Introduction
In women, breast cancer is the most common cancer
type, accounting for 22.9% of all cancer cases. In China, the
incidence of breast cancer has been on the rise (1). The disease is the leading cause of
mortality in patients with distant metastasis, and
chemoradiotherapy for advanced breast cancer is the most effective
treatment method (2,3). As chemotherapy is extremely important
for the treatment of breast cancer, multidrug resistance (MDR) is
likely to be a major obstacle (4).
MDR is a complex process involving multiple mechanisms, including
the overexpression of energy-dependent transporters, which shuttle
anticancer drugs into and out of cells (5,6).
Cytokine-induced apoptosis inhibitor 1 (CIAPIN1) is
a newly identified anti-apoptotic protein; it shares no homology
with Bcl-2, caspase, the IAP family or signal-transduction
molecules that regulate apoptosis (7). CIAPIN1 is associated with MDR in
SGC7901/Adr gastric cancer cells (8,9) and
HL-60/Adr leukemia cells (10), in
which CIAPIN1 upregulates P-glycoprotein (MDR1) expression
(8–10), leading to drug resistance. In a study
examining MCF-7/ADM breast cancer cells, the same MDR mechanism was
reported (11). When RNA interference
(RNAi) was used to downregulate the expression of CIAPIN1 in
MCF-7/ADM breast cancer cells, a significant reduction in drug
resistance was observed and MDR1 expression was inhibited (11). Therefore, we hypothesize that the
CIAPIN1 gene is a putative target for the treatment of
breast cancer MDR (11,12).
Over the last 10 years, a novel class of small RNA
molecules known as microRNAs (miRNAs/miRs) has been recognized as
an important regulator of the initiation and progression of human
cancer types, including breast cancer (13,14).
miR-143 is generally considered to be a tumor suppressor in breast
cancer (15,16), however, the mechanisms by which it is
downregulated here are not yet understood. Although dysregulation
of CIAPIN1 and miR-143 is associated with tumorigenesis in
human breast cancer, little is known about how miR-143 acts on
CIAPIN1. The current study examined whether there are direct
interactions between CIAPIN1, miR-143 and the reversal of
MDR in breast cancer in vitro.
Materials and methods
Cell lines
Human breast carcinoma MCF-7, MDA-MB-231, and
MDA-MB-453 cell lines, as well as HEK-293 cells, were purchased
from the Shanghai Institute of Cell Biology, Chinese Academy of
Sciences (Shanghai, China). The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a 5% CO2
incubator at 37°C, supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and antibiotics (100 µg/ml
streptomycin and 100 U/ml penicillin).
Detection of half maximal inhibitory concentration
(IC50) values of cells against Taxol. Each group of
cells (MCF7, MDA-MB-231 and MDA-MB-453), while in the logarithmic
phase, was seeded (~5,000 cells/well) in culture plates of 96 wells
at 37°C, with 5% CO2, for 24 h. The culture DMEM was
removed when the cells adhered to the plate wall. The cells were
then incubated in 100 µl of medium with Taxol (catalog no.
33069-62-4; Sigma) in different doses at 37°C. The initial
concentration of Taxol was 0.01 µg/ml, and the drug concentration
was increased by 1-fold after 3 passages of culture in
vitro. The concentration of Taxol was as follows: 0.01, 0.02,
0.04, 0.08, 0.16, 0.32, 0.64, 1.28, 2.56, 5.12, 10.24, 20.48 and
40.96 µg/ml. Optical density (OD) values of the cells were tested
using the CCK-8 method after 48 h of incubation. The cells were
then treated with 10 µl CCK-8 reagent for 4 h, and the CCK-8 Cell
Proliferation kit (Beyotime Institute of Biotechnology, Nantong,
China) was used to measure the cell viability according to the
manufacturer's instructions. The absorbance was measured at 450 nm.
The cell growth inhibition ratio = (1 - mean OD value of
experimental group/ mean OD value of control group) × 100. The dose
response curve was drawn in different concentrations to calculate
the IC50 using a probit regression model. Each
concentration, including 5 duplicate wells, was experimentally used
3 times independently to obtain a mean IC50 value.
