Introduction
Acute myeloid leukemia (AML) is a biologically and
clinically heterogeneous disorder, characterized by immature
myeloid cell proliferation and bone marrow failure (1). The most common subtype of AML (M0-M7) is
M2 under FAB classification (2,3). In total
~52% of adult patients with de novo AML are reported to
carry at least one type of chromosomal abnormalities (e.g.
translocations or deletions) (3). A
number of recurrent chromosomal aberrations and gene mutations
involved in hematopoietic proliferation and differentiation have
been used as critical risk stratification tools, including
chromosomal translocations t(8;21) and t(15;17), and internal
tandem duplication of Fms-related tyrosine kinase 3 (FLT3)
mutations (3,4). The 5-year overall survival rates for
each risk group (favorable, intermediate and adverse-risk) are ~55,
~24 and ~5%, respectively (3).
However, prognosis remains markedly different for each risk group
(4). Therefore, there is a
requirement to identify additional genetic alterations that are
associated with prognosis.
The chromosomal translocation t(7;11)(p15;p15) is a
rare genetic lesion in AML, which was initially reported in a
patient with chronic myeloid leukemia (CML) more than 30 years ago
(5). This translocation results in a
fusion of the N-terminal portion of nucleoporin 98 (NUP98)
with the homeodomain of several homeobox A (HOXA) genes,
including HOXA9, HOXA11 and HOXA13; the most common fusion
is HOXA9 (6–8). The association between clinical and
biological characteristics from AML patients with this type of
fusion remains to be fully elucidated. The present study reports
the case of a male patient with AML-M2 and t(7;11) translocation
resulting in a NUP98-HOXA9 fusion gene. The patient
demonstrated dyspoietic alterations, neuroblastoma V-Ras oncogene
homolog (NRAS) and Wilms tumor 1 (WT1) mutations and
a poor prognosis.
Case report
A 30-year-old male was admitted to Affiliated Cancer
Hospital of Zhengzhou University (Zhengzhou, China) following 2
weeks of pain in each leg in August 2015. The patient presented
with pale skin and sternal tenderness. Peripheral blood analysis
identified a white blood cell count of 32.2×109/l (normal range,
