation 1 neighbor, was
identified using the serological analysis of recombinant
complementary DNA (cDNA) expression libraries (SEREX) method and is
encoded by a gene located on the × chromosome (9). NY-SAR-35 is a 255-amino acid multi-pass
membrane protein with a predicted molecular mass of 29.2 kDa and a
trefoil (P-type) domain (9). The
P-type domain is a three-looped clover leaf-shaped domain of ~38
amino acids in length, in which the loops are held together by
highly conserved disulfide bonds (10–12).
NY-SAR-35 has been identified to be expressed in a
number of cancer types, including melanoma, sarcoma, and lung,
breast and ovarian cancer (9).
Despite the aberrant expression of NY-SAR-35 in a range of
malignancies, it is not expressed in certain cancer types,
including colon and renal cancer. Analysis of the methylation
status of the NY-SAR-35 gene indicated that its expression is
regulated by methylation of its promoter region (13).
Hepatocellular carcinoma and lung cancer are the
most common tumor types worldwide and the leading causes of
cancer-associated mortality in Korea (14–17). A
number of germline genes are overexpressed during the development
of these malignancies (5,6,18–23), although whether NY-SAR-35 serves a
role in their tumorigenesis remains undetermined. The present study
hypothesized that NY-SAR-35 may function in oncogenesis. The aim of
the current study was to use in vitro cell models lacking
the NY-SAR-35 gene to assess the function of NY-SAR-35 in
hepatocellular carcinoma and lung cancer. In particular, the
current study aimed to determine whether NY-SAR-35 expression
affects cell proliferation, migration and invasion in cancer
cells.
Materials and methods
Cell culture
Human hepatocellular carcinoma SNU-449 and lung
adenocarcinoma SK-LC-14 cells were obtained from the Korean Cell
Line Bank (Cancer Research Institute, Soul National University,
Seoul, Korea) and the American Type Culture Collection (Manassas,
VA, USA), respectively. Cells were maintained in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.,), 2 mM L-glutamine, 100 U/ml penicillin and 100
µg/ml streptomycin. The cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2.
Total RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from the cells using the
RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer's protocol. Subsequently, cDNA was synthesized from 1
µg total RNA using 5 Units Moloney murine leukemia virus reverse
transcriptase (M-MLV) in 5X M-MLV buffer (both Promega Corporation,
Madison, WI, USA). The reverse transcriptase and reaction buffer
were incubated at 37°C for 50 min, prior to RT. RNA and reverse
transcriptase were subsequently incubated with 100 pmol random
primer (Takara Biotechnology Co., Ltd., Dalian, China) and 1 µl
mixed dNTPs (10 mM; Solgent Co., Ltd., Seoul, Korea) at 65°C for 5
min, and immediately transferred to ice. The NY-SAR-35 primer
sequences used were as follows: Forward, 5′-CTTGGTGCGATCAGCCTTAT-3′
and reverse, 5′-TTGATGCATGAAAACAGAAC-3′. The GAPDH primer sequences
used were as follows: Forward, 5′-GTTTACATGTTCCAATATGATTCCAC-3′ and
reverse, 5′-TCATATTTGGCAGGTTTTTCTAGAC-3′. PCR amplification was
performed using the 2X TOPsimple™ DyeMIX-Tenuto kit (Enzynomics,
Daejeon, Korea) and the following thermocycling conditions:
Denaturation for 5 min at 94°C; 35 cycles of 30 sec at 94°C, 30 sec
at 55°C, and 1 min at 72°C; and a 10 min final extension at 72°C.
PCR products were analyzed by 1.2% agarose gel electrophoresis and
visualized using ethidium bromide. The complementary DNA templates
were normalized using GAPDH.
Construction of stable cell lines
To generate cells stably expressing NY-SAR-35, the
open reading frame of the NY-SAR-35 gene was cloned into the
pcDNA3.1/V5-HisA mammalian expression vector (Invitrogen; Thermo
Fisher Scientific, Inc.), which has a C-terminal fusion tag (V5 and
6-His epitopes) as well as EcoRI and XhoI restriction
sites. Subsequently, cells were seeded into 6-well plates at a
density of 2.5×105 cells/well and transfected with 1 µg
cloned pcDNA3.1/V5-HisA-NY-SAR-35 using Lipofectamine LTX Reagent
(Thermo Fisher Scientific, Inc.). Transfected cells were selected
by supplementing their culture medium with 1 mg/ml G418
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and then being
maintained in culture medium containing 0.3 mg/ml G418.
