Introduction
An estimated 1.8 million new lung cancer cases
occurred in 2012, accounting for ~13% of total cancer diagnoses
(1). During this time period, lung
cancer was the most frequently diagnosed type of cancer and the
leading cause of cancer-associated mortality among males; among
females, it was the leading cause of cancer-associated mortality in
more developed countries and the second leading cause of
cancer-associated mortality in less developed countries (1). In 2011, lung cancer exhibited the
highest morbidity and mortality rates among all cancer types in
China (2).
Abundant supplies of resources and energy are
essential for malignant proliferation. The pentose phosphate
pathway, the only pathway for de novo ribose synthesis, has
been identified as a tumor therapeutic target (3–5). The
pentose phosphate pathway consists of an oxidative pathway and a
non-oxidative pathway (6). In the
oxidative pathway, by oxidative decarboxylation, glucose
6-phosphate is irreversibly converted into ribose 5-phosphate used
for the synthesis of nucleotides, and two NADPH molecules are
produced as reducing equivalents (6).
The non-oxidative pathway joins the pentose phosphate pathway to
glycolysis, and spare ribose or pentose molecules are reversibly
transformed into glyceraldehyde 3-phosphate and fructose
6-phosphate by transketolase (TKT) and transaldolase (TAL), without
restriction of the irreversible oxidative signaling pathway
(6). A previous study revealed the
upregulation of TKT activity in various types of cancer in
vivo and in vitro (3). Our
previous study demonstrated that oxythiamine exerts its anticancer
effect via the inhibition of thiamine-dependent enzymes,
particularly TKT (7).
RNA interference (RNAi) technology provides an
advantageous platform for investigating the function of a given
gene. Prior to RNAi technology, gene knockout was the main research
tool in reverse genetics. However, RNAi technology, using siRNA or
short hairpin RNA expression vectors to knockdown target genes in a
sequence-specific way, may be more time-efficient, economic and
convenient and is therefore an attractive tool for gene function
exploration, particularly within cancer research fields (8). In the present study, siRNA vectors were
applied with cationic liposomes to investigate the influence of the
TKT gene on the metabolism of A549 cells.
Materials and methods
Reagents and equipment
The following reagents and equipment were used in
the present study: Gibco fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA); Dulbecco's modified Eagle's
medium (DMEM; Genom Biotech Pvt., Ltd., Mumbai, India); three types
of TKT-specific siRNA duplex sequences and negative-control siRNA
duplex sequence (catalog no. A10001) and transfection reagents
(siRNA-Mate; catalog no. G04003; Shanghai GenePharma Co., Ltd.,
Shanghai, China); polymerase chain reaction (PCR) primers, as
presented in Table I (Shanghai Sangon
Pharmaceutical Co., Ltd., Shanghai, China); quantitative (q)PCR
reaction kit (UltraSYBR Mixture; catalog no. CW0957; CWBiotech Co.
Ltd., Beijing, China) and cell cycle detection kit (catalog no.
CW2575S, CWBiotech Co. Ltd.); Cell Counting Kit-8 (CCK-8; Beyotime
Institute of Biotechnology, Haimen, China); Transwell chambers and
cell culture dishes (Corning Incorporated, Corning, NY, USA);
Mastercycler ep realplex cycler (Eppendorf, Hamburg, Germany);
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA); NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific,
Inc.).
 | Table I.Polymerase chain reaction primers. |
Table I.
Polymerase chain reaction primers.
