Introduction
Colorectal cancer (CRC) is a common malignant tumor
that occurs worldwide. According to statistics, both the incidence
and mortality of CRC rank third among all forms of cancer in the
United States, even though the mortality rates have declined. The
vast majority of cases (90%) occur in individuals over 50 years of
age (1). Apart from hereditary CRC,
the development of this cancer type is poorly understood (2,3). Both
germline and somatic genetic variations have been suggested as
contributing factors in CRC development (4–6).
MicroRNA (miRNA) refers to a group of small
non-coding RNAs that are ~22 (18–25)
nucleotides in length and which regulate RNA expression at the
translational level (7–9). miRNAs have been associated with a
variety of diseases, including different forms of cancer.
Increasing evidence has confirmed the importance of miRNAs in
regulating common biological characteristics/processes of different
tumors, such as: self-growth signals, insensitivity to anti-growth
signals, abnormal apoptosis, unlimited replication potential,
sustained induction of angiogenesis, invasion, and metastasis
(10). Many researchers have
identified tumor-specific miRNA signatures which accurately
distinguish malignant tumors from several different parts of the
benign tissue, and their results showed that some miRNAs are
carcinogenic depending on other genetic mutations present in tumors
(11). miRNA species can directly
affect cell proliferation and apoptosis of tumor cell lines.
Furthermore, many studies have confirmed the link between abnormal
miRNA expression and abnormalities in intracellular signal
transduction pathways and tumorigenesis (12–16). For
example, miR-9 is activated by YC/MYCN, which induces cancer
metastasis by regulating the expression of metastasis suppressor
protein, E-cadherin (17), and
miR-449a can cause retinoblastoma (Rb)-dependent cell cycle arrest
and cellular senescence of prostate cancer (18).
miRNAs constitute a new class of molecules that
through interaction with oncogenes and/or tumor suppressor genes,
can promote the formation of cancer (19). However, different miRNAs regulate
different signaling pathways and different target proteins/genes
that affect the biological changes characteristic of cancer.
Previous findings showed that human miRNAs with
single-nucleotide polymorphisms (SNPs) can be targets of genetic
variation in human genomic DNA sequences and may be related to
susceptibility of human disease (20). miRNA SNPs have inter-individual
differences in disease diagnosis, treatment, and prognosis. Recent
large-scale studies reported significant risks associated with
different germline variations for development of CRC (5). In addition, novel studies suggest a
potential influence of miRNA SNPs on the risk of cancer
development.
Therefore, the aim of the present study was to
investigate the correlation between miRNA gene polymorphisms and
the susceptibility to CRC by reviewing the literature.
Materials and methods
Screening and identification of
relevant studies
We identified eligible and relevant studies by
performing searches with the terms ‘miRNA/microRNA’, ‘colorectal
cancer’, ‘genotype’, ‘polymorphism’, and ‘variant’ in the PubMed,
Ovid, and Embase databases, as well as the Cochrane Library. The
search was limited to English language studies and only published
studies with full text were included. We evaluated potentially
relevant publications by manually examining their titles and
abstracts. The selected studies in our meta-analysis also matched
the following inclusion criteria: i) Assessment of miRNA
polymorphisms and the risks of suffering from CRC; ii) an
individual case-control study in humans; iii) statistically
sufficient genotype data by odds ratio (OR) values and 95%
confidence intervals (CI); and iv) full-text search. Exclusion
criteria were: i) Lack of controlled studies; ii) repeat of
previous literature; iii) summaries, comments, reviews and
editorials; and iv) focus on benign CRC tumors.
Data extraction and study characteristics. Two
researchers independently extracted the data that met the above
inclusion criteria and differences were resolved through
discussion. For each study, the following information was
extracted: Last name of first author, year of publication,
ethnicity of patients in different studies, miRNA type, SNP ID,
source of research, genotyping method, number of cases and
controls, number of various genotypes among cases and controls, and
the Hardy-Weinberg equilibrium of control subjects. If a study did
not provide complete data, we sent a request to the corresponding
author for the data. A total of 15 eligible studies met the
inclusion criteria (Table I).
 | Table I.Characteristics of the 15 studies
included in the meta-analysis. |
Table I.
Characteristics of the 15 studies
included in the meta-analysis.