Untransfected cells were used as the control.
Cell viability assay
A total of 5×105 cells were seeded into
100-mm culture plates and cultured for 4 days in standard culture
medium supplemented with either 1 or 10% FBS. To test the effect of
growth factor withdrawal on the proliferation of NY-SAR-35
transfectants, the cells were trypsinized and the trypan blue
solution was added on day 4, and incubated for 5 min at room
temperature. Subsequently, samples were counted using a
hemocytometer and the ratio of viable/dead cells was
determined.
Bromodeoxyuridine (BrdU) incorporation
assay
Cell proliferation was measured through BrdU
incorporation using the Cell Proliferation ELISA BrdU kit (Roche
Diagnostics GmbH, Manheim, Germany), according to the
manufacturer's protocol. A total of 20,000 cells/well were grown in
96-well plates for 2 days at 37°C and labeled with 10 µM BrdU for 2
h, prior to fixation and DNA denaturation. Subsequently, anti-BrdU
peroxidase-conjugated fragment-antigen binding fragments and
substrate were added to the medium, and the optical density at 450
nm was determined using an ELISA reader (BioTek Instruments, Inc.,
Winooski, VT, USA) and a reference wavelength of 690 nm.
Immunofluorescence microscopy
Cells were grown at a density of 2×105
cells/dish in 35 mm coverglass-bottom dishes at 37°C for 24 h, and
were washed with PBS and fixed with 4% paraformaldehyde.
Subsequently, cells were blocked with 3% FBS in PBS and incubated
with anti-proliferating cell nuclear antigen (PCNA; 1:150; BD
Biosciences, Franklin Lakes, NJ, USA; cat. no. 555566) or
anti-Ki-67 (1:150; BD Biosciences; cat. no. 556003) primary mouse
antibodies at 37°C for 1 h. Cells were washed three times with PBS
and then incubated with fluorescein isothiocyanate-coupled
secondary goat antibodies (1:500; BD Biosciences; cat. no. 554001)
at 4°C for 30 min in the dark. Cells were visualized and images
were captured using a confocal microscope (Olympus Corporation,
Tokyo, Japan).
Cell migration and invasion
assays
To measure cell migration, transwell chamber assays
were performed using a Corning BioCoat Matrigel Invasion Chamber
(BD Biosciences, Franklin Lakes, NJ, USA). The lower surface of the
filters was coated with 1% gelatin. Cells were suspended in
serum-free RPMI-1640 medium and added to the upper chamber at a
density of 5×104 cells/insert. Culture medium containing
10% FBS was added to the lower chamber and the cells were incubated
at 37°C for 18 h. The number of cells that migrated to the lower
side of the upper chamber was counted following staining with
crystal violet. To measure cell invasion, the BioCoat Matrigel
Invasion chambers (BD Biosciences) were used. The process described
above was performed, with the exception that the cells were
incubated for 24 h. Inserts were then stained with crystal violet
and the number of invading cells was counted. Three fields of cells
on the lower side of the chambers were counted, and the migration
and invasion of cells were expressed in percentage values compared
with those of the mock control cells.
Statistical analysis
Values are presented as the mean ± standard
deviation of three independent experiments. Differences were
analyzed using the Student's t-test. The analysis was
performed using the SPSS statistical package (version 14.0; SPSS
Inc., Chicago, IL, USA). P<0.05 were considered to indicate a
statistically significant difference.