Recombinant lentivirus generation and
lentivirus infection
Lentivirus Package plasmids mix (System Biosciences,
Palo Alto, CA, USA) and selected LV-miR-143 (GenePharma, Shanghai,
China) were cotransfected into the MCF-7, MDA-MB-231 and MDA-MB-453
Taxol-induced drug-resistant (TDR) breast cancer cells to package
and produce lentiviral vector and viral titer using the gradient
dilution method. Lentivirus (1×104 IFU/µl, 20 µl)
packaging of green fluorescent protein (GFP) was infected into the
MCF-7, MDA-MB-231 and MDA-MB-453 TDR cells at various volumes to
obtain the best MOI value corresponding to a concentration at which
no virus toxicity effect on the cells was observed. Subsequently,
viruses were added at an MOI of 20 to the experimental and control
groups, which were then incubated under standard conditions. At 96
h after infection, cells that had the strongest GFP brightness were
selected from the two wells and intermediate clone culturing was
performed in the experimental group.
Luciferase reporter assay
The cDNA sequence of CIAPIN1 (NM_020313.2)
was obtained from Genbank (https://www.ncbi.nlm.nih.gov/genbank/). The entire
3′-untranslated region (3′-UTR) of human CIAPIN1 was
amplified using polymerase chain reaction (PCR) with human genomic
DNA as a template. The PCR products were inserted into the
p-MIR-reporter plasmid (Ambion, Austin, TX, USA). The correct
insertion was confirmed by DNA sequencing. To test the binding
specificity, the sequences that interacted with the seed sequence
of miR-143 were mutated (from TCATCTCA to AATCCTCT for the miR-143
binding site), and the mutant (MT) CIAPIN1 3′-UTR was
inserted into an equivalent luciferase reporter plasmid. For
luciferase reporter assays, HEK-293 cells were seeded in 6-well
plates and co-transfected with 2 µg firefly luciferase reporter
plasmid (pGL, pGL3-promoter vector; E1761; Promega), 2 µg
β-galactosidase (β-gal) expression plasmid (Ambion), and equal
amounts (100 pmol) of miR-143 mimic, inhibitor or scrambled
negative control RNA (RiboBio, Guangzhou, China) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The β-gal plasmid was used as a transfection control. Cells were
harvested at 48 h post-transfection and analyzed for luciferase
activity using a luciferase assay kit (Promega, Madison, WI,
USA).
Western blot analysis
To perform protein analysis, cell lysates were
harvested and measured using western blotting, as previously
described (11). Protein sample (50
µg) was fractionated by 12% SDS-PAGE and transferred to
nitrocellulose membranes. The membranes were blocked in 5% skimmed
milk and PBS for 2 h and then incubated at 4°C overnight. The
following antibodies were used: Anti-CIAPIN1 (1:300 dilution;
catalog no. ab154904; Abcam, Cambridge, UK) and anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH) antibody (1:1,000 dilution;
catalog no. KC-5G4; Kangchen, Shanghai, China). GAPDH served as an
internal control. After washing, the membranes were incubated with
FITC-conjugated secondary antibody (1:5,000 dilution; catalog no.
ab99772; Abcam) at room temperature for 1 h. Images were captured
on the Odyssey CLx Infrared Imaging System (LI-COR Biosciences,
Lincoln, NE, USA). Western blot bands were quantified using Odyssey
CLx v2.1 software.
Quantitative-PCR analysis
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA according to the
manufacturer's protocols. Reverse transcription (RT) was
accomplished by using reverse transcriptase to synthesize cDNA. RT
was performed using 10X RT buffer, Multiscribe Reverse
Transcriptase, 10 mM dNTP and RT-RNA (Roche Diagnostics GmbH,
Mannheim, Germany) at 25°C for 10 min, 37°C for 2 h and 85°C for 5
min. PCR was perfomed using 10X PCR buffer, 10 mM dNTP, Taq enzyme
and primers from the Roche SYBR Green PCR Master Mix kit. The
cycling conditions were as follows: 95°C for 10 min, 95°C for 15
sec and 60°C for 15 sec, and 72°C for 15 sec, for 40 cycles. The
levels of miR-143 mRNA were measured using SYBR Green incorporation
on a Roche LightCycler480 Real Time PCR system (Roche Diagnostics
GmbH), with U6 as an internal control. The PCR primers used were as
follows: hsa-miR-143-3p forward, 5′-UGAGAUGAAGCACUGUAGCUC-3′ and
reverse, 5′-GTCGTATCCAGTGCGTGTCGTG-3′; and U6 forward,
5′-GCTTCGGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. To obtain a relative quantitation
measurement, the data was analyzed using the quantification cycle
(Cq) value compared with U6 mRNA. The amount of target was
determined by normalizing to endogenous reference [using the
2−ΔΔCq method (17)] and
was relative to a calibrator (mean of the control samples).