| miRNA | SNP ID | Author (Refs.) | Year | Ethnicity | Study design | Genotyping
method | Allele | Case no. | Control no. | HWE (P-value) | Jadad |
|---|
| miRNA-146a | rs2910164 | Min et al
(24) | 2012 | Asian | PB | PCR-RFLP | C/G | 446 | 502 | 0.44 | 7 |
|
|
| Hezova et al
(25) | 2012 | Caucasian | HB | TaqMan | C/G | 197 | 212 | 0.41 | 7 |
|
|
| Ma et al
(26) | 2013 | Asian | HB | TaqMan | C/G | 1,147 | 1,203 | 0.075 | 7 |
|
|
| Lv et al
(27) | 2013 | Asian | HB | PCR-RFLP | C/G | 353 | 540 | 0.08 | 7 |
|
|
| Vinci et al
(28) | 2013 | Caucasian | PB | HRM | C/G | 160 | 178 | 0.11 | 6 |
|
|
| Hu et al
(29) | 2014 | Asian | HB | PCR-RFLP | C/G | 276 | 373 | 0.14 | 7 |
|
|
| Parlayan et
al (30) | 2014 | Asian | HB | TaqMan | C/G | 116 | 524 | 0.75 | 7 |
| miRNA-149 | rs2292832 | Min et al
(24) | 2012 | Asian | PB | PCR-RFLP | C/T | 446 | 502 | 0.17 | 7 |
|
|
| Lv et al
(27) | 2013 | Asian | HB | PCR-RFLP | C/T | 353 | 540 | <0.05 | 7 |
|
|
| Vinci et al
(28) | 2013 | Caucasian | PB | HRM | C/T | 160 | 178 | 0.91 | 6 |
| miRNA-196a2 | rs11614913 | Min et al
(24) | 2012 | Asian | PB | PCR-RFLP | T/C | 446 | 502 | 0.63 | 7 |
|
|
| Hezova et al
(25) | 2012 | Caucasian | HB | TaqMan | T/C | 197 | 212 | 0.81 | 7 |
|
|
| Lv et al
(27) | 2013 | Asian | PB | PCR-RFLP | T/C | 353 | 540 | <0.05 | 7 |
|
|
| Vinci et al
(28) | 2013 | Caucasian | PB | HRM | T/C | 160 | 178 | 0.09 | 6 |
|
|
| Parlayan et
al (30) | 2014 | Asian | HB | TaqMan | T/C | 116 | 524 | 0.78 | 7 |
|
|
| Zhu et al
(31) | 2012 | Asian | HB | TaqMan | T/C | 573 | 588 | 0.17 | 7 |
|
|
| Zhan et al
(32) | 2011 | Asian | HB | PCR-RFLP | T/C | 252 | 543 | 0.77 | 7 |
|
|
| Chen et al
(33) | 2012 | Asian | HB | PCR-LDR | T/C | 126 | 407 | 0.82 | 7 |
| miRNA-27a | rs895819 | Hezova et al
(25) | 2012 | Caucasian | HB | TaqMan | G/A | 197 | 212 | 0.78 | 7 |
|
|
| Wang et al
(34) | 2014 | Asian | HB | TaqMan | G/A | 205 | 455 | <0.05 | 7 |
| miRNA-34b/c | rs4938723 | Gao et al
(35) | 2013 | Asian | HB | PCR-RFLP | T/C | 347 | 488 | 0.83 | 7 |
|
|
| Oh et al
(36) | 2014 | Asian | PB | PCR-RFLP | T/C | 545 | 428 | 0.4 | 7 |
| miRNA-let-7 | rs712 | Pan et al
(37) | 2014 | Asian | HB | PCR-RFLP | G/T | 339 | 313 | 0.41 | 7 |
| miRNA-603 | rs11014002 | Wang et al
(38) | 2014 | Asian | HB | Sequenom mass | C/T | 102 | 204 | 0.59 | 7 |
| miRNA-608 | rs4919510 | Ryan et al
(39) | 2012 | Caucasian | PB | TaqMan | C/G | 245 | 446 | 0.94 | 7 |
Quality assessment
Two researchers evaluated the uniform quality of
studies which met the inclusion criteria, and cross-checked in case
of disagreement, which was eventually resolved by discussion. The
quality of the included studies was assessed using a modified Jadad
score (21), where 1–3 points was
considered a low quality study, and 4–7 points was considered
high-quality research. Evaluation included whether to generate a
random sequence by hiding the proper randomized allocation; whether
the study was blinded, and whether the patients were lost or quit
the study (Table I).
Statistical analysis
According to the genotype frequencies of cases and
controls, the correlation between miRNA polymorphisms and risk of
CRC was assessed via OR values with 95% CI. We performed
statistical analyses of OR values and 95% CI by assessing five
different genetic parameters: The allele, the dominant genetic
model, the recessive genetic model, the homozygous comparison, and
the heterozygous comparison. The Chi-square based Q statistic was
used to assess heterogeneity between studies and P<0.05 was
considered to indicate significant heterogeneity between studies.
The I2 index was expressed as a percentage for the total
variability throughout the study. I2 value of 25, 50 and
75% indicated low, medium, and high heterogeneity, respectively.
Funnel plots were used to assess publication bias. When the effects
were assumed to be homogeneous, the fixed-effects model was used
(Mantel-Haenszel method). If heterogeneity was present, the
random-effects model was applied (DerSimonian and Laird method) to
account for inter-study heterogeneity instead of the fixed-effect
model. Data were analyzed with Stata 11.0 software (StataCorp,
College Station, USA) and all P-values were two-sided tests
(22,23).