4–10×109/l), with 44% blasts and 26% monocytes (normal range,
3–8%), a hemoglobin level of 110 g/l (normal range, 120–160 g/l)
and a platelet count of 87×109/l (normal range, 100–300×109/l). A
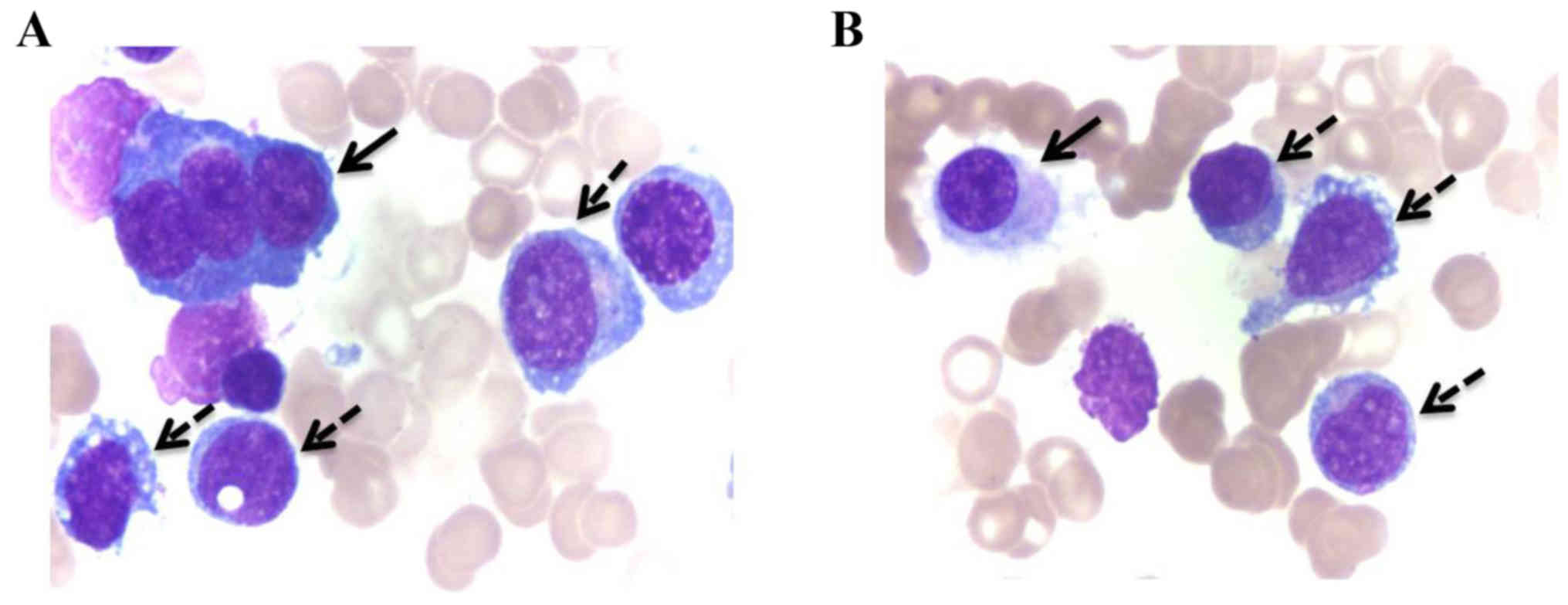
bone marrow (BM) smear demonstrated a markedly hypercellular BM,
with 71% myeloblasts, and a positive result for myeloperoxidase
staining, which suggested an AML-M2 subtype (2). Dyspoietic alterations, including
multinucleated erythrocyte precursors and micromegakaryocytes, were
identified (Fig. 1). Subsequently,
immunophenotyping was performed on a multi-color flow cytometer (BD
FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA) with
monoclonal antibodies in 1:1 dilutions, including cluster of
differentiation (CD) 34-fluorescein isothiocyanate (FITC) (catalog
no. 555821), human leukocyte antigen-antigen D-related
(HLA-DR)-allophycocyanin (APC) (catalog no. 340549),
CD13-phycoerythrin (PE) (catalog no. 347837), cytoplasmic
myeloperoxidase (cMPO)-FITC (catalog no. 340580), CD15-FITC
(catalog no. 332778), CD20-APC (catalog no. 340941) cytoplasmic
(c)CD3-APC (catalog no. 340440), CD5-APC (catalog no. 340583),
CD11b-APC (catalog no. 340937) and CD3-APC (catalog no. 555335),
all of which were purchased from BD Biosciences. Immunophenotyping
was also performed with the following antibodies in 1:1 dilutions:
CD33-PE (catalog no. 555450; BD Pharmingen, San Diego, CA, USA),
CD117-PE (catalog no. 340529; BD Pharmingen), CD56-PE (catalog no.
A07788; Beckman Coulter, Inc., Brea, CA, USA), CD7-PE (catalog no.
IM1429U; Beckman Coulter, Inc.), CD19-FITC (catalog no. A07768;
Beckman Coulter, Inc.), CD4-FITC (catalog no. A07750; Beckman
Coulter, Inc.), CD2-FITC (catalog no. A07743; Beckman Coulter,
Inc.), CD10-PE (catalog no. A07760; Beckman Coulter, Inc.),
CD14-FITC (catalog no. 0645U; Beckman Coulter, Inc.) and cCD79a-PE
(catalog no. IM2221; Beckman Coulter, Inc.).
Blasts were positive for CD34, HLA-DR, CD13, CD33,
CD117 and cMPO antigens, and negative for CD56, CD7, CD15, CD19,
CD4, CD2, CD10, CD20, CD3, CD5, CD11b, CD14, cCD79a and cCD3
antigens. These results confirmed that the blasts were committed to
being myeloid precursors.