Statistical analysis
Data analysis was performed using SPSS 14.0 software
(SPSS Inc., Chicago, IL, USA) and Graphpad software (GraphPad
Software, Inc., La Jolla, CA, USA). The data for the in
vitro experiments were repeated three times and statistical
comparisons between two groups were performed by t-test, and among
multiple groups were performed by one-way ANOVA followed by Tukey's
multiple comparison tests. Data are presented as the mean ±
standard error of the mean. Results with P<0.05 were considered
statistically significant.
Results
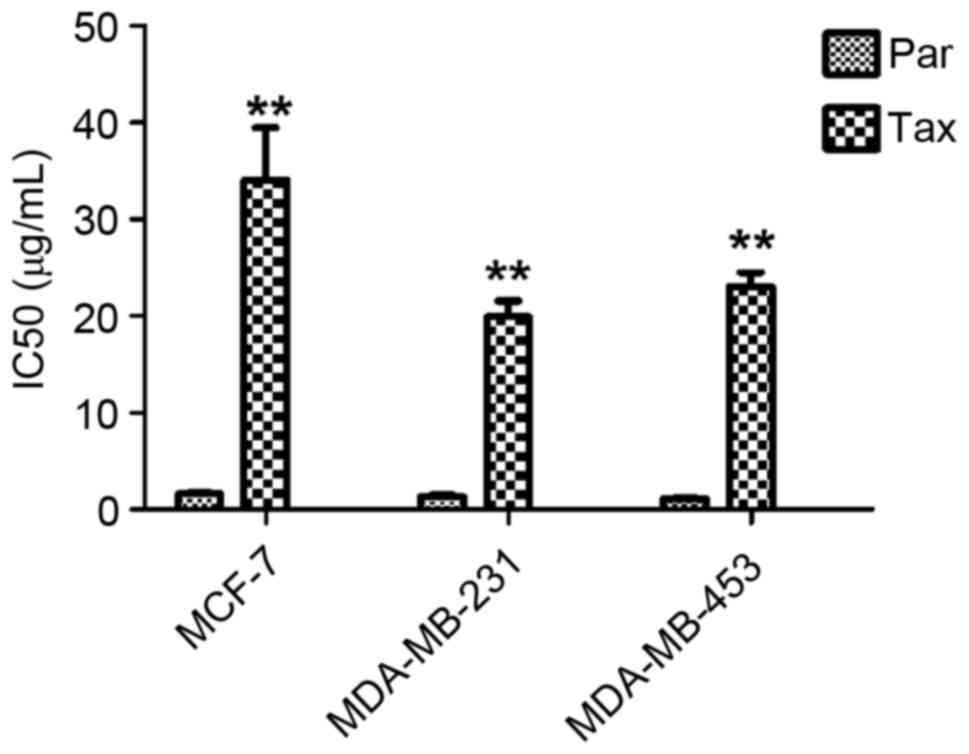
TDR
The IC50 values of MCF-7, MDA-MB-231 and
MDA-MB-453 cells exposed to Taxol were measured, and it was
observed that the values were significantly increased (all
P<0.01) (Fig. 1), indicating a
significant increase in drug resistance.
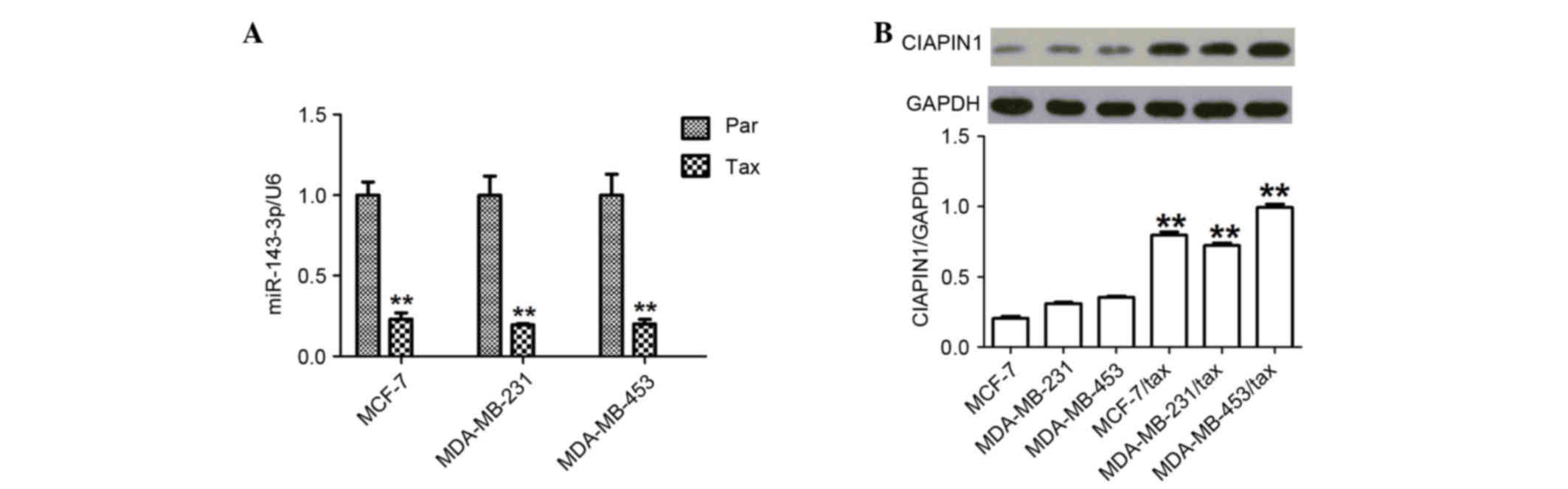
Upregulation of CIAPIN1 protein and
downregulation of miR-143 in TDR breast cancer cells
First, the expression of miR-143 in TDR human breast
cancer cells was examined and it was found that the levels were
significantly decreased (all P<0.01) (Fig. 2A). Next, the study investigated the
differences in CIAPIN1 levels in various TDR breast cancer cell
lines. The amount of CIAPIN1 protein was analyzed using western
