Introduction
Acute myeloid leukemia (AML) is the most common form
of acute leukemia in adults (1). AML
is a genetically heterogeneous disease which results from the
over-proliferation of hematopoietic stem cells and their failure to
differentiate, resulting in an uncontrolled accumulation of
myeloblasts accumulated in the bone marrow as well as the blood
(2,3).
Allogeneic hematopoietic stem cell transplantation has been
demonstrated to be the most effective therapeutic method for acute
leukemia and has been integrated into the standard of care
(4). However, its application has so
far been limited, offering only incomplete prevention of AML
clinical relapse (5,6). Simultaneously, the chemotherapy standard
of care has only changed slightly over the previous decades. AML is
a clinically devastating disease with a 5 year survival rate of
only 25% in adults (7). It remains
associated with high rates of recurrence when treated with
conventional regimens (3). Thus,
novel and more effective therapies that may reduce the risk of
relapse following chemotherapy or stem cell transplantation are
required (5,8).
Previous studies have indicated that AML-associated
antigens may be used as more specific and effective targets for
immunotherapy (8), and may represent
a promising novel treatment option to improve the outcomes of
patients with AML (8,9). Thus far, dozens of tumor
(leukemia)-associated antigens, including hyaluronan-mediated
motility receptor/cluster of differentiation 168, M-phase
phosphoprotein 11, proteinase 3, Wilms' tumor 1, tumor-associated
antigen preferentially expressed antigen in melanoma, oncofectal
antigen-immature laminin receptor protein, B-cell lymphoma-2,
chronic myeloid leukemia (CML) 28 and CML66, survivin, breakpoint
cluster region-abelson murine leukemia, fusion transcript which
results from t (6;9;p23;q34; DEK-CAN), protein which represents
promyelocytic leukemia-retinoic acid receptor (PML-RAR),
runt-related transcription factor 1-myeloid translocation gene 8
(8) and fms related tyrosine kinase 3
(10) have been characterized in
patients with AML, however, novel and more specific antigens remain
rare.
The monocytic leukemia-associated antigen-34
(MLAA-34) gene (GeneBank no. AY288977.2) is one of the novel
identified leukemia-associated antigens and a candidate oncogene
(5,9).
As a novel splice variant of calcium binding protein 39-like
(CAB39L), MLAA-34 exclusively reacts with sera from patients with
allogeneic leukemia but not with normal donor sera (5,8,11,12). In
addition, MLAA-34 has been indicated to be a potent anti-apoptotic
factor associated with carcinogenesis or progression of AML
(5). Downregulation of MLAA-34
expression significantly suppresses the proliferation and increases
the spontaneous apoptosis of U937 cells in vitro (5,12).
Additional studies uncovered that MLAA-34 may be involved in cell
apoptosis through interaction with the Ras, Wnt, calcium and
chemokine signaling pathways in U937 cells (5,9,12). It has been indicated that 13 of the
annotated proteins (PGK1, GAPDH, CRMP1, TBK-1, SEPT7, CLTC, PPP2CA,
SOD2, PARK7, HSPA9, TXN, ESR1 and YWHAE) may interact with MLAA-34
and be directly involved in carcinogenesis (12).
Previous studies have indicated that MLAA-34 may be
a potential candidate for the early diagnosis and therapeutic
application of AML (5,9). However, the association between its
expression level and single nucleotide polymorphisms (SNPs) in the
diagnosis and prognosis of AML remains unclear. Thus, in the
present study, MLAA-34 expression was investigated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
gene mutation by SNP, and associations with the diagnosis
variables. In addition, MLAA-34 gene expression in patients with
AML and healthy donors was examined by fluorescence in situ
hybridization (FISH).
Materials and methods
Patients and sample selection
In the present study, 40 patients with AML in
different clinical stages were enrolled from the Department of
Hematology of the Second Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China) between October 2011 and October 2014,
and 5 healthy donors were assayed. The subtypes of all the patients
with AML were determined according to the French-American-British
(FAB) classification (5) and the
relevant clinical data records are listed in Table I. Conventional and molecular
cytogenetic analysis, as well as other relevant clinical
information, was also investigated in the protocol. Samples of
peripheral blood and bone marrow (3 ml from each donor) were
collected into a syringe with heparin (0.3 ml) for use in MLAA-34
mRNA expression level and MLAA-34 gene mutation or FISH assays,
respectively. The present study was approved by the Ethics
Committee of the School of Medicine, Xi'an Jiaotong University, and
was conducted in accordance with the Declaration of Helsinki.
Informed consent was obtained from all study donors.
 | Table I.Clinical characteristics and MLAA-34
expression levels of patients with acute myeloid leukemia, treated
by standard chemotherapy. |
Table I.
Clinical characteristics and MLAA-34
expression levels of patients with acute myeloid leukemia, treated
by standard chemotherapy.