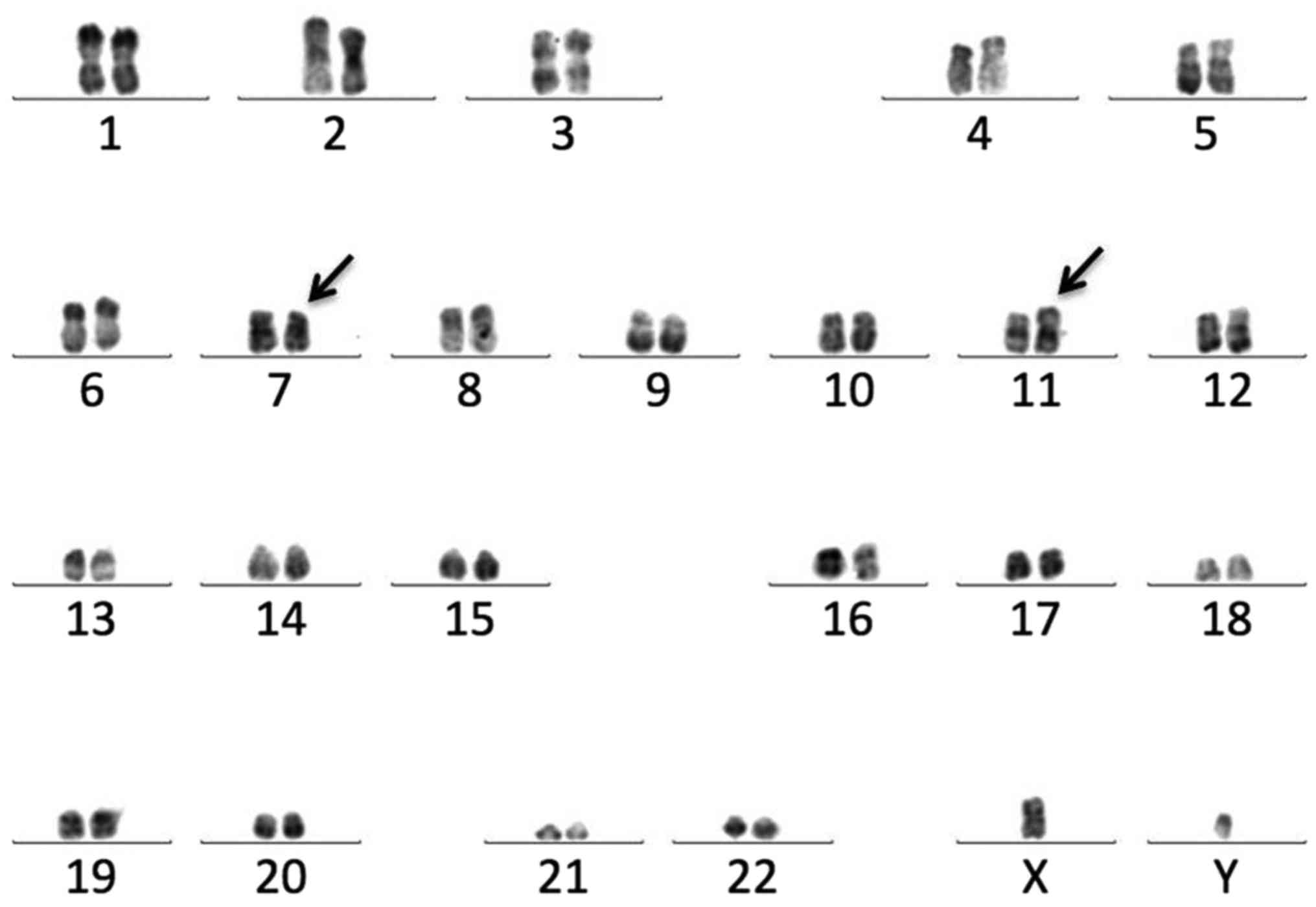
Chromosome analysis using the R-banding method
identified a 46,XY,t(7;11)(p15;p15) karyotype in all 20 metaphase
cells (Fig. 2). NUP98-HOXA9
was identified via a screen of leukemia-associated fusion genes
using a multi-fusion gene detection system (43 Fusion Gene
Screening kit; Shanghai Yuanqi Biopharmaceutical Company, Ltd.,
Shanghai, China) (9). The copy of the
NUP98-HOXA9 fusion transcript was subsequently detected by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) using Leukemia Related Fusion Gene Fluorescent qPCR
Detection kit (Shanghai Shenyou Bio Technology Co., Ltd., Shanghai,
China) with a NUP98-HOXA9/Abelson tyrosine-protein kinase 1
(ABL1) ratio of 0.71. The sequences of primers were designed
as previously reported (10). PCR was
performed on an ABI PRISM 7000 machine (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) under the conditions as
previously described (11) and
quantified using the standard curve method (12). The experiment was repeated
independently three times.