blotting, and a significant increase in CIAPIN1 protein levels was
found in the TDR cells compared with the non-TDR cells (all
P<0.01) (Fig. 2B).
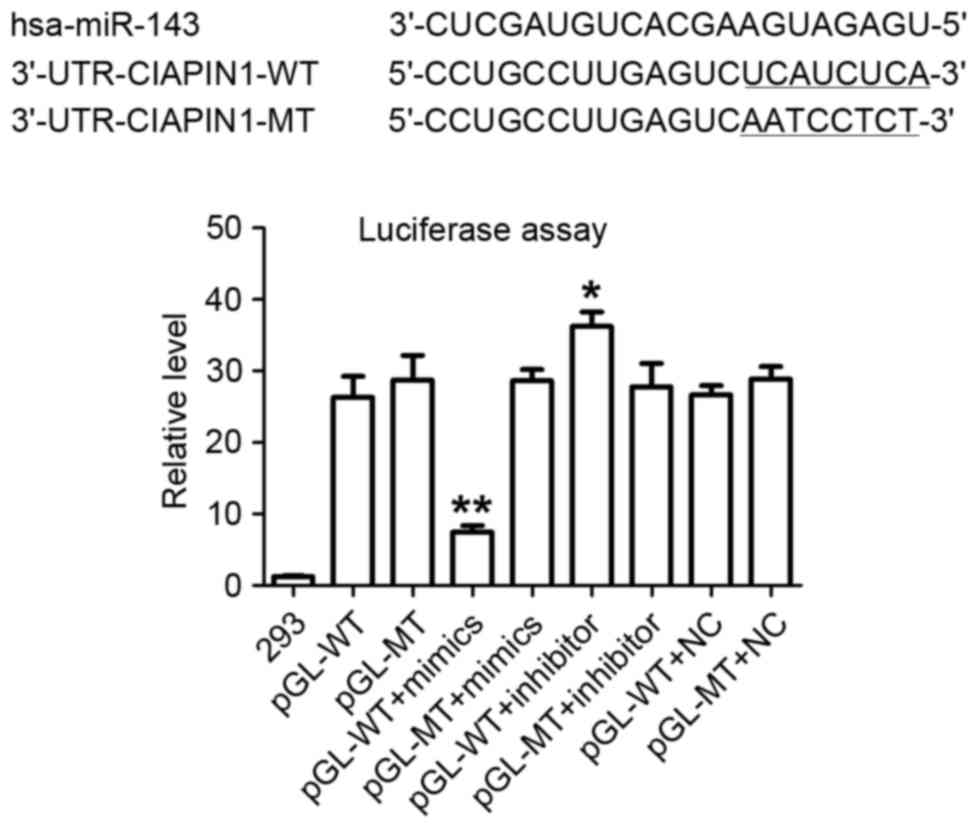
miR-143 targeting of the CIAPIN1
3′-UTR
Two independent informatics software packages
(TargetScan and miRanda; http://www.targetscan.org and http://www.microrna.org) were used to perform a
comprehensive survey for target genes according to consensus
miR-143 binding sites. A sequence was found to be located at bases
373–380 of the CIAPIN1 3′-UTR (NM_020313.2), which was
highly complementary with the seed sequence of miR-143 (Fig. 3). A luciferase assay was performed to
experimentally validate this target. In the presence of miR-143,
the relative luciferase activity of HEK-293 cells with the
wild-type (WT) construct was significantly reduced. However, cells
transfected with the MT construct did not exhibit a significant
suppressive effect in the presence of miR-143 (Fig. 3). In addition, the knockdown of
miR-143 with a miR-143 inhibitor increased luciferase activity in
WT HEK-293 cells. These results suggest a direct and specific
interaction of miR-143 with the CIAPIN1 3′-UTR in HEK-293
cells. Taken together, the data suggest that CIAPIN1 is a
target gene of miR-143.