Results
Study characteristics
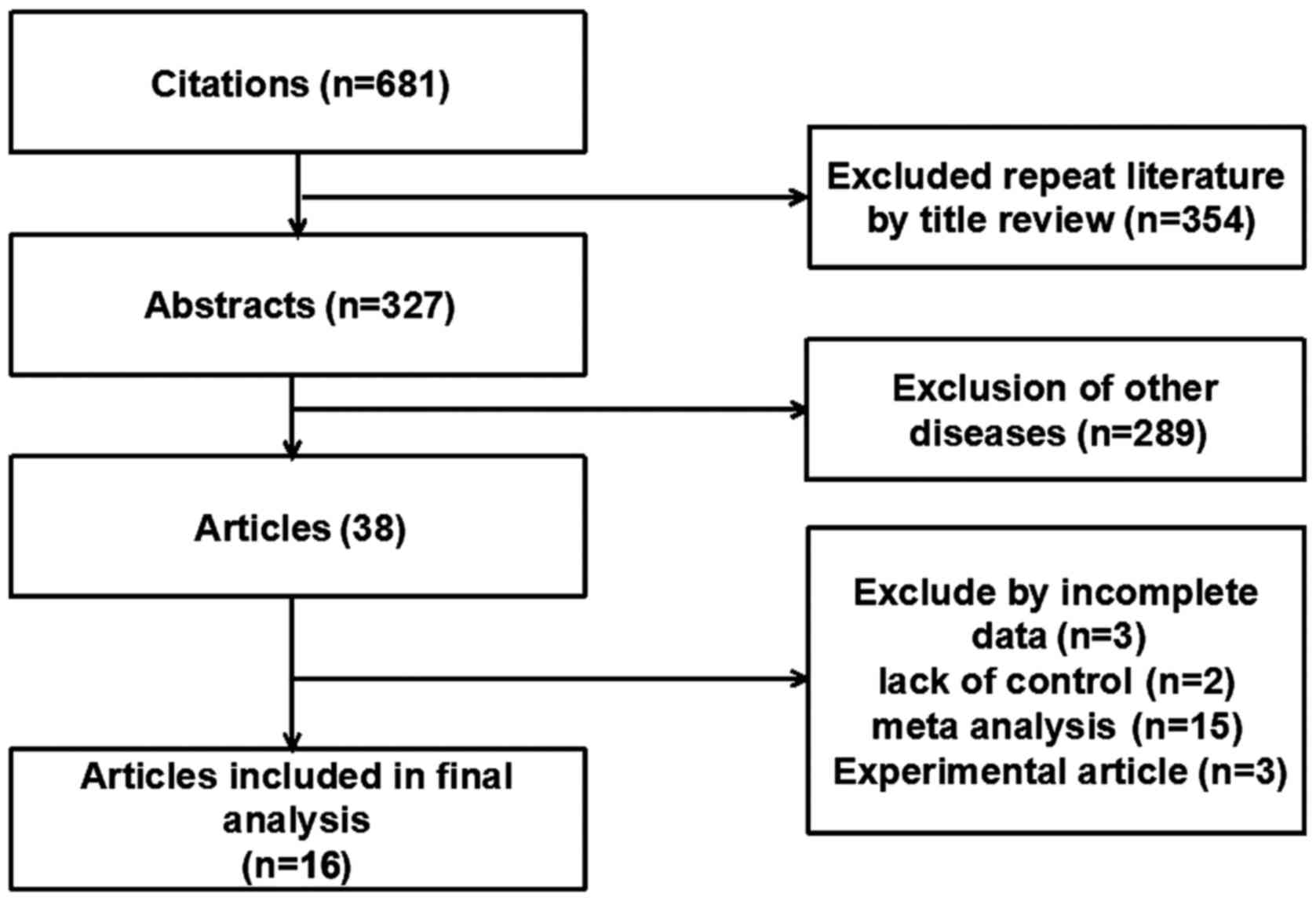
A total of 681 studies relevant to the search words
were identified and only 38 studies were on the association between
CRC and miRNA polymorphisms. According to the aforementioned
inclusion and exclusion criteria, 16 publications (four using
population-based controls and 11 using hospital-based controls)
were included in the final meta-analysis (24–39)
(Fig. 1). Of the 15 articles, seven
related to miR-146a; three were on miRNA-149; eight were on
miRNA-196a2; there were two each related to miRNA-27a and
miRNA-34b/c; and there was one each on miRNA-let-7, miRNA-603, and
miRNA-608. The main characteristics of the studies included in the
meta-analysis are summarized in Table
I.
Overall analyses
Table II shows that
the overall analysis of all the studies revealed a statistically
significant positive association between miRNA-let-7, miR-34b/c,
miR-146a, and miR-149 polymorphisms and the risk of CRC. However,
miR-196a2 (31–33), miR-27 (34), miR-603 (38) and miR-608 (39) did not correlate with the risk of CRC.
Only significantly different results follow.
 | Table II.Analysis of the association between 8
miRNA gene polymorphism and risk of CRC. |
Table II.