Results
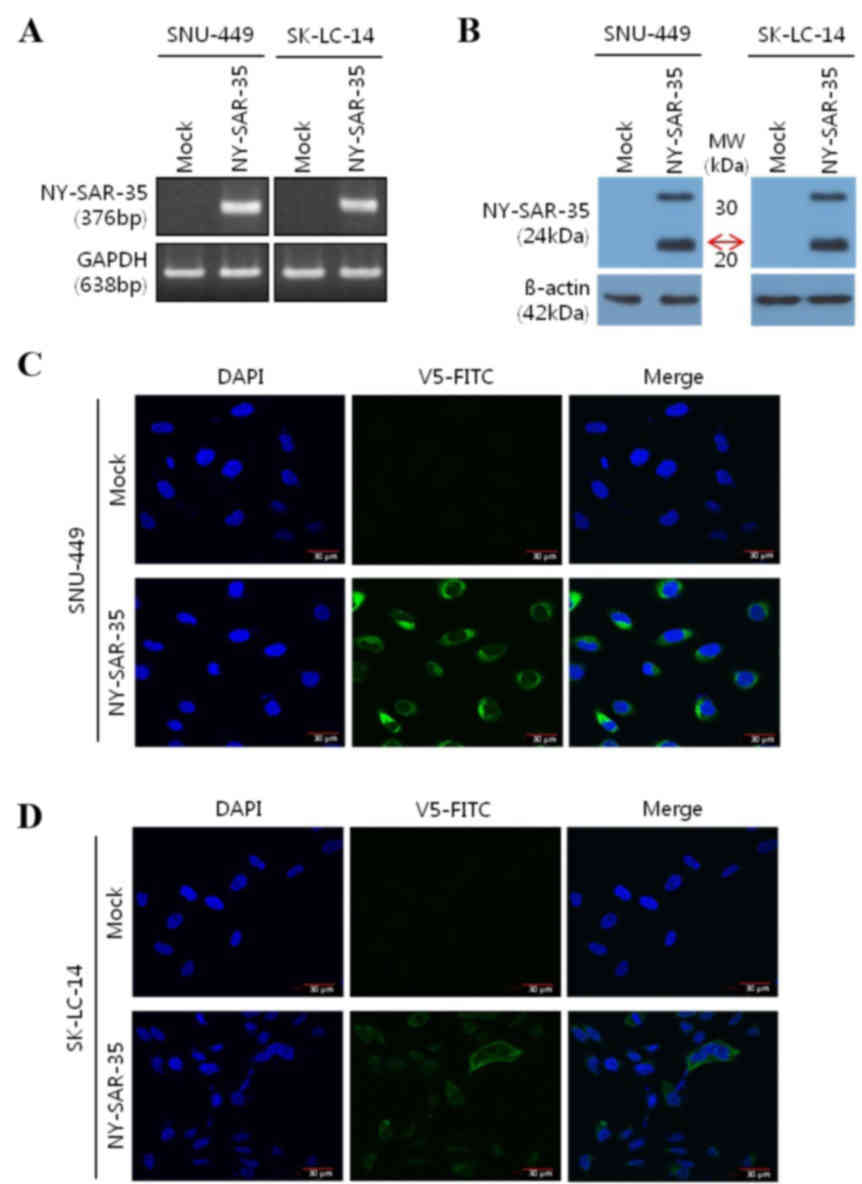
Expression and localization of
NY-SAR-35 in stably transfected SNU-449 and SK-LC-14 cells
To assess the role of NY-SAR-35 in cancer,
NY-SAR-35-positive human hepatocellular carcinoma
(SNU449/NY-SAR-35) and lung adenocarcinoma (SK-LC-14/NY-SAR-35)
cell lines, which do not naturally express NY-SAR-35, were
established through stable transfection. Expression of NY-SAR-35
messenger RNA and protein in the cells was confirmed using RT-PCR
(Fig. 1A) and western blot analysis
(Fig. 1B), respectively. The
subcellular localization of the NY-SAR-35 was analyzed using
immunofluorescence miscopy. This revealed that NY-SAR-35 was
located in the cytoplasm of the cells (Fig. 1C and D).