Lv-miR-143 suppresses proliferation of
breast cancer MDR cells
First, the expression levels of miR-143 were
measured to evaluate the effectiveness of Lv-miR-143 in the MCF-7,
MDA-MB-231 and MDA-MB-453 TDR cells. As expected, following
transfection of Lv-miR-143, the expression of miR-143 mRNA levels
was significantly increased in the MCF-7, MDA-MB-231 and MDA-MB-453
TDR cells (Fig. 4A).
Lentiviral-expressed vector miR-143 was infected into the MCF-7,
MDA-MB-231 and MDA-MB-453 TDR cells (MOI=20, best MOI) and a stably
expressed cell line was obtained subsequent to screening (Fig. 4B). The expression of GFP and miR-143
was observed, demonstrating that the MCF-7, MDA-MB-231 and
MDA-MB-453 MDR LV-miR-143 cells were of good quality (Fig. 4B). At 24 and 48 h after Lv-miR-143
transfection, Lv-miR-143 was found to have decreased the
proliferation of the MCF-7, MDA-MB-231, and MDA-MB-453 TDR cells
(Fig. 4C).
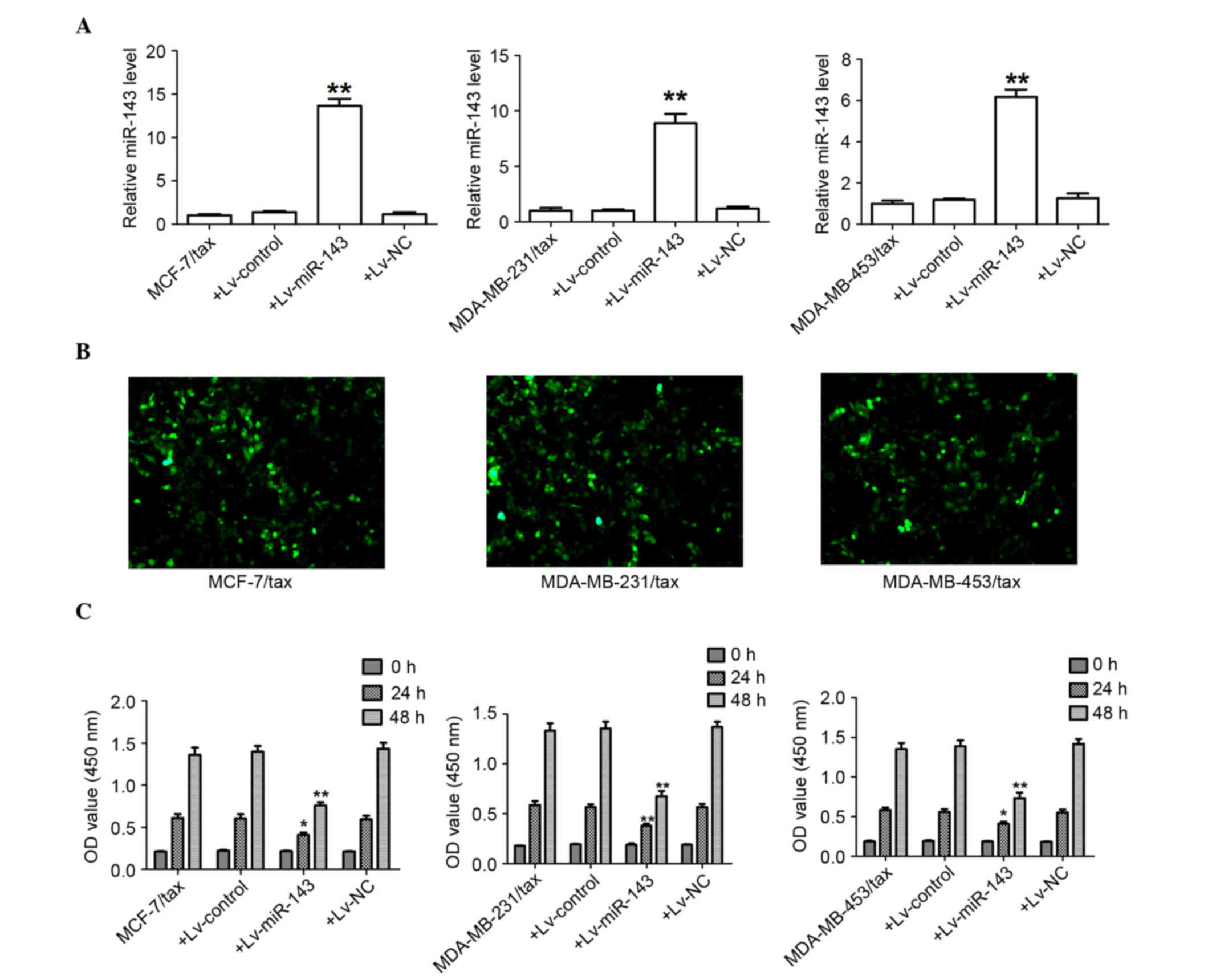 | Figure 4.miR-143 attenuates the growth of
breast cancer TDR cells. (A) Transcript level of miR-143 determined
using quantitative polymerase chain reaction. **P<0.01 for
MCF7/Tax+Lv-miR-143 versus MCF7/Tax, MDA-MB-231/Tax+Lv-miR-143
versus MDA-MB-231/Tax, and MDA-MB-453/Tax+Lv-miR-143 versus
MDA-MB-453/Tax. (B) Fluorescence microscopy results of MCF7/Tax,
MDA-MB-231/Tax and MDA-MB-453/Tax 96 h after lentiviral-miR-143
infection (MOI=20×120). The condition of the lentiviral-miR-143
infected cells was good and the percentage of cells expressing
green fluorescent protein was greater than 90% (magnification,
×120). (C) miR-143 significantly attenuated cell proliferation of
MCF-7, MDA-MB-231 and MDA-MB-453 TDR cells in vitro.
*P<0.05 for MCF7/Tax+Lv-miR-143 versus MCF7/Tax (24h),
MDA-MB-453/Tax+Lv- miR-143 versus MDA-MB-453/Tax (24h), and