Analysis of the association between 8
miRNA gene polymorphism and risk of CRC.
| miRNA | Contrast | Na |
P-valueb | OR (95% CI) | I2
(%) |
P-valuec |
P-valued |
|---|
| miRNA-146a |
| 7 |
|
|
|
|
|
|
| G/C |
| 0.02 | 1.09
(1.01–1.01) | 26.4 | 0.227 | 0.036 |
|
| GC+GG/CC |
| 0.068 | 1.13
(0.99–1.99) | 75.1 | 0.001 | 0.527 |
|
| GG/CC+GC |
| 0.047 | 1.13
(1.00–1.00) | 28.9 | 0.207 | 0.155 |
|
| GG/CC |
| 0.042 | 1.19
(1.01–1.01) | 25.2 | 0.237 | 0.141 |
|
| GC/CC |
| 0.191 | 1.09
(0.94–1.94) | 79.8 | <0.001 | 0.646 |
| miRNA-149 |
| 3 |
|
| T/C |
| 0.082 | 1.13
(0.96–1.96) | 0 | 0.533 | 0.805 |
|
| TT+CT/CC |
| 0.871 | 1.02
(0.79–1.79) | 0 | 0.655 | 0.06 |
|
| TT/TC+CC |
| 0.025 | 1.24
(1.03–1.03) | 0 | 0.582 | 0.332 |
|
| TT/CC |
| 0.241 | 1.19
(0.89–1.89) | 0 | 0.591 | 0.4 |
|
| TC/CC |
| 0.382 | 0.88
(0.67–1.67) | 0 | 0.89 | 0.191 |
| miRNA-196a2 |
| 8 |
|
| T/C |
| 0.747 | 1.01
(0.94–1.94) | 89.2 | <0.001 | 0.595 |
|
| TT+TC/CC |
| 0.656 | 1.03 (0.9–1.9) | 87.6 | <0.001 | 0.011 |
|
| TT/TC+CC |
| 0.898 | 1.01
(0.89–1.89) | 83 | <0.001 | 0.636 |
|
| TT/CC |
| 0.763 | 1.03
(0.87–1.87) | 90.2 | <0.001 | 0.061 |
|
| TC/CC |
| 0.49 | 1.05
(0.91–1.91) | 84.7 | <0.001 | 0.023 |
| miRNA-27a |
| 2 |
|
| G/A |
| 0.057 | 1.19
(0.99–1.99) | 66.4 | 0.085 | – |
|
| (GG+GA)/AA |
| 0.221 | 1.14 (0.9–1.9) | 48.1 | 0.165 | – |
|
| GG/(GA+AA) |
| 0.082 | 1.3
(0.97–1.97) | 3.3 | 0.309 | – |
|
| GG/AA |
| 0.071 | 1.38
(0.97–1.97) | 39 | 0.201 | – |
|
| GA/AA |
| 0.569 | 1.09
(0.81–1.81) | 0 | 0.408 | – |
| miRNA-34b/c |
| 2 |
|
| T/C |
| 0.058 | 1.15 (1–1.33) | 50.9 | 0.154 | – |
|
| TT+TC/CC |
| 0.016 | 1.49
(1.08–2.08) | 0 | 0.519 | – |
|
| TT/(TC+CC) |
| 0.288 | 1.11
(0.92–1.92) | 50 | 0.157 | – |
|
| TT/CC |
| 0.015 | 1.52
(1.08–2.08) | 0 | 0.342 | – |
|
| TC/CC |
| 0.032 | 1.46
(1.03–2.03) | 0 | 0.812 | – |
| miRNA-let-7 |
| 1 |
|
| T/G |
| 0.003 | 1.49
(1.15–1.15) | – | – | – |
|
| TT+TG/GG |
| 0.015 | 1.48
(1.08–2.08) | – | – | – |
|
| TT/TG+GG |
| 0.015 | 2.52
(1.19–5.19) | – | – | – |
|
| TT/GG |
| 0.007 | 2.81
(1.32–5.32) | – | – | – |
|
| TG/GG |
| 0.074 | 1.35
(0.97–1.97) | – | – | – |
| miRNA-603 |
| 1 |
|
| C/T |
| 0.154 | 0.74
(0.50–1.50) | – | – | – |
|
| CC+CT/TT |
| 0.517 | 1.05
(0.59–1.59) | – | – | – |
|
| CC/CT+TT |
| 0.152 | 0.94
(0.69–1.69) | – | – | – |
|
| CC/TT |
| 0.398 | 1.02
(0.56–1.56) | – | – | – |
|
| CT/TT |
| 0.778 | 1.10
(1.60–2.60) | – | – | – |
| miRNA-608 |
| 1 |
|
| C/G |
| 0.829 | 0.97
(0.76–1.76) | – | – | – |
|
| CG+CC/GG |
| 0.869 | 1.05
(0.59–1.59) | – | – | – |
|
| CC/CG+GG |
| 0.716 | 0.94
(0.69–1.69) | – | – | – |
|
| CC/GG |
| 0.956 | 1.02
(0.56–1.56) | – | – | – |
|
| CG/GG |
| 0.769 | 1.10
(0.59–2.59) | – | – | – |
Analysis of miR-146a polymorphisms and
susceptibility to CRC
i) Seven studies (24–30)
reported the association between G/C alleles and susceptibility to
CRC, there was no significant heterogeneity between studies
(I2=26.4%, P=0.227), and the fixed-effect model was
used. The total analysis showed that individuals with the G allele
are more susceptible to CRC than with C (OR=1.09, 95% CI=1.01–1.18,
P=0.02). ii) Seven studies (24–30)
reported the association between the recessive genetic model,
GG/(CC+GC), and susceptibility to CRC, there was no significant
heterogeneity between studies (I2=28.9%, P=0.207), and
the fixed-effect model was used. The total analysis showed that
people with the recessive genetic model, GG, are more susceptible
to CRC than (CC+GC) (OR=1.13, 95% CI=1.00–1.28, P=0.047). iii)
Seven studies (24–30) reported the association between
homozygous GG/CC and the susceptibility to CRC, there was no
significant heterogeneity between studies (I2=25.2%,
P=0.237), and the fixed-effect model was used. The total analysis
showed that people with homozygous GG are more susceptible to CRC
than CC (OR=1.19, 95% CI=1.01–1.41, P=0.042).