| Sex | Age (years) | Relative MLAA-34
level | Positive response to
chemotherapy | Leukocyte number
(≥50×109/2) | Extra-medullary
lesions | Abnormal
karyotype | MLAA-34 mutation | Risk
stratification |
|---|
| M | 58 | 331.00 | N | Y | N | Y | − | IR |
| M | 35 | 22.40 | Y | N | N | Y | − | IR |
| F | 45 | 128.00 | Y | N | N | N | − | IR |
| M | 65 | 9.69 | Y | N | N | N | − | LR |
| F | 36 | 774.00 | N | Y | N | Y | + | HR |
| M | 47 | 37.60 | Y | N | N | Y | − | IR |
| M | 28 | 4380.00 | N | N | N | Y | + | HR |
| F | 66 | 55.40 | Y | N | N | Y | − | HR |
| F | 54 | 5560.00 | N | Y | Y | Y | + | HR |
| M | 42 | 221.00 | N | N | N | N | − | IR |
| F | 18 | 88.70 | Y | N | N | Y | − | LR |
| M | 70 | 1120.00 | N | Y | N | Y | + | HR |
| F | 39 | 8.82 | Y | N | N | Y | − | IR |
| F | 21 | 11.20 | Y | N | N | N | − | IR |
| F | 28 | 886.00 | N | Y | N | Y | + | IR |
| F | 54 | 7.85 | Y | N | N | N | − | IR |
| M | 64 | 11.20 | Y | N | N | Y | − | IR |
| M | 35 | 101.00 | N | N | Y | Y | − | IR |
| F | 59 | 2230.00 | N | N | N | Y | + | HR |
| M | 33 | 362.00 | Y | Y | N | Y | − | HR |
| M | 31 | 99.40 | Y | N | N | Y | − | IR |
| M | 42 | 3850.00 | N | Y | N | Y | + | IR |
| M | 52 | 452.00 | Y | N | N | N | − | IR |
| M | 31 | 66.20 | N | N | N | Y | − | IR |
| M | 19 | 537.00 | N | Y | Y | Y | + | HR |
| M | 23 | 7.98 | Y | N | N | N | − | HR |
| M | 24 | 1.01 | Y | N | N | Y | − | IR |
| M | 31 | 886.00 | N | N | N | Y | + | HR |
| M | 25 | 2240.00 | N | Y | Y | Y | + | IR |
| M | 37 | 9.66 | Y | N | N | N | − | LR |
| F | 39 | 8.87 | Y | N | N | Y | − | IR |
| F | 24 | 458.00 | N | N | N | Y | + | HR |
| F | 26 | 10.80 | Y | N | N | Y | − | IR |
| M | 33 | 238.00 | N | Y | Y | Y | + | HR |
| F | 35 | 8.86 | Y | N | N | N | − | IR |
| F | 43 | 8.84 | N | N | N | Y | − | IR |
| M | 12 | 7.96 | Y | N | N | N | − | LR |
| F | 30 | 7.88 | N | N | N | Y | − | IR |
| F | 40 | 10.50 | N | N | N | Y | − | IR |
| F | 35 | 2.21 | Y | N | N | N | − | HR |
| Normal control |
|
|
|
|
|
|
|
|
| F | 35 | 0.00150 | − | N | N | N |
|
|
| M | 26 | 0.00885 | − | N | N | N |
|
|
| F | 37 | 0.00042 | − | N | N | N |
|
|
| M | 24 | 0.00135 | − | N | N | N |
|
|
| F | 29 | 0.00100 | − | N | N | N |
|
|
Protocols and therapies
Patients with AML were treated according to the
protocols and therapies which were described in a previously
published study (9).
RNA extraction, cDNA synthesis and
RT-qPCR
Total RNA was extracted from mononuclear cells by
TRIzol (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the standard protocol. The extracted RNA of 1 µl
(about 2 µg) added to each lane was verified for integrity by 1.5%
agarose gel electrophoresis and estimated for purity at 260 and 280
nm wavelengths, as determined by an ultraviolet spectrophotometer
(ZF1 Shanghai Jia Peng Technology Co., Ltd., Shanghai, China),
samples were used for cDNA synthesis with the first strand cDNA
synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's protocol.
TaqMan-based PCR technology was performed on an ABI
5700 FAST instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.) in a total volume of 50 µl. MLAA-34 primers as well as the
TaqMan probe sequence were as follows: (forward,
3′-AAGCCGGAGAACCTGAAACTC-5′ and reverse,
3′-TGAGGACTGGCCACAAACAC-5′) and probe
[FAM-TGA-GAACCTCCTTCGGGATAAAAG-tetramethylrhodamine (TAMRA)] (DaAn
Gene Co., Ltd., Guangzhou, China). Expression of β-actin was used
as a reference gene control. Primers and probes for β-actin were as
follows: Forward, 3′-TCCTTCCTGGGTATGGAATC-5′ and reverse,
3′-GCACTGTGTTGGCATAGAGG-5′; probe,
FAM-CGGATGTCAACGTCACACACTTCATGA-TAM RA (DaAn Gene Co., Ltd.). The
reaction procedure was performed as follows: The reaction was
performed in triplicate with a total volume of 50 µl supplemented
with 10 µl 5x TaqMan Universal PCR buffer, 1.0 µl (300 nM)
forward/reverse primer, 1 µl (200 nM) probe, 1 µl dNTP, 1 µl Taq
DNAase, 5 µl cDNA and 30 µl nuclease-free water (Shanghai GeneCore
BioTechnologies Co., Ltd., Shanghai, China). PCR protocol was
performed as follows: 93°C for 2 min, 93°C for 30 sec, 55°C for 1
min for 40 cycles. The cycle threshold (CT) was determined
automatically. The samples without DNA were routinely included as a
no template control (H2O). Relative quantification of
the MLAA-34 gene was conducted with three independent experiments
by the 2−ΔΔCq method as described in a previous study
(9).
DNA sequence analysis gene mutation
for exons
Genomic DNA was extracted from peripheral blood
using the QIAamp DNA blood kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocol, and then stored at −20°C
until use. Primers for 12 exons of the MLAA-34 gene were designed
with Primer Express Software Version 2.0 (Application Binary
Interface of China Branch Office, Shanghai, China) and are listed
in Table II. Simultaneously, primers
used for genes which are closely associated with AML, including
FMS-like tyrosine kinase 3 (Flt3), DNA methyl-transferase 3A
(DNMT3A), c-ki, CCAAT-enhancer-binding protein α (CEBPα) and
nucleophosmin-1 (NMP1), were synthesized and are listed as follows:
FLT3 forward 5′-GCAATTTAGGTATGAAAGCCAGC-3 and reverse
5-CTTTCAGCATTTTGACGGCAACC-3′; DNMT3a forward
5′-CTGCTGTGTGGTTAGACG-3′ and reverse 5′-TATTTCCGCCTCTGTGGTTT-3′;
NMP1 forward NMP1F 5′-TCGGGAGATGAAGTTGGAAG-3′ and reverse
5′-AACATTTATCAAACACGGTAG-3′; C-KIT (exon 17) forward
5′-CAGCCAGAAATATCCTCCTTACT-3′ and reverse
5′-TGTCAAGCAGAGAATGGGTACTC-3′; CEB PA: PP1F
5′-TCGCCATGCCGGGAGAACTCTAAC-3′ (nucleotides 120–143) and PP1R
5′-CTGGTAAGGGAAGAGGCCGGCCAG-3′ (nucleotides 692–669), PP2F
5′-CCGCTGGTGATCAAGCAGGA-3′ (nucleotides 615–634) and PP2R
5′-CACGGTCTGGGCAAGCCTCGAGAT-3′ (nucleotides 1317-1294). All samples
were genotyped using PCR and direct sequencing. PCR amplification
was performed using the Takara Ex Taq kit (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. The PCR
thermocycling conditions were: 95°C for 3 min; 32 cycles of 95°C
for 30 sec; 58°C for 30 sec; 72°C for 1 min; and a final extension
at 72°C for 10 min. Subsequent to purifying the PCR products with
an AxyPrep DNA purification kit (Qiagen GmbH), direct sequencing
was performed on the ABI 5700 DNA Analyzer using a Big Dye
Terminator kit v3.1 (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the corresponding forward primer as the sequencing
primer. The analytical approach, which applied the sequencing of
data by Vector NTI 8.0 analysis software (Invitrogen; Thermo Fisher
Scientific, Inc.), focused on base pair substitutions (SNPs).