NY-SAR-35 increases SK-LC-14 and
SNU-449 cell viability and proliferation
Expression of NY-SAR-35 in SNU-449 cells cultured in
medium containing 1 or 10% FBS significantly increased cell
viability by a mean of 1.3- (P=0.049) and 1.9-fold (P=0.036),
respectively, compared with that of mock cells (Fig. 2A). However, no significant difference
in viability was noticed between SK-LC-14/NY-SAR-35 and mock cells
(Fig. 2B). In addition, BrdU
incorporation assays determined that cell proliferation was
significantly increased in SNU449/NY-SAR-35 (P=0.002) and
SK-LC-14-NY-SAR-35 cells (P=0.021) compared with that of mock cells
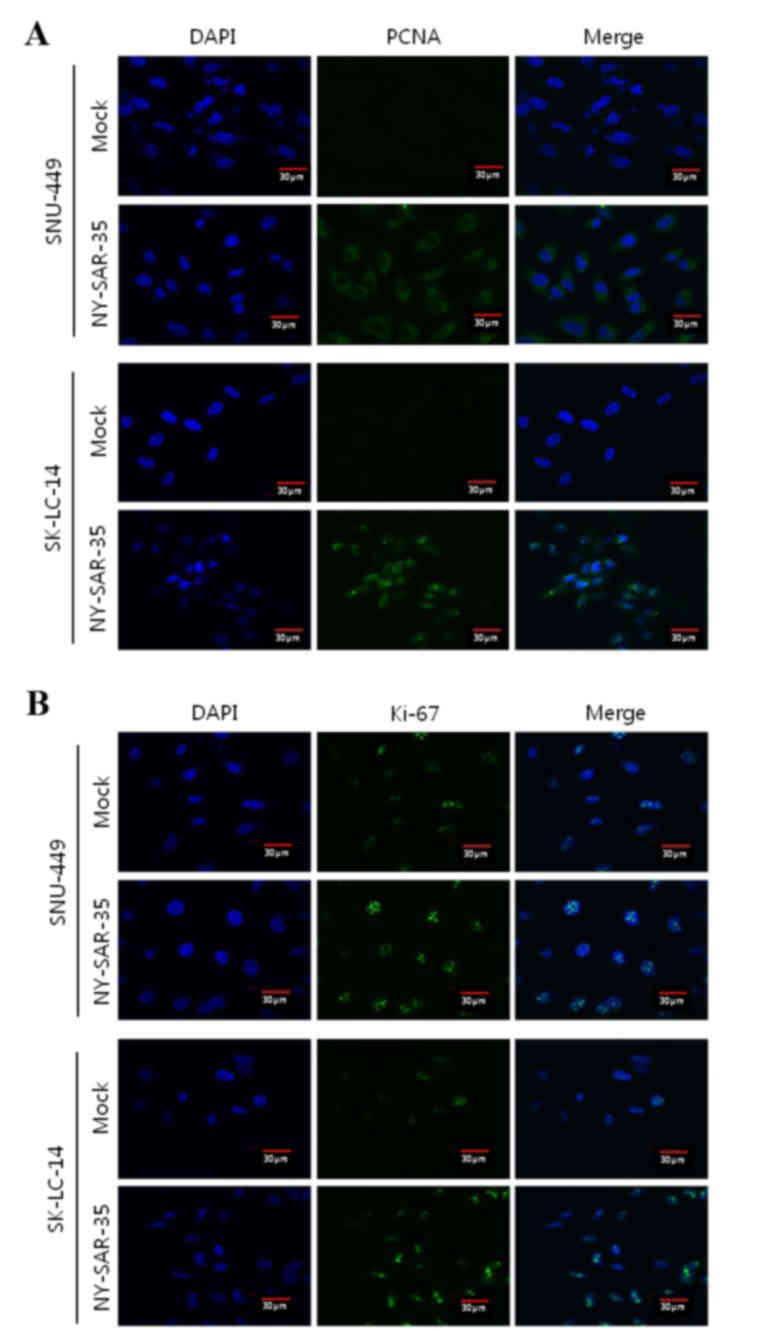
(Fig. 2C and D). Furthermore, the
effect of NY-SAR-35 expression on SNU449 and SK-LC-14 cell
proliferation was analyzed by immunofluorescence staining for PCNA
(Fig. 3A) and Ki-67 (Fig. 3B). SNU449/NY-SAR-35 and
SK-LC-14/NY-SAR-35 cells had increased PCNA and markedly increased
Ki-61 staining compared with those in the mock cells, indicating
that NY-SAR-35 stimulates cancer cell proliferation.