Analysis of miRNA-149 polymorphisms
and susceptibility to CRC
Five studies (24–28)
reported the association between the recessive genetic model,
TT/(TC+CC), and susceptibility to CRC, there was no significant
heterogeneity between studies (I2=0, P=0.582), and the
fixed-effect model was used. The total analysis showed that people
with the recessive genetic model, TT, are more susceptible to CRC
than (TC+CC) (OR=1.24, 95% CI=1.03–1.5, P=0.025).
Analysis of miRNA-34b/c polymorphisms
and susceptibility to CRC
i) Two studies (35,36)
reported the association between the dominant genetic model,
TT+TC/CC, and susceptibility to CRC, there was no significant
heterogeneity between studies (I2=0, P=0.519), and the
fixed-effect model was used. The total analysis showed that people
with the dominant model, TT+TC, are more susceptible to CRC than CC
(OR=1.49, 95% CI=1.08–2.06, P=0.016). ii) Two studies (34,35)
reported the association between the homozygous model, TT/CC, and
susceptibility to CRC, there was no significant heterogeneity
between studies (I2=0, P=0.342), and the fixed-effect
model was used. The total analysis showed that people with the
homozygous model, TT, are more susceptible to CRC than CC (OR=1.52,
95% CI=1.08–2.13, P=0.015). iii) Two studies (34,35)
reported the association between the heterozygous model, TC/CC, and
susceptibility to CRC, there was no significant heterogeneity
between studies (I2=0, P=0.812), and the fixed-effect
model was used. The total analysis showed that people with the
heterozygous model, TC, are more susceptible to CRC than CC
(OR=1.46, 95% CI=1.03–2.05, P=0.032).
Analysis of miRNA-let-7 polymorphisms
and susceptibility to CRC
i) One study (37)
reported the association between the allele, T/G, and
susceptibility to CRC. The analysis showed that people with allele
T are more susceptible to CRC than G (OR=1.49, 95% CI=1.15–1.94,
P=0.003). ii) One study (37)
reported the association between the dominant genetic model,
TT+TG/GG, and susceptibility to CRC. The analysis showed that
people with the dominant genetic model, TT+TG, are more susceptible
to CRC than GG (OR=1.48, 95% CI=1.08–2.03, P=0.015). iii) One study
(37) reported the association
between the recessive genetic model, TT/GG+TG, and susceptibility
to CRC. The analysis showed that people with the recessive genetic
model, TT, are more susceptible to CRC than GG+TG (OR=2.52, 95%
CI=1.19–5.31, P=0.015). iv) One study (37) reported the association between the
homozygous model, TT/GG, and susceptibility to CRC. The analysis
showed that people with the homozygous model, TT, are more
susceptible to CRC than GG (OR=2.81, 95% CI=1.32–5.98,
P=0.007).
These results indicate that miRNA-let-7, miR-34b/c,
miR-146a, and miR-149 polymorphisms were positively correlated with
CRC.
Statistical sensitivity
Statistical sensitivity where one study was removed
and the rest were analyzed, the pooled relative risks (RRs) were
similar with the overall pooled RRs (data not shown), supporting
the robustness of our results.
Publication bias
Egger's test was used to assess the publication bias
of the studies (Table II). The
results show some evidence of publication bias in some
comparisons.
Discussion
At present, miRNA gene polymorphisms and
susceptibility to a variety of tumors have been reported. De Ruyck
et al found that miRNA-let-7 polymorphisms in the KRAS
3′-UTR are a prognostic factor of oropharyngeal cancer (40). Another study showed that
miR-146aG>C and miR-196a2C>T polymorphisms were associated
with the risk of hepatocellular carcinoma (HCC) in patients in
China, especially in patients with hepatitis B virus infection
(41). Palmieri et al found
that miR-146a polymorphisms are not associated with tumor
development in oral squamous cell carcinoma. However, a slight
increase in the frequency of the variant allele was observed in
stage II tumors (42).