 | Table II.Primers of 12 exons for monocytic
leukemia-associated antigen-34. |
Table II.
Primers of 12 exons for monocytic
leukemia-associated antigen-34.
| Exon no. | Product length | Forward primer | Reverse primer |
|---|
| 1 | 68 |
5′-CAGGCCGACCTACCTAAACC-3′ |
5′-CACCATTCCTCGCTCTCTCT-3′ |
| 2 | 138 |
5′-CTTGCAGCTGTACATTGAGACC-3′ |
5′-GAAAACCCATGCCTGCTAGA-3′ |
| 3 | 76 |
5′-TTGAAAGGTCTGCCACTTGA-3′ |
5′-GGGAGGAATTCAGGCTCTCT-3′ |
| 4 | 138 |
5′-AAGCAAGGCTTGGAATCTGA-3′ |
5′-AACCTCTCCTAGTAACAGCAATTCA-3′ |
| 5 | 142 |
5′-AAATTTGGCATAAAACTTGAAACT-3′ |
5′-GTTGCATAAAACCTGAAATCAAC-3′ |
| 6 | 165 |
5′-TCCCCTCACTGTTTTTGTTTG-3′ |
5′-GTTTGGCTTTTTGCTTTTGT-3′ |
| 7 | 119 |
5′-TGCAAGCACAGCTTGTTAGG-3′ |
5′-TGCAAAGAAAGGATTTTGCTG-3′ |
| 8 | 169 |
5′-CAGTGGATATTGAATGAATCGTG-3′ |
5′-CAGACTGGCCTCATAGACTGC-3′ |
| 9 | 60 |
5′-ATTTTGTGGCGCAAATGAA-3′ |
5′-CGAAGAGATGTGAAAAAGGTGA-3′ |
| 10 | 66 |
5′-GTCCCCCAGTGTCTTCACAT-3′ |
5′-AGCAGGACAGGACACTTACATT-3′ |
| 11 | 144 |
5′-TTGCTTTTATGCCTGTGCTTT-3′ |
5′-TGGGCATTCATTAAGATAACTCTG-3′ |
| 12 | 342 |
5′-TCAGGGGCTTCTACGCATTA-3′ |
5′-GGGCTCACATCTGCAAGTTA-3′ |
FISH
Metaphase chromosomes were prepared from the
cultivated bone marrow cells according to the method described
previously (13). R-banding of
chromosomes stained with Giemsa-staining was performed according to
the standard procedures and was analyzed according to the
International System for Human Cytogenetic Nomenclature 2013 using
a light microscope and magnification, ×100 (Olympus Corporation,
Tokyo, Japan). FISH was performed on metaphase cells of bone marrow
samples using a MLAA-34 and RB1 probes for chromosome 13 labeled in
red spectrums, provided by An Biping Pharmaceutical Co., Ltd.
(Guangzhou, China, http://www.gzlbp.com/). The RB1 probe located at
chromosome 13q14 and labeled in green spectrums was used to confirm
MLAA-34 location. The clone numbers of RB1 probe for FISH were
RP11-755M4 (chr13:113686037-113853591 bp), CTD-3019N20
(chr13:113755798-114014009 bp), CTD-2147J22
(chr13:114010132-114154734 bp) and RP11-281G7
(chr13:114081588-114285555 bp), respectively. The clone numbers of
MLAA-34 for FISH were CTD-2503J7 (chr13: 49250000-49400000 bp) and
RP11-803H8 (chr13: 49450000-49600000 bp). Nuclei were
counterstained with DAPI (Vysis; Abbott Laboratories, Abbott Park,
IL, USA) and signals from 200 nuclei were counted under a
fluorescent microscope of magnification, ×100. All FISH procedures
were performed according to the manufacturer's protocol (An Biping
Pharmaceutical Co., Ltd.).
Statistical analysis
Statistical analyses were performed using SPSS
version 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference. The
χ2 test was applied for baseline clinical variables
between groups for categorical data and the significance of
difference of homologous chromosomes signals. The probabilities of
OS and PFS were estimated with the Kaplan-Meier method. The Cox
model and regression analysis were used to analyze the effect of
exon 2 (E2) mutation in patients with AML.