NY-SAR-35 increases SK-LC-14 and
SNU-449 cell migration and invasion
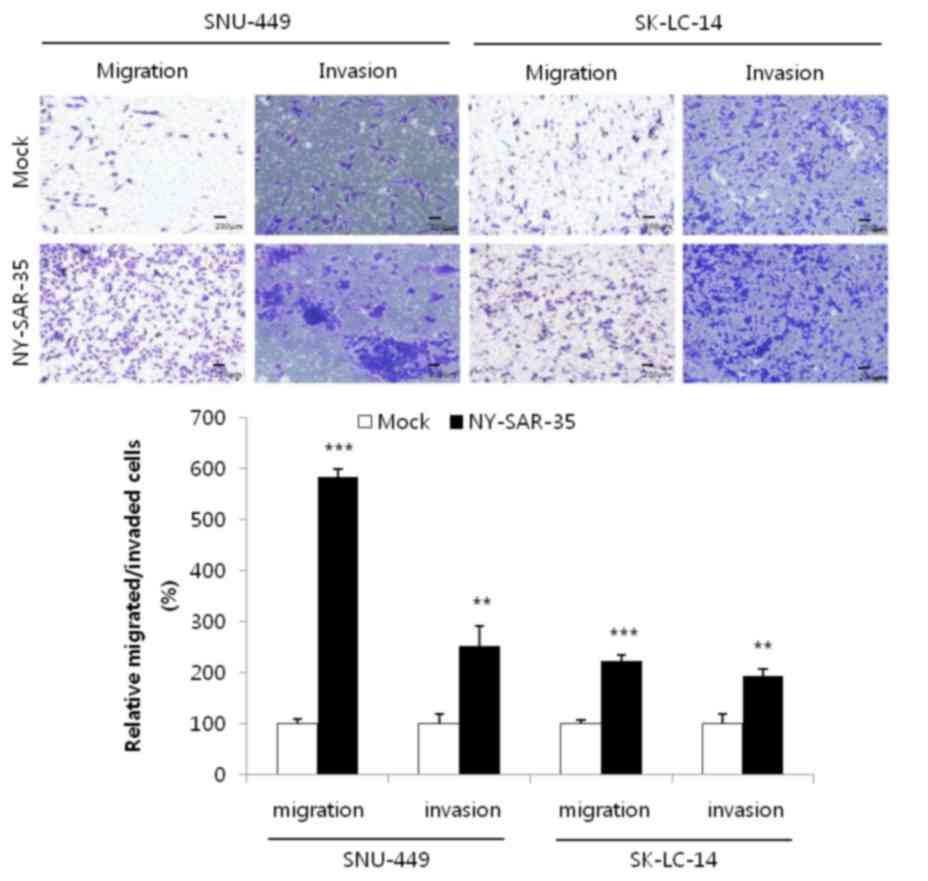
The migratory and invasive capacities of
NY-SAR-35-transfected SK-LC-14 and SNU-449 cells were examined
using transwell assays. This demonstrated that the migration of
SK-LC-14/NY-SAR-35 and SNU449/NY-SAR-35 cells was significantly
increased by 2.2- (P=0.00000137) and 6.2-fold (P=0.0000976),
respectively, compared with that of mock cells (Fig. 4). In addition, cell invasion was
increased by 2.5- (P=0.0044) and 1.9-fold (P=0.0024) in each cell
line, compared with that in mock cells (Fig. 4). These results indicate that
NY-SAR-35 increases the migration and invasion of cancer cells.
Discussion
During carcinogenesis, numerous genes that are
typically expressed during the embryonic developmental stage are
re-expressed in cancer cells, including a number of proto-oncogenes
and CT antigens (2,8,24–27). CT antigens are aberrantly expressed in
variable proportions of a wide range of different types of tumor;
however, not in normal tissues, excluding germ cells. As these
cells do not express major histocompatibility class I complexes,
cluster of differentiation (CD)8+ T cells are not able to recognize
CT antigens expressed on these cells, suggesting that CT antigens
expressed in tumors are targets for vaccine-based immunotherapy.
SEREX-derived CT antigens have been demonstrated to induce CD8+
cytotoxic T lymphocytes (CTLs), and a positive association was
observed between serum positivity for immunoglobulin G antibody and
induction of CD8+ CTLs against several CT antigens, specifically
NY-ESO-1 (2,25). Due to these features of CT antigens,
numerous studies have analyzed their potential use in cancer
immunotherapy (1,3). NY-SAR-35 was identified as encoding a CT
antigen by SEREX (9), and another
study suggested that the NY-SAR-35 gene is subject to epigenetic
regulation (13). However, the role
of NY-SAR-35 in carcinogenesis remains unclear. The results of the
present study demonstrated that NY-SAR-35 expression promoted the
viability, proliferation, migration and invasion of hepatocellular
carcinoma and lung adenocarcinoma cells, suggesting that NY-SAR-35
promotes carcinogenesis.
Immunofluorescence microscopy in the current study
identified increased PCNA and markedly increased Ki-67 levels in
SK-LC-14/NY-SAR-35 and SNU-449/NY-SAR-35 cells. PCNA is increased
in late G1 and S phases of the cell cycle, and is
correlated with the rate of DNA synthesis and cellular
proliferation (28). In addition,
Ki-67 is associated with cell proliferation and is detected
throughout the cell cycle (29,30). The
differences observed between the extent of increase of PCNA and
Ki-67 levels suggest that these antigens may be differentially
affected by NY-SAR-35 expression, which may be due to the fact that
they are expressed differently in different cell types (31). Furthermore, NY-SAR-35 expression
significantly increased the migration and invasion of
hepatocellular and lung carcinoma cells in the present study. This
further suggests that NY-SAR-35 promotes cancer progression.