| Primer | Sequence (5 to
3) | Product length |
|---|
| β-actin | F:
TGACGTGGACATCCGCAAAG | 205 kb |
|
| R:
CTGGAAGGTGGACAGCGAGG |
|
| TKT | F:
TACCCAAGTGATGGCGTTGC | 129 kb |
|
| R:
GACCTGGAAGTCCTCATTGTTGTTAT |
|
| G6PDH | F:
TGTTGTCCCGGTTCCAGATG | 217 bp |
|
| R:
TGCATGAGCCAGATAGGCTG |
|
| TAL | F:
ATGGTGAGGAAGTCACAGCC | 201 bp |
|
| R:
ATTTGTTGGGCGCATCCTTG |
|
| SORD | F:
AAGACCTCATTTGGGCCTGG | 92 bp |
|
| R:
AAGCCCAACAACCTTTCCCT |
|
| HK1 | F:
TTATCACGCAGGCGGTTCA | 147 bp |
|
| R:
TGCCAAAGAAATCCTGACCC |
|
| PRPS1 | F:
TGCGAGAAATAGCAGGACCG | 263 bp |
|
| R:
CCAGGAGACCTGAGTGACCT |
|
| TKTL1-1 | F:
ACTAATGGCGGATGCTGAGG | 153 bp |
|
| R:
CATGATGTAGGGTGGCCGGA |
|
| TKTL1-2 | F:
ATGGCTCGGACAAGGACTGG | 191 bp |
|
| R:
GGCGGTTCACATCAAAGATTGC |
|
| TKTL1-3 | F:
AAACTTGCCCCGAGTCCAC | 125 bp |
|
| R:
GGTAAGGAATGCAGGGCTCA |
|
| TKTL2 | F:
ATCCATTCCATCAGGGCCAC | 175 bp |
|
| R:
GGATAGGAGCAGCATGTCCC |
|
Cell culture
The A549 lung cancer cell line was provided by
Zhongshan Hospital Central Laboratory of Fudan University
(Shanghai, China). A549 cells were cultured in DMEM supplemented
with 10% FBS, penicillin (100 kU/l) and streptomycin (100 kU/l) in
a humidified incubator at 37°C and 5% CO2.
siRNAs
The sequences of the siRNAs used in the present
study were as follows: TKT-siRNA-A sense,
5′-CCGGCAAAUACUUCGACAATT-3′; antisense,
5′-UUGUCGAAGUAUUUGCCGGTT-3′. TKT-siRNA-B sense,
5′-GCAUCUAUAAGCUGGACAATT-3′; antisense,
5′-UUGUCCAGCUUAUAGAUGCTT-3′. TKT-siRNA-C sense,
5′-CCAGCCAACAGCCAUCAUUTT-3′; antisense,
5′-AAUGAUGGCUGUUGGCUGGTT-3′. Negative control siRNA and fluorescein
amidite (FAM)-negative control siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′; antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Transfection
FAM-labelled siRNA (0, 25, 50 and 75 nM) was
transfected into A549 cells to determine the optimal transfection
concentration. Cells were transfected with siRNAs when cell
confluence 30–50%, according to the manufacturer's protocol.
FAM-siRNA (25 µM; 0, 2.2, 4.4 and 6.6 µl) was added into 200 µl
DMEM solution for 5 min and, subsequently, 8 µl siRNA-mate was
mixed into the solution for 30 min. During this time,
siRNA-siRNA-mate complexes were generated. During this procedure,
cell culture medium was replaced with 2 ml DMEM with serum (Gibco;
Thermo Fisher Scientific, Inc.). The siRNA-siRNA-mate complexes
were added to the cells and incubated for 6–8 h at 37°C. Cells were
digested with trypsin (Genom Biotech Pvt., Ltd.) for 2 min at room
temperature, centrifuged at 1,000 × g for 5 min at room temperature
and washed with ice-cold PBS (Genom Biotech Pvt., Ltd.) twice at
room temperature. For each cell suspension, ≥10,000 events were
analyzed using a FACSCalibur flow cytometer and FlowJo software
(version 7.6.1; Tree Star, Inc., Ashland, OR, USA) for data
analysis. To determine the most effective transfection sequence,
six groups were assessed and compared: Blank control (DMEM only),
mock control (DMEM and siRNA mate), negative control (negative
control siRNA), TKT-siRNA-A, TKT-siRNA-B and TKT-siRNA-C. Reverse
transcription (RT-)qPCR was used to determine the TKT mRNA
expression levels.