MDA-MB-231/Tax+Lv-miR-143 versus MDA-MB-231/Tax (24h). **P<0.01
for MCF7/Tax+Lv-miR-143 versus MCF7/Tax (48h),
MDA-MB-231/Tax+Lv-miR-143 versus MDA-MB-231/Tax (48h) and
MDA-MB-453/Tax+Lv-miR-143 versus MDA-MB-453/Tax (48h). miR-143,
microRNA-143; Tax, Taxol; TDR, Taxol-induced drug-resistant; Lv,
lentivirus. |
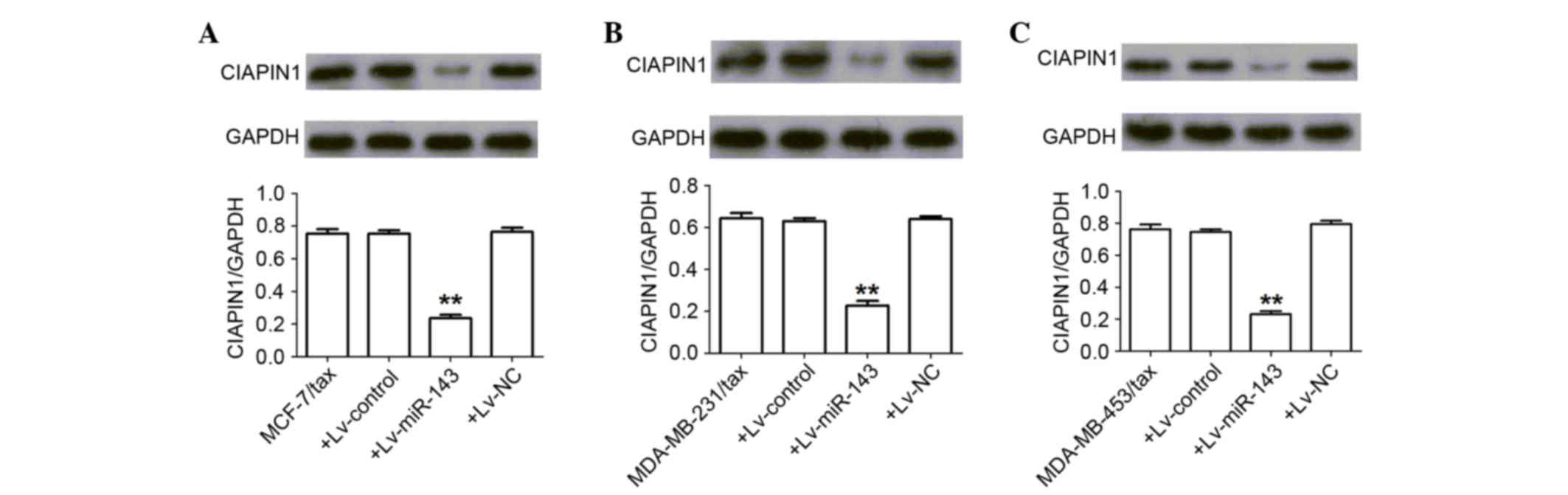
miR-143 represses CIAPIN1 expression
in breast cancer TDR cells
The aforementioned miRNA databases predicted that
CIAPIN1 contains a putative binding site for miR-143,
suggesting that it is a potential target for miR-143. Consistent
with this prediction, the present results confirmed that miR-143
post-transcriptionally repressed CIAPIN1 expression by binding to
the CIAPIN1 3′-UTR (P<0.01) (Fig. 5).
Discussion
MDR remains a major obstacle to achieving curative
breast cancer chemotherapy treatment, and causes >90% of
metastatic breast cancer patients to experience treatment failure
(18). For this reason, the
development of novel avenues for overcoming MDR is urgent. MDR in
breast cancer involves multiple genes and mechanisms. Current
cancer research has become increasingly focused on gene therapy,
and the present study aimed to improve the effectiveness of
chemotherapy treatment by targeting the genes associated with
breast cancer MDR. Shibayama et al (7) first identified CIAPIN1, a novel
apoptosis inhibitor, in 2004. More recently, studies of MDR in
gastric cancer, leukemia cell lines and breast cancer cells
(8–11)
have provided evidence that CIAPIN1 plays a role in the MDR process
in malignant cancer. Thus, CIAPIN1 is a potential target for
the improvement of therapeutic efficacy. In a study using nude mice
with MDR breast cancer tumors, the combination of chemotherapy with
the knockdown of CIAPIN1 levels using lentivirus-vector-based RNAi