Results
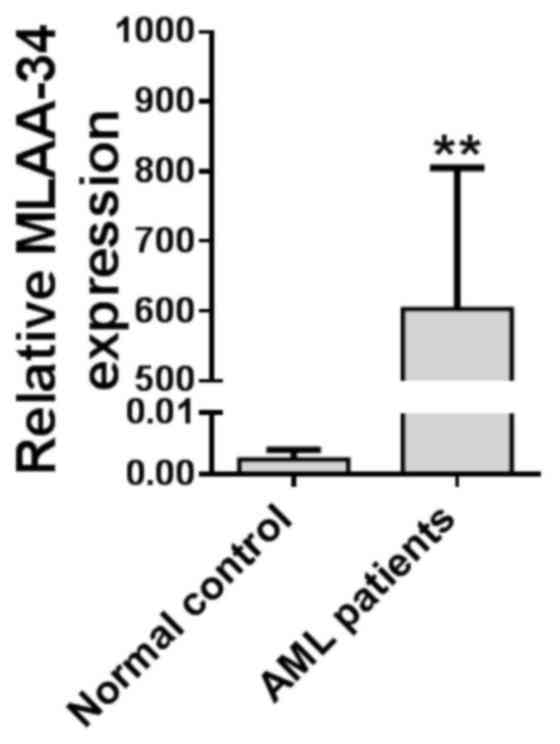
MLAA-34 is upregulated in patients
with AML
In order to investigate the function of MLAA-34 in
AML, the expression level of MLAA-34 was detected in 40 patients
with AML and 5 healthy volunteers, and the results demonstrated
that MLAA-34 was significantly upregulated in 40 patients with AML
when compared with healthy volunteers (Fig. 1; Table
I). When all the patients received standard chemotherapy, it
was evident that the patients with increased MLAA-34 levels had
poor or no response to the treatment (Table I). In addition, MLAA-34 mRNA was
associated with peripheral white blood cell (WBC) numbers and was
prone to overexpression in the high-WBC group (WBC count,
≥50×109/l) compared with the low-WBC group (WBC count,
<50×109/l). Of the 40 patients with AML, abnormal
karyotypes were observed in 29 patients (Table I). No significant differences were
observed when the subjects were categorized according to age and
sex.
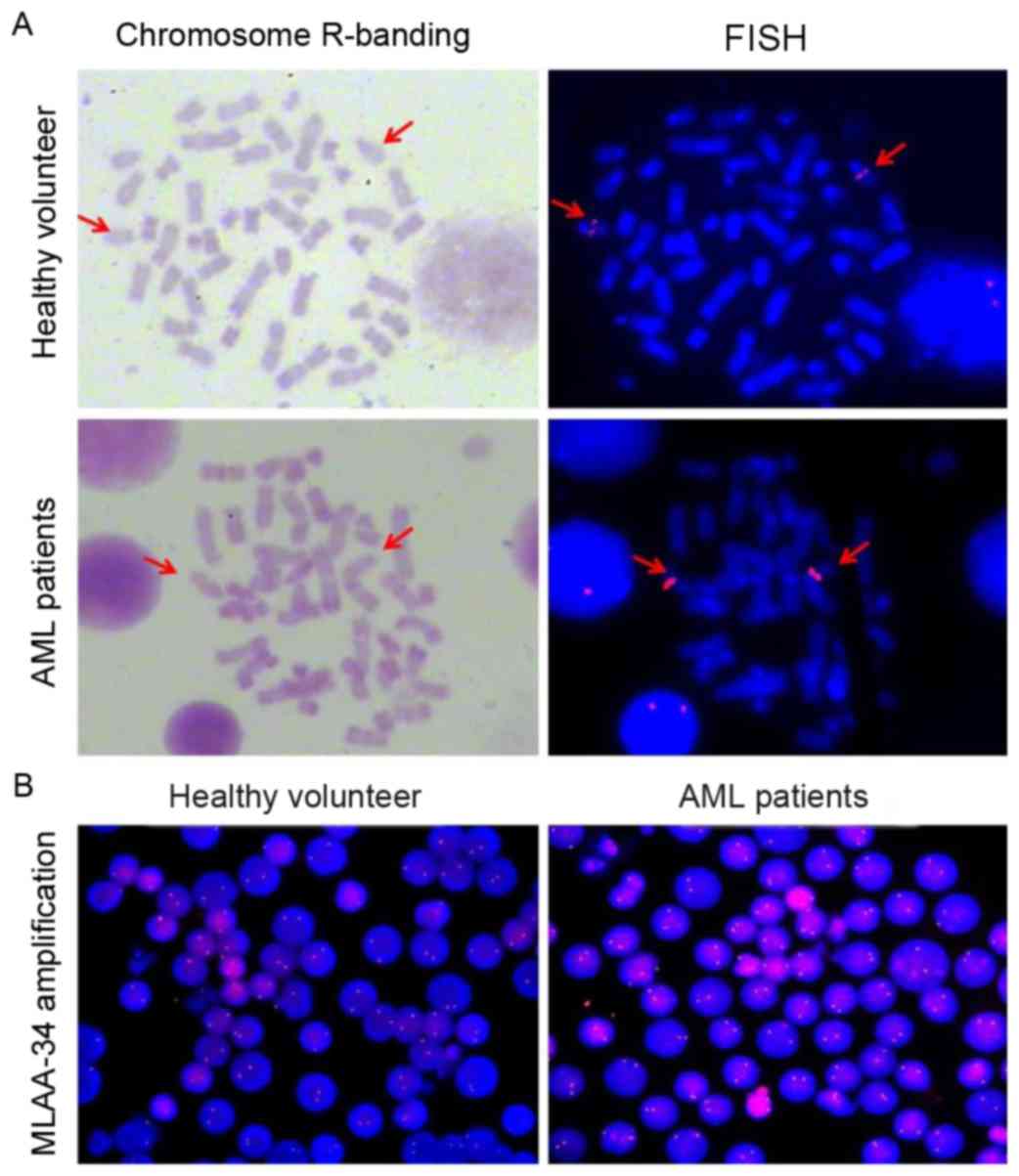
MLAA-34 is mapped to 13q14.2 and there
is no translocation in patients with AML
FISH was used to determine whether there was a
difference in MLAA-34 localization and gene copy number between
patients with AML and healthy controls. The MLAA-34 gene was
localized at chromosome 13q14.2 (Fig.
2A). No differences were observed for MLAA-34 gene location
between healthy volunteers and patients with AML. To confirm this
data, the tumor suppressor gene retinoblastoma (RB1), located at
chromosome 13q14, was selected as a positive control. The results
revealed that RB1 and MLAA-34 were co-localized (data not shown).
In addition, the fluorescence intensity of patients with AML was
relatively increased compared with that of healthy controls, but no
significant differences were observed between patients with AML and
healthy controls (Fig. 2B). This
indicated that the MLAA-34 gene copy number of patients with AML
was inconsistent with that of normal controls.