In conclusion, the results of the present study
demonstrate that NY-SAR-35 increases cancer cell viability,
proliferation, migration and invasion. These results are similar to
those observed in human embryonic kidney 293 cells (32). However, the mechanisms underlying the
effect of NY-SAR-35 in cancer have not been determined. The results
of the current study indicate that this area warrants further
study. In addition, further investigation of the effects and
mechanisms of NY-SAR-35 may elucidate the functions of other CT
antigens, and may provide novel approaches for cancer diagnosis and
therapy.
Acknowledgements
The present study was supported by the Basic Science
Research Program of the National Research Foundation of Korea,
which is funded by the Ministry of Education of Korea (grant no.
NRF-2012R1A1A2041573).
References
|
1
|
Scanlan MJ, Simpson AJ and Old LJ: The
cancer/testis genes: Review, standardization, and commentary.
Cancer Immun. 4:12004.PubMed/NCBI
|
|
2
|
Fratta E, Coral S, Covre A, Parisi G,
Colizzi F, Danielli R, Nicolay HJ, Sigalotti L and Maio M: The
biology of cancer testis antigens: Putative function, regulation
and therapeutic potential. Mol Oncol. 5:164–182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scanlan MJ, Gure AO, Jungbluth AA, Old LJ
and Chen YT: Cancer/testis antigens: An expanding family of targets
for cancer immunotherapy. Immunol Rev. 188:22–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hofmann O, Caballero OL, Stevenson BJ,
Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A,
et al: Genome-wide analysis of cancer/testis gene expression. Proc
Natl Acad Sci USA. 105:20422–20427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Simpson AJ, Caballero OL, Jungbluth A,
Chen YT and Old LJ: Cancer/testis antigens, gametogenesis and
cancer. Nat Rev Cancer. 5:615–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng YH, Wong EW and Cheng CY:
Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis.
Spermatogenesis. 1:209–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kulkarni P, Shiraishi T, Rajagopalan K,
Kim R, Mooney SM and Getzenberg RH: Cancer/testis antigens and
urological malignancies. Nat Rev Urol. 9:386–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whitehurst AW: Cause and consequence of
cancer/testis antigen activation in cancer. Annu Rev Pharmacol
Toxicol. 54:251–272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee SY, Obata Y, Yoshida M, Stockert E,
Williamson B, Jungbluth AA, Chen YT, Old LJ and Scanlan MJ:
Immunomic analysis of human sarcoma. Proc Natl Acad Sci USA.
100:2651–2656. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thim L: Trefoil peptides: From structure
to function. Cell Mol Life Sci. 53:888–903. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sands BE and Podolsky DK: The trefoil
peptide family. Annu Rev Physiol. 58:253–273. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Braun BC, Ringleb J, Waurich R, Viertel D
and Jewgenow K: Functional role of feline zona pellucida protein 4
trefoil domain: A sperm receptor or structural component of the
domestic cat zona pellucida? Reprod Domest Anim. 44:(Suppl 2).
S234–S238. 2009. View Article : Google Scholar
|
|
13
|
Park JH, Song MH, Lee CH, Lee MK, Park YM,
Old L and Lee SY: Expression of the human cancer/testis antigen
NY-SAR-35 is activated by CpG island hypomethylation. Biotechnol
Lett. 33:1085–1091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Davis GL, Dempster J, Meler JD, Orr DW,
Walberg MW, Brown B, Berger BD, O'Connor JK and Goldstein RM:
Hepatocellular carcinoma: Management of an increasingly common
problem. Proc (Bayl Univ Med Cent). 21:266–280. 2008.PubMed/NCBI
|
|
15
|
Yoon SK and Chun HG: Status of
hepatocellular carcinoma in South Korea. Chin Clin Oncol.