To date, four types of fusion transcripts have been
reported, which appear to result from alternative splicing of
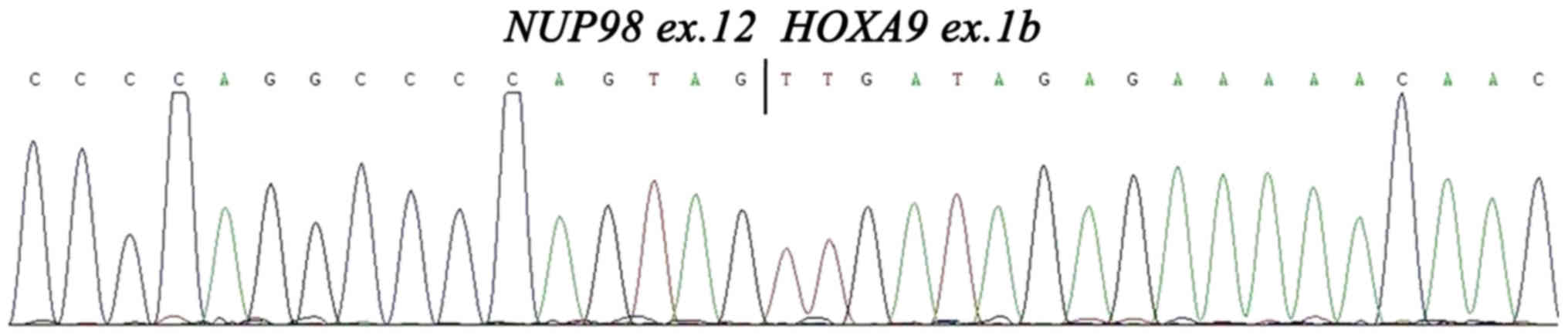
NUP98 and HOXA9 and each fusion is in-frame (10). In this case, detection of the
NUP98-HOXA9 breakpoint was performed by RT-PCR and
sequencing as previously described (10). Analysis demonstrated a type I fusion
transcript, which means that the breakpoint of NUP98-HOXA9
is between NUP98 exon 12 and HOXA9 exon 1b (10) (Fig. 3).
Subsequently, 1 point mutation in NRAS and 2 point mutations
in WT1 were detected using the next generation sequencing
(NGS)-based multigene mutational screening system on an Ion-Torrent
Personal Genome machine (Thermo Fisher Scientific, Inc.) (13). The gene screening profile and results
are summarized in Table I.
 | Table I.Leukemia-associated fusion genes and
mutation-screened genes. |
Table I.
Leukemia-associated fusion genes and
mutation-screened genes.
| Fusion and mutation
gene screening profile |
|---|
| Fusion genes for
screening |
|
AML1-ETO, AML1-MDS1/EV11,
AML1-MTG16, BCR-ABL, CBFβ-MYH11, DEK-CAN, E2A-HLF, E2A-PBX1,
ETV6-PDGFRA, FIP1L1-PDGFRA, MLL-AF4, MLL-AF9, NPM-MLF, PML-RARα,
PLZF-RARα, SET-CAN, SIL-TAL1, STAT5b-RARα, TEL-ABL, TEL-JAK2,
TEL-PDGFRβ, TLS-ERG, MLL-(AF6, AF10, AF17, AF1q, AF1p, AFX, ELL,
ENL, SEPT6), NUP98-(HOXA13, HOXA11, HOXA9a, HOXC11, HOXD13, PMX1), (NPM,
FIP1L1, PRKAR1A, NUMA1)-RARα |
| Genes for
mutational detection |
| AKT3,
ASXL1, ATRX, BCOR, CBL, CCND1, CDKN2A, CEBPA, CREBBP, CSF1R, CSF3R,
DNMT3A, EP300, ETV6, EZH2, FLT3, GATA2, IDH1, IDH2, IKZF1, JAK3,
KIT, KRAS, MLH1, MPL, NOTCH1, NOTCH2, NPM1, NRASb, PHF6, PTPN11, RAD21, RUNX1,
SETBP1, SRSF2, STAG2, STAT3, TET2, TP53, U2AF1, WT1c, ZRSR2 |
Following diagnosis, the patient was treated with
induction chemotherapy for 7 days using a combination of
daunorubicin (80 mg/day for 3 days) and cytosine arabinoside
(Ara-c; 200 mg/day for 7 days). After 3 weeks, the patient
demonstrated hematological complete remission (CR), with 1.6%
myeloblasts in the BM smear. The repeat RT-qPCR revealed a decrease
in the NUP98-HOXA9/ABL1 ratio to 0.0076.