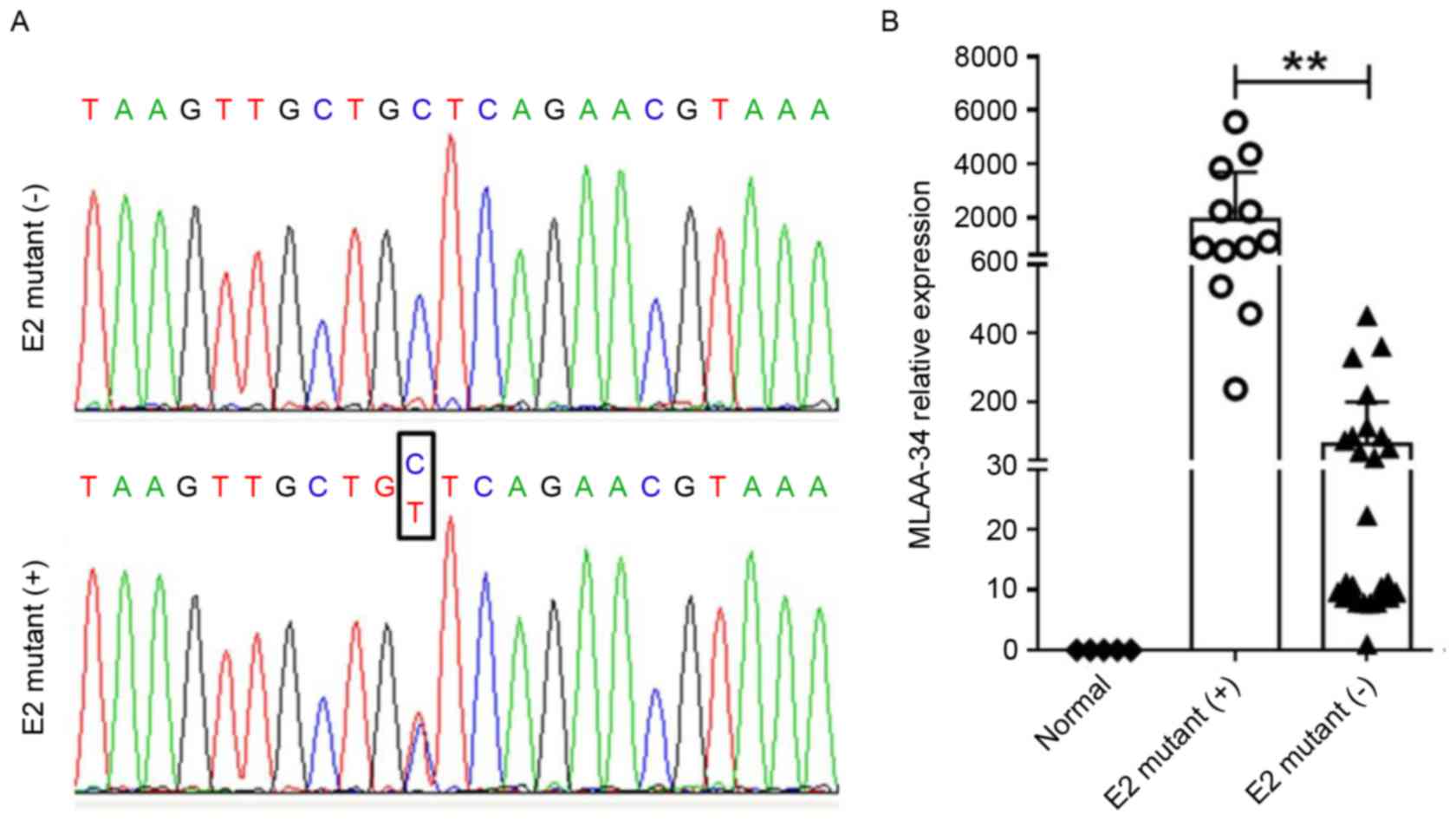
MLAA-34 contains a C59T SNP site in
patients with AML
To uncover the mechanism for MLAA-34 overexpression,
genomic DNA samples were prepared from 40 patients with AML and 5
healthy controls, and all 12 exons of the MLAA-34 gene were
amplified by PCR. PCR products were genotyped and a SNP site was
identified in 12 acute mynocytic leukemia (AML-M5) patients
(Table I, Fig. 3A). In these patients with mutations, 9
patients were identified as high risk (HR) and 3 were identified as
intermediate risk (IR). In 28 patients without mutation, 4, 19 and
5 were identified as HR, IR and low risk (LR), respectively
(Table I). This CC/CT allele was
located at the 59th bp of E2 of MLAA-34. Although this site belongs
to the 5′ untranslated region, it is associated with MLAA-34
overexpression in patients with AML M5 (3). MLAA-34 was significantly upregulated in
12 E2-mutant (+) patients with AML when compared with 28 E2-mutant
(−) patients (P<0.01; Fig. 3B). In
addition, patients with AML containing E2 mutations usually had
unfavorable therapeutic effects and were prone to recurrence
(Table III).
 | Table III.Comparisons of the MLAA-34 gene
mutations in subgroups stratified by genotypes of Flt3, DNMT3A,
C-kit, CEBPA and NPM1 in the control and exposed groups. |
Table III.
Comparisons of the MLAA-34 gene
mutations in subgroups stratified by genotypes of Flt3, DNMT3A,
C-kit, CEBPA and NPM1 in the control and exposed groups.