There are also several meta-analyses that
statistically correlated with miRNAs and cancer risk. Some
meta-analysis studies evaluated the correlation between one miRNA
and a single cancer susceptibility. Wang et al confirmed the
association of the polymorphism, miR-196a2 rs11614913, with the
risk of CRC, but not with tumor stage and grade (43). Some meta-analysis studies evaluated
the correlation between one miRNA and a variety of cancer
susceptibilities. Tao et al suggested that the hsa-miR-34b/c
rs4938723 polymorphism may play opposite roles in different types
of cancer based on the present studies, and a subgroup analysis
revealed that the variant CT genotypes were associated with an
increased risk of HCC compared with the wild-type TT genotype.
However, a decreased risk of CRC was found in the genetic model of
CC/TT and CC/CT+TT (44). Li et
al reported that the miR-146a rs2910164 polymorphism may
decrease the susceptibility of digestive system cancers, especially
in the Asian population (45). Some
meta-analysis studies evaluated the correlation between a variety
of miRNAs and a variety of cancer susceptibilities. For example,
one meta-analysis provided evidence that the miR-196a2 rs11614913
polymorphism is associated with an increased cancer risk and
rs2910164 in miR-146a may be associated with susceptibility to
papillary thyroid carcinoma and cervical cancer (46). In addition, some studies analyzed the
correlation between a variety of miRNAs and a specific cancer
susceptibility. Dikeakos et al investigated the association
of the miR-146aC>G, miR-149T>C, and miR-196a2T>C
polymorphisms with the risk of gastric cancer and survival in the
Greek population (47). They found
that the risk of gastric cancer was significantly higher in
carriers of miR-149 rs2292832CC and miR-196a2 rs11614913CC
genotypes, as well as for carriers of the
rs2910164/rs2292832/rs11614913 CCC and GTC haplotype. The
rs2910164/rs2292832/rs11614913 CTT and CCT haplotypes appear to
have a protective role against the development of gastric cancer.
Their data demonstrate that specific miRNA SNPs are associated with
gastric cancer susceptibility in the Greek population (47).
In the present study, we performed statistical
analyses of the relationships between miRNA polymorphisms and CRC.
We investigated the effects of miRNA-196a2, miRNA-146a, miRNA-27a,
miRNA-34b/c, miRNA-let-7, miRNA-603, miRNA-608 and miRNA-149 on CRC
susceptibility. This was a more comprehensive statistical analysis
of the various miRNAs that affect the risk of CRC. However,
regardless of whether the aforementioned miRNA and risk of CRC risk
are highly relevant, they are combined with other factors such as
Helicobacter pylori (48),
smoking (49,50), age (49,50), and
drugs, which can lead to increased cancer risk. Therefore,
stratified analysis is necessary. In addition, the limitation of
this study is that most enrolled studies involved Asian poplations,
and the results may not be generalized to the global population.
Therefore, we need a larger number of samples for statistical
analysis, and more data on the relationship between miRNA gene
polymorphisms and CRC among non-Asian populations, to make more
reliable conclusions.
In conclusion, we statistically analyzed the
relationship between miRNA gene polymorphisms and CRC prevalence
through a meta-analysis of several publications. miRNA-let-7,
miR-34b/c, miR-146a, miR-603, and miR-149 gene polymorphisms can
significantly increase the risk of CRC, while miR-192a and miR-27a
polymorphisms are not related to the risk of CRC. Analyses of a
larger number of samples are required.
Acknowledgements
This study was partially supported by the National
Natural Science Foundation of China (grant nos. 81672970 and
81301933), Health Research Projects in Jiangsu Province (H201313),
the projects of Suzhou Technology Bureau (SYS201552), the focus of
clinical disease treatment technology special funds of Suzhou City
(LCZX201505), and the Second Affiliated Hospital of Soochow
University Preponderant Clinic Discipline Group Project
funding.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferrari P, Jenab M, Norat T, Moskal A,
Slimani N, Olsen A, Tjønneland A, Overvad K, Jensen MK,
Boutron-Ruault MC, et al: Lifetime and baseline alcohol intake and
risk of colon and rectal cancers in the European prospective
investigation into cancer and nutrition (EPIC). Int J Cancer.
121:2065–2072. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Corté H, Manceau G, Blons H and
Laurent-Puig P: MicroRNA and colorectal cancer. Dig Liver Dis.
44:195–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peters U, Jiao S, Schumacher FR, Hutter
CM, Aragaki AK, Baron JA, Berndt SI, Bézieau S, Brenner H,
Butterbach K, et al: Colon Cancer Family Registry and the Genetics
and Epidemiology of Colorectal Cancer Consortium: Identification of
genetic susceptibility loci for colorectal tumors in a genome-wide
meta-analysis. Gastroenterology. 144:799–807.e724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greenman C, Stephens P, Smith R, Dalgliesh
GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C,
et al: Patterns of somatic mutation in human cancer genomes.