Reverse transcription (RT)-qPCR
Total RNA was extracted from the cells using
TRIzol® reagent (CWBiotech Co. Ltd.). The quantity and
integrity of mRNA was validated using a NanoDrop 2000c
spectrophotometer (Thermo Fisher Scientific, Inc.) and gDNA Eraser
was used to erase the genomic DNA at 42°C for 2 min. Subsequently,
HiFiScript cDNA Synthesis kit (catalog no. CW2569; CWBiotech Co.
Ltd.) was used to reverse transcribe 1 µg total RNA at 37°C for 15
min and 85°C for 5 sec into cDNA. qPCR was performed using a
Mastercycler ep realplex cycler with SYBR® Green Master
Mix (catalog no. CW0957; CWBiotech Co. Ltd.). Primers for TKT,
glucose-6-phosphate dehydrogenase (G6PDH), TAL, phosphoribosyl
pyrophosphate synthetase 1 (PRPS1), sorbitol dehydrogenase (SORD),
hexokinase 1 (HK1), transketolase-like 1 (TKTL1),
transketolase-like 2 (TKTL2) and β-actin were used, and the details
are listed in Table I. The
amplification procedure consisted of 95°C for 10 min, followed by
40 cycles of 95°C for 15 sec and 60°C for 60 sec. Each sample was
performed in triplicate. The quantification cycle (Cq) values of
the target genes were normalized to the housekeeping gene β-actin,
and relative expression levels were compared according to the
2−ΔΔCq method (9).
CCK-8 assay
Following a 72-h transfection, cells were seeded
into 96-well plates (2,000 cells/per well) and incubated for 24, 48
and 72 h at 37°C in an atmosphere containing 5% CO2.
Following incubation, 10 µl CCK-8 solution was added to the wells,
according to the manufacturer's protocol. Following a further 2 h
incubation, optical density (OD) was evaluated at 450 nm, which
provided an indirect indication of the number of viable cells.
Cell cycle analysis
Following 72 h transfection, cells (when confluence
reached more than 90%) were digested by trypsin (Genom Biotech
Pvt., Ltd.) for 2 min at room temperature, centrifuged at 1,000 × g
for 5 min at room temperature and washed with ice-cold PBS (Genom
Biotech Pvt., Ltd.) twice at room temperature. Cells were fixed
with 75% ethanol overnight at 4°C and subsequently dyed with
propidium iodide for 30 min at 37°C in the dark. For each cell
suspension, ≥10,000 events were analyzed using a FACSCalibur flow
cytometer and FlowJo software (version 7.6.1; Tree Star, Inc.,
Ashland, OR, USA) for data analysis.
Wound healing
Following 72 h transfection, 2×106 cells
were seeded into 6-well plates. When the confluence reached 100%,
linear wounds were created using a sterile 10 µl plastic pipette
tip and serum-free medium was added. Following 0, 6, 12 and 24 h
incubations at 37°C in an atmosphere containing 5% CO2,
the wounds were imaged using an inverted microscope (Olympus,
Japan) under bright-field illumination. ImageJ software (version
1.48u; National Institutes of Health) was used to calculate the
width of wounds.
Transwell migration assay
Following a 72 h transfection, 1.0×105
cells in 100 µl serum-free medium were added to the upper
compartment of each Transwell chamber and 600 µl DMEM supplemented
with 10% FBS was added into each of the lower compartments.
Following a 12 h incubation at 37°C in an atmosphere containing 5%
CO2, cells that had penetrated the membrane were dyed
with 0.1% crystal violet for 20 min. Five randomly selected fields
of view were observed using a microscope (magnification, ×10 and
×20; DP72; Olympus, Japan) under bright-field illumination and
cells that penetrated the Transwell chamber membrane (violet dying)
were counted. The mean number of cells of each image was calculated
for each group including the blank group, negative-siRNA group and
siRNA-C group.
Statistical analysis
Data are presented as the mean ± standard deviation.