| Variables | MLAA-34 Mutation
(−), n (%) | MLAA-34 Mutation
(+), n (%) | OR (95%
CI)a |
P-valueb |
|---|
| Flt3 (−) | 27 (96.4) | 6 (50.0) |
|
|
| Flt3 (+) | 1 (3.6) | 6 (50.0) | 27.000
(2.722–267.796) | 0.000 |
| DNMT3A (−) | 26 (92.9) | 7 (58.3) |
|
|
| DNMT3A (+) | 2 (7.1) | 5 (41.7) | 9.286
(1.475–58.467) | 0.008 |
| C-kit (−) | 25 (89.3) | 10 (83.3) |
|
|
| C-kit (+) | 3 (10.7) | 2 (16.7) | 1.667
(0.241–11.525) | 0.602 |
| CEBP (−) | 22 (78.6) | 11 (91.7) |
|
|
| CEBP (+) | 6 (21.4) | 1 (8.3) | 0.333
(0.036–3.123) | 0.318 |
| NPM1 (−) | 22 (78.6) | 12 (100.0) |
|
|
| NPM1 (+) | 6 (21.4) | 0 (0.0) | 0.786
(0.648–0.953) | 0.082 |
| Extramedullary
disease (−) | 27 (96.4) | 8 (66.7) |
|
|
| Extramedullary
disease (+) | 1 (3.6) | 4 (33.3) | 13.500
(1.315–138.615) | 0.009 |
| Leukocyte
<50×109 | 25 (89.3) | 5 (41.7) |
|
|
| Leukocyte
≥50×109 | 3 (10.7) | 7 (58.3) | 11.667
(2.221–61.278) | 0.001 |
| Remission (+) | 20 (71.4) | 0 (0.0) |
|
|
| Remission (−) | 8 (28.6) | 12 (100.0) | 0.286
(0.159–0.513) | 0.000 |
| Abnormal karyotype
(−) | 10 (35.7) | 0 (0.0) |
|
|
| Abnormal karyotype
(+) | 18 (64.3) | 12 (100.0) | 0.643
(0.488–0.847) | 0.017 |
| Male | 15 (53.6) | 7 (58.3) |
|
|
| Female | 13 (46.4) | 5 (41.7) | 0.824
(0.210–3.234) | 0.781 |
MLAA-34 C59T mutation is associated
with Flt3, DNMT3A mutations and other clinical features
Flt3 (14,15), DNMT3A (16), c-kit (13), CCAAT-enhancer-binding protein α
(17) and nucleophosmin-1 (18) mutations represent the most frequent
gene alterations detectable in AML. In order to know whether there
are associations between MLAA-34 and these molecular markers, the
mutations of these genes were analyzed by direct sequencing.
MLAA-34 mutation in AML is associated with Flt3 and DNMT3A
mutations (P<0.01; Table III),
but no apparent links between MLAA-34 and other markers were
observed. In addition, mutation of MLAA-34 gene in patients with
AML was associated with extramedullary disease, periphery leukocyte
numbers, remission and cytogenetic abnormalities. Patients without
this SNP site in MLAA-34 usually had a lower number of leukocytes
(P=0.001), and indicated a relative higher percentage (35.7%) of
normal karyotypes, which means an increased success rate for
hematopoietic stem cell transplantation treatment. No significant
differences of MLAA-34 mutation were observed between males and
females.
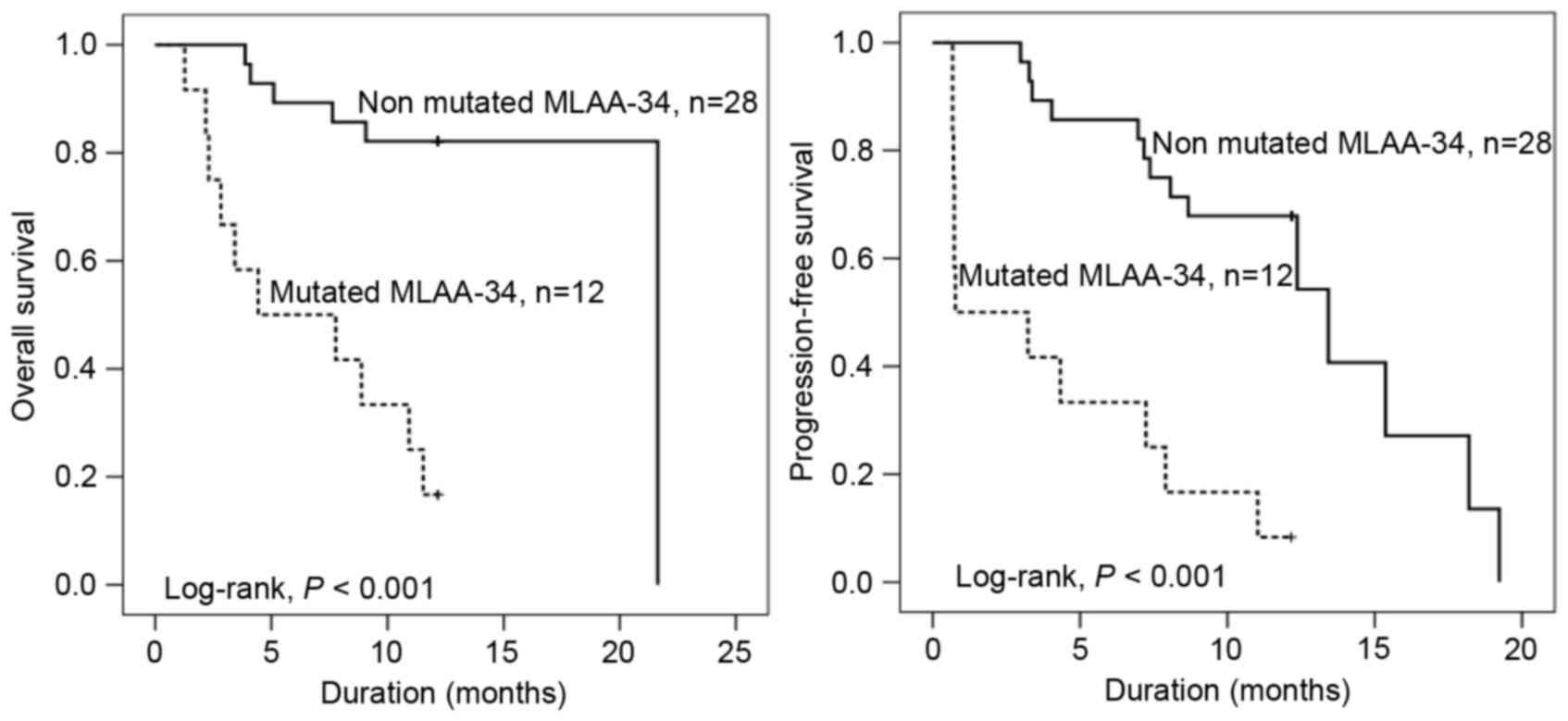
MLAA-34 C59T mutation indicates short
OS, PFS and survival function
To assess the prognostic potential of the MLAA-34
C59T mutation in 40 patients with AML, additional analyses of OS,
PFS and survival function were performed. The OS and PFS times of
patients with MLAA-34 C59T mutation were significantly shorter when
compared with that of patients without C59T mutations (P<0.001;
Fig. 4). The median OS times with
MLAA-34 C59T mutation and without MLAA-34 C59T mutation were 4.4
and 21.6 months, respectively. The median PFS times with or without
MLAA-34 C59T mutation were 0.8 or 13.4 months, respectively. In
addition, the results revealed that E2 mutation and extramedullary
disease indicated a significant association (P=0.333 and P=0.007,
respectively). For relative risk [RR; Exp(B)], the RR in patients
with AML with E2 mutation was 5.034 times that of patients with AML
without E2 mutation. In addition, the RR in AML with extramedullary
disease was 6.165 times that of patients with AML without
extramedullary disease (Table IV).