Subsequently, the patient underwent 4 courses of consolidation
chemotherapy with a high-dose Ara-c (HD-Ara-c) regimen (1.5 g/m2
q12 h on days 1, 3 and 5) at the same time as seeking an
HLA-matched sibling or unrelated donor for allogenic-hematopoietic
stem cell transplantation (HSCT). During this period, repeated BM
and fusion examinations were performed 4 times. Hematological CR
was observed in the 4 tests, however, the
NUP98-HOXA9/ABL1 ratio remained detectable, with a
lowest level of 0.0023.
Due to the failure to find an HLA-matched donor, it
was suggested that the patient should receive autogenic-HSCT.
However, following treatment with a further course of HD-Ara-c
chemotherapy (1.5 g/m2 q12 h on days 1, 3 and 5) as a hematopoietic
stem cell mobilization method, the disease reoccurred 11 months
after the original diagnosis (June 2016). A BM smear indicated 46%
myeloblasts, with the presence of micromegakaryocytes. The
NUP98-HOXA9/ABL1 ratio was markedly increased to
0.079. Cytogenetic analysis and NGS demonstrated the same
46,XY,t(7;11)(p15;p15) karyotype and NRAS and WT1
mutations as the diagnostic tests, which indicates that there was
no genetic evolution of the tumor. The disease became refractory,
and the patient contracted severe pneumonia and respiratory
failure, which caused mortality 1 month after recurrence.
Discussion
The role of NUP98-HOXA9 in leukemogenesis has
been repeatedly demonstrated in the past two decades. The
transforming potential for the development of leukemia may require
other cooperative factors (10).
Using transgenic mouse models, breakpoint cluster region
(BCR)/ABL was observed to interact with
NUP98-HOXA9 and promote AML development in CML progression
(14). Using retroviral insertional
mutagenesis, myeloid ecotropic viral integration site 1 homolog
(Meis1), dynein axonemal light chain 4, Fc fragment of IgG
receptor IIb, Fc receptor-like and Con1 glycoprotein were
identified as co-factors that associate with NUP98-HOXA9 in
myeloid leukemogenesis (15). The
most frequent of these associations was with Meis1, and the
interaction between Meis1 and NUP98-HOXA9 reduces the
latency of AML development (16).
Furthermore, microarray analysis of human CD34-positive
hematopoietic cells also identified oncogenes that may potentially
associate with the NUP98-HOXA9, including the homeobox
transcription factors, FLT3, c-kit proto-oncogene and
WT1 (17). The following four
gene mutations, FLT3-internal duplications, NRAS, Kirsten
rat sarcoma viral oncogene homolog (KRAS) and WT1
have been identified with the NUP98-HOXA9 in a total of 7
patients with AML and t(7;11) (10,18).
KRAS and WT1 mutations demonstrated a significantly
association with the NUP98-HOXA9 fusion gene (10). In the present study, a novel gene
mutation group of NRAS and WT1 mutations was found to
coexist with NUP98-HOXA9 at the initial diagnosis and at the
relapse. As RAS and WT1 mutations are key components
of the proliferative drive of AML (19,20), taken
together with previous studies, the present study indicates these
gene alterations may cooperate with NUP98-HOXA9 in promoting
the development and relapse of leukemia.
Previous studies have established that patients with
AML and t(7;11) have distinct epidemic and clinical
characteristics. A literature review was performed using the PubMed
database to search for the keywords ‘t(7;11)’, ‘acute myeloid
leukemia’ (www.ncbi.nlm.nih.gov/pubmed/?term=t(7%3B11)+AND+
acute+myeloid+leukemia). Patient characteristics including gender,
age (years) and French-American-British (FAB) classification
subtypes (2) were recorded. To date,
only 57 adult patients (7,10,18,21–34)
were recorded with de novo AML in the literature. The
majority of the case studies (98.2%, 56/57) originated in Japan or
China. The incidence rate of t(7;11) on de novo adult
patients with AML was low at 0.68–2.23% (10,18,27,28,30).