Nature. 446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kusenda B, Mraz M, Mayer J and Pospisilova
S: MicroRNA biogenesis, functionality and cancer relevance. Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub. 150:205–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santarpia L, Nicoloso M and Calin GA:
MicroRNAs: a complex regulatory network drives the acquisition of
malignant cell phenotype. Endocr Relat Cancer. 17:F51–F75. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattie MD, Benz CC, Bowers J, Sensinger K,
Wong L, Scott GK, Fedele V, Ginzinger D, Getts R and Haqq C:
Optimized high-throughput microRNA expression profiling provides
novel biomarker assessment of clinical prostate and breast cancer
biopsies. Mol Cancer. 5:242006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grady WM, Parkin RK, Mitchell PS, Lee JH,
Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz
AM, et al: Epigenetic silencing of the intronic microRNA
hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene.
27:3880–3888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lanza G, Ferracin M, Gafà R, Veronese A,
Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM and Negrini M:
mRNA/microRNA gene expression profile in microsatellite unstable
colorectal cancer. Mol Cancer. 6:542007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gaur A, Jewell DA, Liang Y, Ridzon D,
Moore JH, Chen C, Ambros VR and Israel MA: Characterization of
microRNA expression levels and their biological correlates in human
cancer cell lines. Cancer Res. 67:2456–2468. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saito Y, Suzuki H and Hibi T: The role of
microRNAs in gastrointestinal cancers. J Gastroenterol. 44:(Suppl
19). 18–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
18
|
Noonan EJ, Place RF, Basak S, Pookot D and
Li LC: miR-449a causes Rb-dependent cell cycle arrest and
senescence in prostate cancer cells. Oncotarget. 1:349–358.
2010.PubMed/NCBI
|
|
19
|
Panarelli NC and Yantiss RK: Microrna
expression in selected carcinomas of the gastrointestinal tract.
Pathol Res Int. 2011:1246082011. View Article : Google Scholar
|
|
20
|
Wu D, Yang G, Zhang L, Xue J, Wen Z and Li
M: Genome-wide association study combined with biological context
can reveal more disease-related SNPs altering microRNA target seed
sites. BMC Genomics. 15:6692014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jadad AR, Moore RA, Carroll D, Jenkinson
C, Reynolds DJ, Gavaghan DJ and McQuay HJ: Assessing the quality of
reports of randomized clinical trials: is blinding necessary?
Control Clin Trials. 17:1–12. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Bi J, Liu X, Li K, Di J and Wang
B: Has-miR-146a polymorphism (rs2910164) and cancer risk: a
meta-analysis of 19 case-control studies. Mol Biol Rep.
39:4571–4579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu X, Yang X, Ru G, Wu Y, Zhang S, Xing C,
Wu Y and Cao J: miR-146a gene polymorphism rs2910164 and the risk
of digestive tumors: a meta-analysis of 21 case-control studies.
Oncol Rep. 31:472–479. 2014.PubMed/NCBI
|
|
24
|
Min KT, Kim JW, Jeon YJ, Jang MJ, Chong
SY, Oh D and Kim NK: Association of the miR-146aC>G, 149C>T,
196a2C>T, and 499A>G polymorphisms with colorectal cancer in
the Korean population. Mol Carcinog. 51:(Suppl 1). E65–E73. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hezova R, Kovarikova A, Bienertova-Vasku
J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I,
Vyzula R, et al: Evaluation of SNPs in miR-196-a2, miR-27a and
miR-146a as risk factors of colorectal cancer. World J
Gastroenterol. 18:2827–2831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma L, Zhu L, Gu D, Chu H, Tong N, Chen J,
Zhang Z and Wang M: A genetic variant in miR-146a modifies
colorectal cancer susceptibility in a Chinese population. Arch
Toxicol. 87:825–833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lv M, Dong W, Li L and Zhang L, Su X, Wang
L, Gao L and Zhang L: Association between genetic variants in
pre-miRNA and colorectal cancer risk in a Chinese population. J
Cancer Res Clin Oncol. 139:1405–1410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vinci S, Gelmini S, Mancini I, Malentacchi
F, Pazzagli M, Beltrami C, Pinzani P and Orlando C: Genetic and
epigenetic factors in regulation of microRNA in colorectal cancers.
Methods. 59:138–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu X, Li L, Shang M, Zhou J, Song X, Lu X,
Wang J, Ying B and Wang L: Association between microRNA genetic
variants and susceptibility to colorectal cancer in Chinese
population. Tumour Biol. 35:2151–2156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Parlayan C, Ikeda S, Sato N, Sawabe M,
Muramatsu M and Arai T: Association analysis of single nucleotide
polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer
in elderly Japanese: a case-control study. Asian Pac J Cancer Prev.
15:2101–2107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu L, Chu H, Gu D, Ma L, Shi D, Zhong D,
Tong N, Zhang Z and Wang M: A functional polymorphism in
miRNA-196a2 is associated with colorectal cancer risk in a Chinese
population. DNA Cell Biol. 31:350–354. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie
GZ, Sun AM and Liu Y: A functional variant in microRNA-196a2 is
associated with susceptibility of colorectal cancer in a Chinese
population. Arch Med Res. 42:144–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen H, Sun LY, Chen LL, Zheng HQ and
Zhang QF: A variant in microRNA-196a2 is not associated with
susceptibility to and progression of colorectal cancer in Chinese.