significantly inhibited tumor growth in vivo (19). In agreement with these previous
studies, the present results demonstrated that CIAPIN1 protein
levels significantly increased in TDR breast cancer cells.
In numerous human tumor types, including prostate
cancer, hepatocellular carcinoma, non-small cell lung cancer and
gastric cancer, miR-143 is significantly dysregulated (20,21).
However, the role of miR-143 in breast cancer remains relatively
uninvestigated. In the present study, it was found that miR-143
mRNA levels were significantly decreased in the TDR breast cancer
cells. The association between miR-143 and CIAPIN1 in MCF-7,
MDA-MB-231 and MDA-MB-453 TDR cells was also investigated.
Bioinformatic methods were used to identify whether CIAPIN1
is a target gene of miR-143. As predicted, in the CIAPIN1
3′UTR, a highly complementary potential binding site of miR-143 was
found at the bases of the CIAPIN1 3′UTR (NM_020313.2).
Luciferase activity assay showed that the expression of a reporter
vector containing the WT sequence of the CIAPIN1 3′UTR was
inhibited by ectopic expression of miR-143. In addition, after
transfecting the miR-143 mimic into TDR breast cancer cells,
functional assays were performed. It was found that the expression
levels of miR-143 increased, whereas the protein levels of CIAPIN1
decreased in the MCF-7, MDA-MB-231 and MDA-MB-453 TDR cells. These
findings demonstrate that miR-143 functions as a CIAPIN1
suppressor. Abnormal growth is a fundamental characteristic of
tumor cells, and the main reason for uncontrolled growth is due to
an imbalance in cell cycle regulation (11). Experiments on the cell proliferation
inhibition of tumor cells revealed that Lv-miR-143 decreases the
proliferation of MCF-7, MDA-MB-231 and MDA-MB-453 TDR cells.