For these 57 patients, the median onset age was 36 years (range,
31–38 years) and 63.1% (36/57) were female. A total of 54 patients
had FAB classification subtypes and 68.5% (37/54) of these patients
has an M2-subtype (7,10,18,21–34).
These characteristics of being predominantly younger, being female
and having an M2-subtype were statistically proven in a large
sample study by Chou et al (10) by comparing 11 patients carrying
t(7;11) translocations with another 482 adult patients with AML.
The case identified by the present study was also of a patient of a
young age and with an M2 subtype, which is concordant with these
previous studies.
In the present study, marked concomitant abnormal
myelopoiesis, including dysplastic erythrocytopoiesis and
megakaryocytopoiesis, were established from a de novo bone
morrow smear. Similarly, previous studies have identified t(7;11)
in AML patients to be associated with myelodysplasia (MDS) or
myeloid maturation (10,21,27,30,31).
A transgenic murine model with the NUP98-HOXA9 fusion gene
also acquired myeloproliferative disorder and subsequently
developed AML (35). Although it is
primarily observed in AML, t(7;11) has also been identified in
patients with MDS (36), CML
(5,37)
and juvenile myelomonocytic leukemia (8). Therefore, the present study is
concordant with the hypothesis that t(7;11) may affect multipotent
myeloid stem and progenitor cells.
Patients with AML that have t(7;11) exhibit
aggressive clinical progression and a poor prognosis. Using
multivariate analysis, Chou et al established that this
translocation was an independent factor for predicting a decrease
in overall, relapse-free and disease-free survival rates (10). The patient presented in the present
study achieved a CR following 2 courses of induction chemotherapy,
but relapsed and became refractory after a short duration, with 12
months of overall survival. Via the monitoring of
NUP98-HOXA9 transcript copies, fusion protein signals were
consistently detected throughout treatment courses and after a
hematological CR was achieved. This suggested that residual
leukemia cells were not eradicated. This is concordant with the
majority of cases reported by Chou et al (10), indicating that this fusion gene may
promote survival signals and may be a useful prognostic marker for
monitoring minimal residual disease.
The present study and literature review demonstrated
that patients with AML and t(7;11) translocation are rare, and may
have distinct genetic, molecular and clinical characteristics.
NRAS and WT1 gene mutations are able to coexist with
NUP98-HOXA9 in a number of patients, indicating that there
may be an association between NUP98-HOXA9 and
leukemogenesis. Patients with AML and t(7;11) are predominantly
from Asia, typically young, female and have a FAB M2-subtype, and
may present with concomitant BM dyspoiesis. These patients exhibit
an aggressive clinical course and have a poor prognosis. Further
studies are required with regard to this distinct entity.
Acknowledgements
The present study was supported by the Science and
Technology Project of Henan Province (grant no. 162102310338).
References
|
1
|
Saultz JN and Garzon R: Acute myeloid
leukemia: A concise review. J Clin Med. 5:pii: E33. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposals for the
classification of the acute leukaemias. French-American-British
(FAB) co-operative group. Brit J Haemat. 33:451–458. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Byrd JC, Mrózek K, Dodge RK, Carroll AJ,
Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS,
et al: Pretreatment cytogenetic abnormalities are predictive of
induction success, cumulative incidence of relapse, and overall
survival in adult patients with de novo acute myeloid leukemia:
Results from Cancer and Leukemia Group B (CALGB 8461). Blood.