Intern Med J. 42:e115–e119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Z, Sun X, Wang Y, Liu X, Xuan Y and
Hu S: Association between miR-27a genetic variants and
susceptibility to colorectal cancer. Diagn Pathol. 9:1462014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao LB, Li LJ, Pan XM, Li ZH, Liang WB,
Bai P, Zhu YH and Zhang L: A genetic variant in the promoter region
of miR-34b/c is associated with a reduced risk of colorectal
cancer. Biol Chem. 394:415–420. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oh J, Kim JW, Lee BE, Jang MJ, Chong SY,
Park PW, Hwang SG, Oh D and Kim NK: Polymorphisms of the
pri-miR-34b/c promoter and TP53 codon 72 are associated with risk
of colorectal cancer. Oncol Rep. 31:995–1002. 2014.PubMed/NCBI
|
|
37
|
Pan XM, Sun RF, Li ZH, Guo XM, Zhang Z,
Qin HJ, Xu GH and Gao LB: A let-7 KRAS rs712 polymorphism increases
colorectal cancer risk. Tumour Biol. 35:831–835. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang FJ, Ding Y, Mao YY, Jing FY, Zhang
ZY, Jiang LF, Guo JF, Sun XJ, Jin MJ and Chen K: Associations
between hsa-miR-603 polymorphism, lifestyle-related factors and
colorectal cancer risk. Cancer Biomark. 14:225–231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ryan BM, McClary AC, Valeri N, Robinson D,
Paone A, Bowman ED, Robles AI, Croce C and Harris CC: rs4919510 in
hsa-mir-608 is associated with outcome but not risk of colorectal
cancer. PLoS One. 7:e363062012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Ruyck K, Duprez F, Ferdinande L, Mbah
C, Rios-Velazquez E, Hoebers F, Praet M, Deron P, Bonte K, Speel
EJ, et al: A let-7 microRNA polymorphism in the KRAS 3′-UTR is
prognostic in oropharyngeal cancer. Cancer Epidemiol. 38:591–598.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou B, Dong LP, Jing XY, Li JS, Yang SJ,
Wang JP and Zhao LF: Association between miR-146aG>C and
miR-196a2C>T polymorphisms and the risk of hepatocellular
carcinoma in a Chinese population. Tumour Biol. 35:7775–7780. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Palmieri A, Carinci F, Martinelli M,
Pezzetti F, Girardi A, Cura F, Rubini C and Scapoli L: Role of the
MIR146A polymorphism in the origin and progression of oral squamous
cell carcinoma. Eur J Oral Sci. 122:198–201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang N, Li Y, Zhu LJ, Zhou RM, Jin W, Guo
XQ, Wang CM, Chen ZF and Liu W: A functional polymorphism
rs11614913 in microRNA-196a2 is associated with an increased risk
of colorectal cancer although not with tumor stage and grade.
Biomed Rep. 1:737–742. 2013.PubMed/NCBI
|
|
44
|
Tao T, Chen S, Xu B, Liu C, Wang Y, Huang
Y and Chen M: Association between hsa-miR-34b/c rs4938723 T > C
promoter polymorphism and cancer risk: a meta-analysis based on
6,036 cases and 6,204 controls. Chin J Cancer Res. 26:315–322.
2014.PubMed/NCBI
|
|
45
|
Li YJ, Zhang ZY, Mao YY, Jin MJ, Jing FY,
Ye ZH and Chen K: A genetic variant in MiR-146a modifies digestive
system cancer risk: a meta-analysis. Asian Pac J Cancer Prev.
15:145–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Wang Q, Liu H, Shao N, Tan B,
Zhang G, Wang K, Jia Y, Ma W, Wang N, et al: The association of
miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with
cancer risk: a meta-analysis of 32 studies. Mutagenesis.
27:779–788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dikeakos P, Theodoropoulos G, Rizos S,
Tzanakis N, Zografos G and Gazouli M: Association of the
miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms
with gastric cancer risk and survival in the Greek population. Mol
Biol Rep. 41:1075–1080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song MY, Su HJ, Zhang L, Ma JL, Li JY, Pan
KF and You WC: Genetic polymorphisms of miR-146a and miR-27a, H.
pylori infection, and risk of gastric lesions in a Chinese
population. PLoS One. 8:e612502013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zeng Y, Sun QM, Liu NN, Dong GH, Chen J,
Yang L and Wang B: Correlation between pre-miR-146a C/G
polymorphism and gastric cancer risk in Chinese population. World J
Gastroenterol. 16:3578–3583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou F, Zhu H, Luo D, Wang M, Dong X, Hong
Y, Lu B, Zhou Y, Zhou J, Zhang Z, et al: A functional polymorphism
in Pre-miR-146a is associated with susceptibility to gastric cancer
in a Chinese population. DNA Cell Biol. 31:1290–1295. 2012.
View Article : Google Scholar : PubMed/NCBI
|