For analyses of survival function, the survival rate (Cum survival)
was lower in patients with E2 mutations compared with patients
without mutation (Fig. 4).
 | Table IV.Cox regression analysis of E2
mutation and extramedullary disease in patients with acute myeloid
leukemia. |
Table IV.
Cox regression analysis of E2
mutation and extramedullary disease in patients with acute myeloid
leukemia.
|
|
|
|
|
|
| 95% CI for
Exp(B) |
|---|
|
|---|
| Variables | B | SE | Wald | P-value | Exp(B) | Lower | Upper |
|---|
| E2 mutation | 1.616 | 0.553 | 8.533 | 0.003 | 5.034 | 1.702 | 14.890 |
| Extramedullary
disease | 1.819 | 0.675 | 7.257 | 0.007 | 6.165 | 1.841 | 23.155 |
Discussion
MLAA-34 is one of the newly identified monocytic
leukemia-associated antigens (11,19). This
gene is homologous to the known human CAB39L gene and has been
confirmed to be a novel splice variant of CAB39L (5,9). The
authors of the present study previously reported that MLAA-34 may
act as an anti-apoptosis factor in vitro via interacting
with Ras, Wnt or calcium and chemokine signaling pathways, and
lentivirus-mediated ectopic expression of MLAA-34 in U937 cells
markedly suppressed the spontaneous apoptosis of U937 cells
(5,12). Additional clinical studies uncovered
that high MLAA-34 expression levels usually indicated unfavorable
clinical features of patients with AML, and may be used as an early
biomarker for detection of relapse (5,9,12).
In the present study, it was revealed that MLAA-34
is upregulated in patients with AML when compared with healthy
volunteers. A previous study reported that MLAA-34 was
significantly induced in patients with acute monocytic leukemia
(AML-M5), and MLAA-34 overexpression was associated with an
unfavorable day 7 response to induction chemotherapy, and was also
associated with a poor survival rate (5,9). In
addition, increased MLAA-34 levels were independently associated
with poorer relapse-free survival and overall survival in patients
with AML-M5 (9).
In attempt to uncover the mechanism of MLAA-34
overexpression, no gene translocation or copy number variance was
identified. Finally, an SNP site was identified in the exon 2 of
MLAA-34 when the gene was analyzed by DNA sequencing. Although this
mutation site was located in the untranslated region, an
association between the mutation and expression level of MLAA-34
was observed. In addition, MLAA-34 mutation was also associated
with the molecular markers of AML, namely Flt3 and DNMT3A (20,21).
However, the detailed underlying mechanism remains to be
elucidated.
Regarding the prognostic potential of MLAA-34 C59T
mutation in AML, it was revealed that MLAA-34 C59T mutation
provided shorter survival durations for OS and PFS. In the present
study, MLAA-34 C59T mutation was associated with extramedullary
disease, periphery leukocyte numbers, remission and cytogenetic
abnormalities. Studies have reported that these clinical lesions
may result in shorter life for patients with AML (22,23). For
risk stratification, the HR was relatively increased in E2 mutation
patients compared with patients without E2 mutation. This was in
line with the RR in patients with E2 mutations. Thus, the survival
rate was relatively lower in patients with E2 mutation compared
with patients lacking E2 mutation. Therefore, MLAA-34 C59T mutation
may indicate a risk recurrence and a prognostic factor for patients
with AML.
E2 mutated positive in a total of 12 patients with
AML-M5, potentially for the following reasons: in contrast to the
other subtypes of AML, only a few leukemia-associated antigens have
been characterized in patients of AML-M5, a distinct subtype of
acute myeloid leukemia with characteristic clinical features
(3); clinically, the disease is
associated with hyperleukocytosis (5), extramedullary involvement (6), and coagulation abnormalities (7); and identification of immunogenic
leukemia-associated antigens as target structures is mandatory for
specific immunotherapy of AML-M5. For other FAB subsets and E2
mutations of the MLAA-34 gene, MLAA-34 mutation in patients with
AML was associated with extramedullary disease, periphery leukocyte
numbers, remission and cytogenetic abnormalities (Table III).
In conclusion, the evidence that the wild-type
MLAA-34 is an anti-apoptotic factor (12,24) and
that overexpression of MLAA-34 was observed in patients with AML
with poor response to standard chemotherapy indicated that MLAA-34
is a candidate oncogene. Thus, the present study shed light on the
diagnosis and treatment of AML, and MLAA-34 may be a novel marker
for AML therapy. This may be further demonstrated in the future by
additional studies with a larger sample size.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 8153000580 and
81270597).
References
|
1
|
Lindblad O, Chougule RA, Moharram SA,
Kabir NN, Sun J, Kazi JU and Rönnstrand L: The role of HOXB2 and
HOXB3 in acute myeloid leukemia. Biochem Biophys Res Commun.
467:742–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stahnke B, Thepen T, Stöcker M, Rosinke R,
Jost E, Fischer R, Tur MK and Barth S: Granzyme B-H22(scFv), a
human immunotoxin targeting CD64 in acute myeloid leukemia of
monocytic subtypes. Mol Cancer Ther. 7:2924–2932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tettamanti S, Marin V, Pizzitola I,
Magnani CF, Attianese GM Giordano, Cribioli E, Maltese F,
Galimberti S, Lopez AF, Biondi A, et al: Targeting of acute myeloid
leukaemia by cytokine-induced killer cells redirected with a novel
CD123-specific chimeric antigen receptor. Br J haematol.