100:4325–4336. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Kouchkovsky I and Abdul-Hay M: Acute
myeloid leukemia: A comprehensive review and 2016 update. Blood
Cancer J. 6:e4412016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tomiyasu T, Sasaki M, Kondo K and Okada M:
Chromosome banding studies in 106 cases of chronic myelogenous
leukemia. Jinrui Idengaku Zasshi. 27:243–258. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borrow J, Shearman AM, Stanton VP Jr,
Becher R, Collins T, Williams AJ, Dubé I, Katz F, Kwong YL, Morris
C, et al: The t(7;11)(p15;p15) translocation in acute myeloid
leukaemia fuses the genes for nucleoporin NUP98 and class I
homeoprotein HOXA9. Nat Genet. 12:159–167. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taketani T, Taki T, Ono R, Kobayashi Y,
Ida K and Hayashi Y: The chromosome translocation t(7;11)(p15;p15)
in acute myeloid leukemia results in fusion of the NUP98 gene with
a HOXA cluster gene, HOXA13, but not HOXA9. Genes Chromosomes
Cancer. 34:437–443. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mizoguchi Y, Fujita N, Taki T, Hayashi Y
and Hamamoto K: Juvenile myelomonocytic leukemia with
t(7;11)(p15;p15) and NUP98-HOXA11 fusion. Am J Hematol. 84:295–297.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Mao S, Su S, Wei L, Li T and Liu
Y: Detection of concurrent TEL-AML1/TEL-ABL fusion genes in a
patient with B-acute lymphoblastic leukemia using a multi-fusion
gene qRT-PCR screening method. Pathol Int. 66:475–477. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chou WC, Chen CY, Hou HA, Lin LI, Tang JL,
Yao M, Tsay W, Ko BS, Wu SJ, Huang SY, et al: Acute myeloid
leukemia bearing t(7;11)(p15;p15) is a distinct cytogenetic entity
with poor outcome and a distinct mutation profile: Comparative
analysis of 493 adult patients. Leukemia. 23:1303–1310. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lyu X, Yang J, Wang X, Hu J, Liu B, Zhao
Y, Guo Z, Liu B, Fan R and Song Y: A novel BCR-ABL1 fusion gene
identified by next-generation sequencing in chronic myeloid
leukemia. Mol Cytogenet. 9:472016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bookout AL and Mangelsdorf DJ:
Quantitative real-time PCR protocol for analysis of nuclear
receptor signaling pathways. Nucl Recept Signal. 1:e0122003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanagal-Shamanna R, Portier BP, Singh RR,
Routbort MJ, Aldape KD, Handal BA, Rahimi H, Reddy NG, Barkoh BA,
Mishra BM, et al: Next-generation sequencing-based multi-gene
mutation profiling of solid tumors using fine needle aspiration
samples: Promises and challenges for routine clinical diagnostics.
Mod Pathol. 27:314–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dash AB, Williams IR, Kutok JL, Tomasson
MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J,
Housman D, et al: A murine model of CML blast crisis induced by
cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci
USA. 99:7622–7627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iwasaki M, Kuwata T, Yamazaki Y, Jenkins
NA, Copeland NG, Osato M, Ito Y, Kroon E, Sauvageau G and Nakamura
T: Identification of cooperative genes for NUP98-HOXA9 in myeloid
leukemogenesis using a mouse model. Blood. 105:784–793. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kroon E, Krosl J, Thorsteinsdottir U,
Baban S, Buchberg AM and Sauvageau G: Hoxa9 transforms primary bone
marrow cells through specific collaboration with Meis1a but not
Pbx1b. EMBO J. 17:3714–3725. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takeda A, Goolsby C and Yaseen NR:
NUP98-HOXA9 induces long-term proliferation and blocks
differentiation of primary human CD34+ hematopoietic cells. Cancer
Res. 66:6628–6637. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei S, Wang S, Qiu S, Qi J, Mi Y, Lin D,
Zhou C, Liu B, Li W, Wang Y, et al: Clinical and laboratory studies
of 17 patients with acute myeloid leukemia harboring
t(7;11)(p15;p15) translocation. Leuk Res. 37:1010–1015. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bowen DT, Frew ME, Hills R, Gale RE,
Wheatley K, Groves MJ, Langabeer SE, Kottaridis PD, Moorman AV,
Burnett AK and Linch DC: RAS mutation in acute myeloid leukemia is
associated with distinct cytogenetic subgroups but does not
influence outcome in patients younger than 60 years. Blood.
106:2113–2119. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rampal R and Figueroa ME: Wilms tumor 1
mutations in the pathogenesis of acute myeloid leukemia.
Haematologica. 101:672–679. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sato Y, Abe S, Mise K, Sasaki M, Kamada N,
Kouda K, Musashi M, Saburi Y, Horikoshi A and Minami Y: Reciprocal
translocation involving the short arms of chromosomes 7 and 11,
t(7p-; 11p+), associated with myeloid leukemia with maturation.