Together, these findings demonstrate that miR-143
participates in the reversal of TDR in human breast cancer cells by
inhibiting CIAPIN1, and that it suppresses the growth of
breast cancer TDR cells. These data provide compelling evidence for
making the miR-143/CIAPIN1 pathway a novel target for breast
cancer therapy.
Acknowledgements
This study was supported by the Natural Science
Foundation of Heilongjiang Province of China (grant no.
H201391).
References
|
1
|
Guo P, Huang ZL, Yu P and Li K: Trends in
cancer mortality in China: An update. Ann Oncol. 23:2755–2762.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seruga B, Hertz PC, Le LW and Tannock IF:
Global drug development in cancer: A cross-sectional study of
clinical trial registries. Ann Oncol. 21:895–900. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marquette C and Nabell L:
Chemotherapy-resistant metastatic breast cancer. Curr Treat Options
Oncol. 13:263–275. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuo MT: Roles of multidrug resistance
genes in breast cancer chemoresistance. Adv Exp Med Biol.
608:23–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cascorbi I: Role of pharmacogenetics of
ATP-binding cassette transporters in the pharmacokinetics of drugs.
Pharmacol Ther. 112:457–473. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shibayama H, Takai E, Matsumura I, Kouno
M, Morii E, Kitamura Y, Takeda J and Kanakura Y: Identification of
a cytokine-induced antiapoptotic molecule anamorsin essential for
definitive hematopoiesis. J Exp Med. 199:581–592. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hao Z, Li X, Qiao T, Du R, Hong L and Fan
D: CIAPIN1 confers multidrug resistance by upregulating the
expression of MDR-1 and MRP-1 in gastric cancer cells. Cancer Biol
Ther. 5:261–266. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Fan R, Zou X, Hong L, Gao L, Jin H,
Du R, He L, Xia L and Fan D: Reversal of multidrug resistance of
gastric cancer cells by down-regulation of CIAPIN1 with CIAPIN1
siRNA. Mol Biol (Mosk). 42:102–109. 2008.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Hong L, Zhao Y, Jin H, Fan R, Du R,
Xia L, Luo G and Fan D: A new apoptosis inhibitor, CIAPIN1
(cytokine-induced apoptosis inhibitor1), mediates multidrug
resistance in leukemia cells by regulating MDR-1, Bcl-2, and Bax.
Biochem Cell Biol. 85:741–750. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu D, Xiao Z, Wang W, Xu Y, Gao S, Deng L,
He W, Yang Y, Guo X and Wang X: Down regulation of CIAPIN1 reverses
multidrug resistance in human breast cancer cells by inhibiting
MDR1. Molecules. 17:7595–7611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Wu K and Fan D: CIAPIN1 as a
therapeutic target in cancer. Expert Opin Ther Targets. 14:603–610.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andorfer CA, Necela BM, Thompson EA and
Perez EA: MicroRNA signatures: Clinical biomarkers for the
diagnosis and treatment of breast cancer. Trends Mol Med.
17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi M and Guo N: MicroRNA expression and
its implications for the diagnosis and therapeutic strategies of
breast cancer. Cancer Treat Rev. 35:328–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH,
Liang S, Li B, Li Y, Li D, Wang ED and Liu MF: A novel
miR-155/miR-143 cascade controls glycolysis by regulating
hexokinase 2 in breast cancer cells. EMBO J. 31:1985–1998. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Egerton N: Ixabepilone (ixempra), a
therapeutic option for locally advanced or metastatic breast
cancer. P T. 33:523–531. 2008.PubMed/NCBI
|
|
19
|
Wang XM, Gao SJ, Guo XF, Sun WJ, Yan ZQ,
Wang WX, Xu YQ and Lu D: CIAPIN1 gene silencing enhances
chemosensitivity in a drug-resistant animal model in vivo. Braz J
Med Biol Res. 47:273–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang ZQ, Meng H, Wang N, Liang LN, Liu
LN, Lu SM and Luan Y: Serum microRNA 143 and microRNA 215 as
potential biomarkers for the diagnosis of chronic hepatitis and
hepatocellular carcinoma. Diagn Pathol. 9:1352014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhuang M, Shi Q, Zhang X, Ding Y, Shan L,
Shan X, Qian J, Zhou X, Huang Z, Zhu W, et al: Involvement of
miR-143 in cisplatin resistance of gastric cancer cells via
targeting IGF1R and BCL2. Tumour Biol. 36:2737–2745. 2015.
View Article : Google Scholar : PubMed/NCBI
|