161:389–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mensen A, Oh Y, Becker SC, Hemmati PG,
Jehn C, Westermann J, Szyska M, Göldner H, Dörken B, Scheibenbogen
C, et al: Apoptosis susceptibility prolongs the lack of memory B
cells in acute leukemic patients after allogeneic hematopoietic
stem cell transplantation. Biol Blood Marrow Transplant.
21:1895–1906. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang PY, Zhang WG, He AL, Wang JL and Li
WB: Identification and functional characterization of the novel
acute monocytic leukemia associated antigen MLAA-34. Cancer Immunol
Immunother. 58:281–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goswami M and Hourigan CS: Novel antigen
targets for immunotherapy of acute myeloid leukemia. Curr Drug
Targets. 18:296–303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sykes DB, Haynes MK, Waller A, Garcia M,
Urso U, Gouveia KE, Sklar L, Lewis TA, Dandapani S, Munoz B, et al:
Identifying small molecules that overcome differentiation arrest in
acute myeloid leukemia. Oncologist. 17:32012.PubMed/NCBI
|
|
8
|
Greiner J, Döhner H and Schmitt M: Cancer
vaccines for patients with acute myeloid leukemia-definition of
leukemia-associated antigens and current clinical protocols
targeting these antigens. Haematologica. 91:1653–1661.
2006.PubMed/NCBI
|
|
9
|
Zhao J, He A, Zhang W, Meng X and Gu L:
Quantitative assessment of MLAA-34 expression in diagnosis and
prognosis of acute monocytic leukemia. Cancer Immunol Immunother.
60:587–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kottaridis PD, Gale RE, Frew ME, Harrison
G, Langabeer SE, Belton AA, Walker H, Wheatler K, Bowen DT, Burnett
AK, et al: The presence of a FLT3 internal tandem duplication in
patients with acute myeloid leukemia (AML) adds important
prognostic information to cytogenetic risk group and response to
the first cycle of chemotherapy: Analysis of 854 patients from the
United Kingdom Medical Research Council AML 10 and 12 trials.
Blood. 98:1752–1759. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen G, Zhang W, Cao X, Li F, Liu X and
Yao L: Serological identification of immunogenic antigens in acute
monocytic leukemia. Leuk Res. 29:503–509. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang WJ, Zhang WG, Zhang PY, Cao XM, He
AL, Chen YX and Gu LF: The expression and functional
characterization associated with cell apoptosis and proteomic
analysis of the novel gene MLAA-34 in U937 cells. Oncol Rep.
29:491–506. 2013.PubMed/NCBI
|
|
13
|
Park IK, Mundy-Bosse B, Whitman SP, Zhang
X, Warner SL, Bearss DJ, Blum W, Marcucci G and Caligiuri MA:
Receptor tyrosine kinase Axl is required for resistance of leukemic
cells to FLT3-targeted therapy in acute myeloid leukemia. Leukemia.
29:2382–2389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park SH, Lee HJ, Kim IS, Kang JE, Lee EY,
Kim HJ, Kim YK, Won JH, Bang SM, Kim H, et al: Incidences and
prognostic impact of c-KIT, WT1, CEBPA, and CBL mutations, and
mutations associated with epigenetic modification in core binding
factor acute myeloid leukemia: A multicenter study in a Korean
population. Ann Lab Med. 35:288–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sawyers CL: Finding the next Gleevec: FLT3
targeted kinase inhibitor therapy for acute myeloid leukemia.
Cancer Cell. 1:413–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Berenstein R, Blau IW, Suckert N, Baldus
C, Pezzutto A, Dörken B and Blau O: Quantitative detection of
DNMT3A R882H mutation in acute myeloid leukemia. J Exp Clin Cancer
Res. 34:552015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tawana K, Wang J, Renneville A, Bödör C,
Hills R, Loveday C, Savic A, van Delft FW, Treleaven J, Georgiades
P, et al: Disease evolution and outcomes in familial AML with
germline CEBPA mutations. Blood. 126:1214–1223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoon JH, Kim HJ, Jeon YW, Lee SE, Cho BS,
Eom KS, Kim YJ, Lee S, Min CK, Cho SG, et al: Outcome of allogeneic
hematopoietic stem cell transplantation for cytogenetically normal
AML and identification of high-risk subgroup using WT1 expression
in association with NPM1 and FLT3-ITD mutations. Genes Chromosomes
Cancer; 2015
|
|
19
|
Fulton KM and Twine SM: Immunoproteomics:
Methods and protocols. Methods Mol Biol New York: Humana Press,
Springer; pp. 10612013
|
|
20
|
Yohe S: Molecular genetic markers in acute
myeloid leukemia. J Clin Med. 4:460–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Annesley CE and Brown P: Novel agents for
the treatment of childhood acute leukemia. Ther Adv Hematol.
6:61–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chessells JM, Harrison CJ, Kempski H, Webb
DK, Wheatley K, Hann IM, Stevens RF, Harrison G and Gibson BE: MRC
Childhood Leukaemia working party: Clinical features, cytogenetics
and outcome in acute lymphoblastic and myeloid leukaemia of
infancy: Report from the MRC childhood leukaemia working party.
Leukemia. 16:776–784. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bolkun L, Lemancewicz D, Jablonska E,
Szumowska A, Bolkun-Skornicka U, Ratajczak-Wrona W, Dzieciol J and
Kloczko J: The impact of TNF superfamily molecules on overall
survival in acute myeloid leukaemia: Correlation with biological
and clinical features. Ann Hematol. 94:35–43. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qian L, Zhang W, Zhang P, Lei B, Wang X,
Wang M, Bai J and He A: The anti-apoptosis effect of MLAA-34 in
leukemia and the β-catenin/T cell factor 4 protein pathway. Am J
Transl Res. 7:2270–2278. 2015.PubMed/NCBI
|