Blood. 70:1654–1658. 1987.PubMed/NCBI
|
|
22
|
Mise K, Abe S, Sato Y, Miura Y and Sasaki
M: Localization of c-Ha-ras-1 oncogene in the t(7p-; 11p+)
abnormality of two cases with myeloid leukemia. Cancer Genet
Cytogenet. 29:191–199. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takahashi E, Kaneko Y, Ishihara T,
Minamihisamatsu M, Murata M and Hori T: A new rare distamycin
A-inducible fragile site, fra(11)(p15. 1), found in two acute
nonlymphocytic leukemia (ANLL) patients with t(7;11)(p15-p13;p15).
Hum Genet. 80:124–126. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morris CM, Fitzgerald PH, Kennedy MA,
Hollings PE, Garry M and Corbett GM: HRAS1 and INS genes are
relocated but not structurally altered as a result of the
t(7;11)(p15;p15) in a clone from a patient with acute myeloid
leukaemia (M4). Br J Haematol. 71:481–486. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujimura T, Ohyashiki K, Ohyashiki JH,
Kawakubo K, Iwabuchi A, Kodama A and Toyama K: Two additional cases
of acute myeloid leukemia with t(7;11)(p15;p15) having low
neutrophil alkaline phosphatase scores. Cancer Genet Cytogenet.
68:143–146. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abe A, Tanimoto M, Towatari M, Matsuoka A,
Kitaori K, Kato H, Toyozumi H, Takeo T, Adachi K and Emi N: Acute
myeloblastic leukemia (M2) with translocation (7;11) followed by
marked eosinophilia and additional abnormalities of chromosome 5.
Cancer Genet Cytogenet. 83:37–41. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang SY, Tang JL, Liang YJ, Wang CH, Chen
YC and Tien HF: Clinical, haematological and molecular studies in
patients with chromosome translocation t(7;11): A study of four
Chinese patients in Taiwan. Br J Haematol. 96:682–687. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwong YL and Pang A: Low frequency of
rearrangements of the homeobox gene HOXA9/t(7;11) in adult acute
myeloid leukemia. Genes Chromosomes Cancer. 25:70–74. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawakami K, Miyanishi S, Nishii K, Usui E,
Murata T, Shinsato I and Shiku H: A case of acute myeloid leukemia
with t(7;11)(p15;p15) mimicking myeloid crisis of chronic
myelogenous leukemia. Int J Hematol. 76:80–83. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kwong YL and Chan TK: Translocation
(7;11)(p15;p15) in acute myeloid leukemia M2: Association with
trilineage myelodysplasia and giant dysplastic myeloid cells. Am J
Hematol. 47:62–64. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inaba T, Shimazaki C, Yoneyama S, Hirai H,
Kikuta T, Sumikuma T, Sudo Y, Yamagata N, Ashihara E, Goto H, et
al: t(7;11) and trilineage myelodysplasia in acute myelomonocytic
leukemia. Cancer Genet Cytogenet. 86:72–75. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ohyashiki K: Nonrandom cytogenetic changes
in human acute leukemia and their clinical implications. Cancer
Genet Cytogenet. 11:453–471. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Okada M, Mizoguchi H, Kubota K and Nomura
Y: Quantitative analysis of chromosomal G-bands in human
hematopoietic disorders by methotrexate synchronization technique.
Cancer Genet Cytogenet. 13:225–237. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ishihara T, Minamihisamatsu M and Yokoyama
Y: Translocation t(7;11)(p13;p15) in acute nonlymphocytic leukemia.
Cancer Genet Cytogenet. 19:363–364. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kroon E, Thorsteinsdottir U, Mayotte N,
Nakamura T and Sauvageau G: NUP98-HOXA9 expression in hemopoietic
stem cells induces chronic and acute myeloid leukemias in mice.
EMBO J. 20:350–361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hatano Y, Miura I, Nakamura T, Yamazaki Y,
Takahashi N and Miura AB: Molecular heterogeneity of the
NUP98/HOXA9 fusion transcript in myelodysplastic syndromes
associated with t(7;11)(p15;p15). Br J Haematol. 107:600–604. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ahuja HG, Popplewell L, Tcheurekdjian L
and Slovak ML: NUP98 gene rearrangements and the clonal evolution
of chronic myelo-genous leukemia. Genes Chromosomes Cancer.
30:410–415. 2001. View Article : Google Scholar : PubMed/NCBI
|