2:392013.PubMed/NCBI
|
|
16
|
Bae JM, Lee MS, Shin MH, Kim DH, Li ZM and
Ahn YO: Cigarette smoking and risk of lung cancer in Korean men:
The Seoul male cancer cohort study. J Korean Med Sci. 22:508–512.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG
and Lee JS: Cancer statistics in Korea: Incidence, mortality,
survival and prevalence in 2010. Cancer Res Treat. 45:1–14. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen YT, Hsu M, Lee P, Shin SJ,
MhawechFauceglia P, Odunsi K, Altorki NK, Song CJ, Jin BQ, Simpson
AJ and Old LJ: Cancer/testis antigen CT45: Analysis of mRNA and
protein expression in human cancer. Int J Cancer. 124:2893–2898.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gure AO, Chua R, Williamson B, Gonen M,
Ferrera CA, Gnjatic S, Ritter G, Simpson AJ, Chen YT, Old LJ and
Altorki NK: Cancer-testis genes are coordinately expressed and are
markers of poor outcome in non-small cell lung cancer. Clin Cancer
Res. 11:8055–8062. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng JR, Chen HS, Mou DC, Cao J, Cong X,
Qin LL, Wei L, Leng XS, Wang Y and Chen WF: Expression of
cancer/testis (CT) antigens in Chinese hepatocellular carcinoma and
its correlation with clinical parameters. Cancer Lett. 219:223–232.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawa K, Noguchi Y, Uenaka A, Sato S,
Okumura H, Tanaka M, Shimono M, Ali Eldib AM, Ono T, Ohara N, et
al: XAGE-1 expression in non-small cell lung cancer and antibody
response in patients. Clin Cancer Res. 11:5496–5503. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kurashige T, Noguchi Y, Saika T, Ono T,
Nagata Y, Jungbluth A, Ritter G, Chen YT, Stockert E, Tsushima T,
et al: Ny-ESO-1 expression and immunogenicity associated with
transitional cell carcinoma: Correlation with tumor grade. Cancer
Res. 61:4671–4674. 2001.PubMed/NCBI
|
|
23
|
Scanlan MJ, Altorki NK, Gure AO,
Williamson B, Jungbluth A, Chen YT and Old LJ: Expression of
cancer-testis antigens in lung cancer: Definition of bromodomain
testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett.
150:155–164. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ayyoub M, Taub RN, Keohan ML, Hesdorffer
M, Metthez G, Memeo L, Mansukhani M, Hibshoosh H, Hesdorffer CS and
Valmori D: The frequent expression of cancer/testis antigens
provides opportunities for immunotherapeutic targeting of sarcoma.
Cancer Immun. 4:72004.PubMed/NCBI
|
|
25
|
Dobrynin P, Matyunina E, Malov SV and
Kozlov AP: The novelty of human cancer/testis antigen encoding
genes in evolution. Int J Genomics. 2013:1051082013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pandey A, Kurup A, Shrivastava A, Radhi S,
Nguyen DD, Arentz C, D'Chuna N, Hardwick F, D'Souza MJ, Jenkins M,
et al: Cancer testes antigens in breast cancer: Biological role,
regulation, and therapeutic applicability. Int Rev Immunol.
31:302–320. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shiraishi T, Terada N, Zeng Y, Suyama T,
Luo J, Trock B, Kulkarni P and Getzenberg RH: Cancer/testis
antigens as potential predictors of biochemical recurrence of
prostate cancer following radical prostatectomy. J Transl Med.
9:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kelman Z: PCNA: Structure, functions and
interactions. Oncogene. 14:629–640. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gerdes J, Schwab U, Lemke H and Stein H:
Production of a mouse monoclonal antibody reactive with a human
nuclear antigen associated with cell proliferation. Int J Cancer.
31:13–20. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ihmann T, Liu J, Schwabe W, Häusler P,
Behnke D, Bruch HP, Broll R, Windhovel U and Duchrow M: High-level
mRNA quantification of proliferation marker pKi-67 is correlated
with favorable prognosis in colorectal carcinoma. J Cancer Res Clin
Oncol. 130:749–756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bologna-Molina R, MosquedaTaylor A,
MolinaFrechero N, MoriEstevez AD and Sánchez-Acuña G: Comparison of
the value of PCNA and Ki-67 as markers of cell proliferation in
ameloblastic tumors. Med Oral Patol Oral Cir Bucal. 1:e174–e179.
2013. View Article : Google Scholar
|
|
32
|
Song MH, Kim YR, Lee JW, Lee CH and Lee
SY: Cancer/testis antigen NY-SAR-35 enhances cell proliferation,
migration and invasion. Int J Oncol. 48:569–576. 2016.PubMed/NCBI
|