Each experiment was repeated three times at least. The results were
analyzed using SPSS version 19.0 software (IBM Corp., Armonk, NY,
USA). The significance of differences was determined using one-way
analysis of variance with post hoc least significant difference, or
Dunnett's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Optimization of the transfection
sequence and concentration
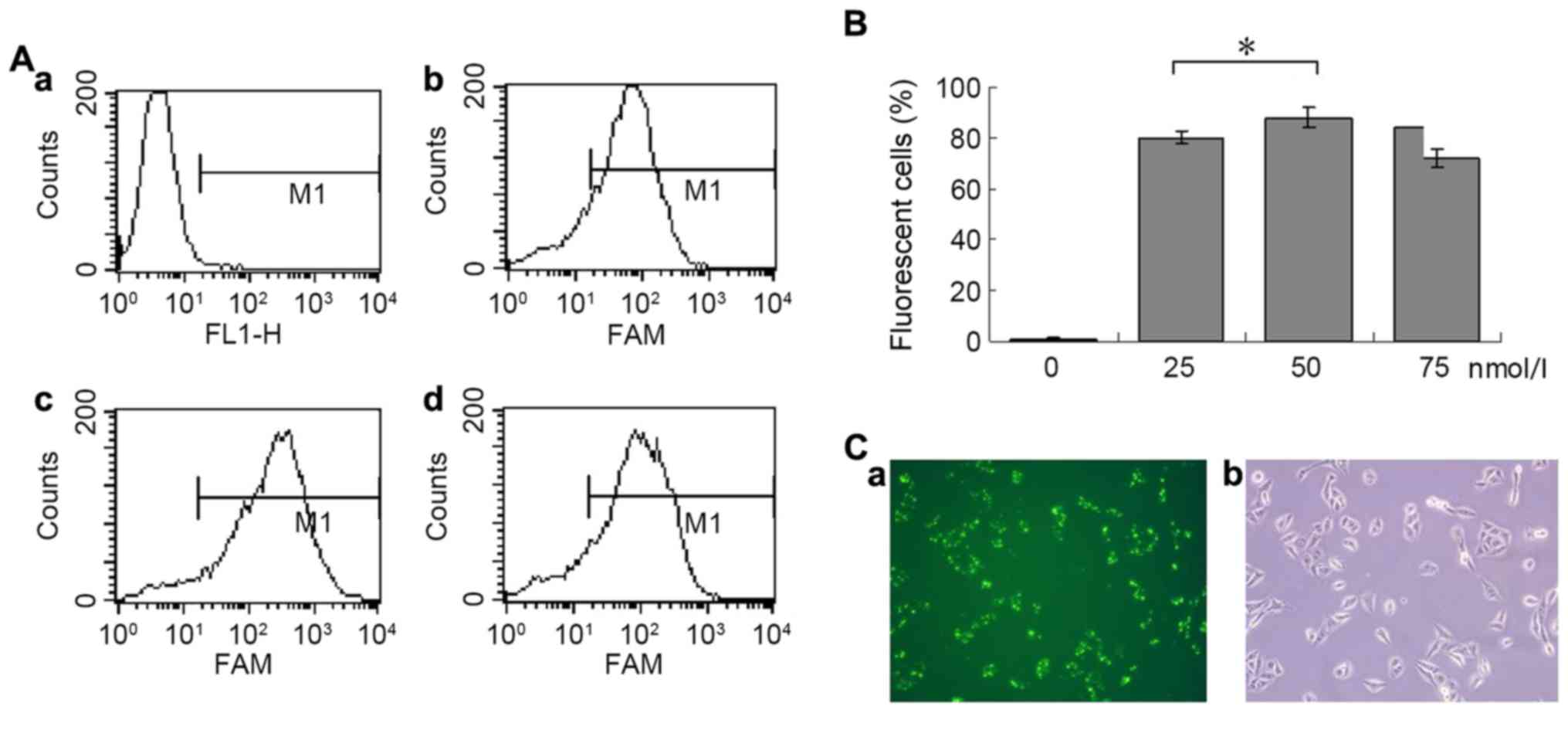
Fig. 1A presents the
flow cytometry results from cells transfected with FAM-labelled
negative control siRNA at four different concentrations. The
maximum transfection efficiency (88%) was achieved at a
concentration of 50 nmol/l (Fig. 1B).
Fig. 1C presents the fluorescence and
light microscopic images of the same field of view in cells
transfected with 50 nmol/l siRNA.
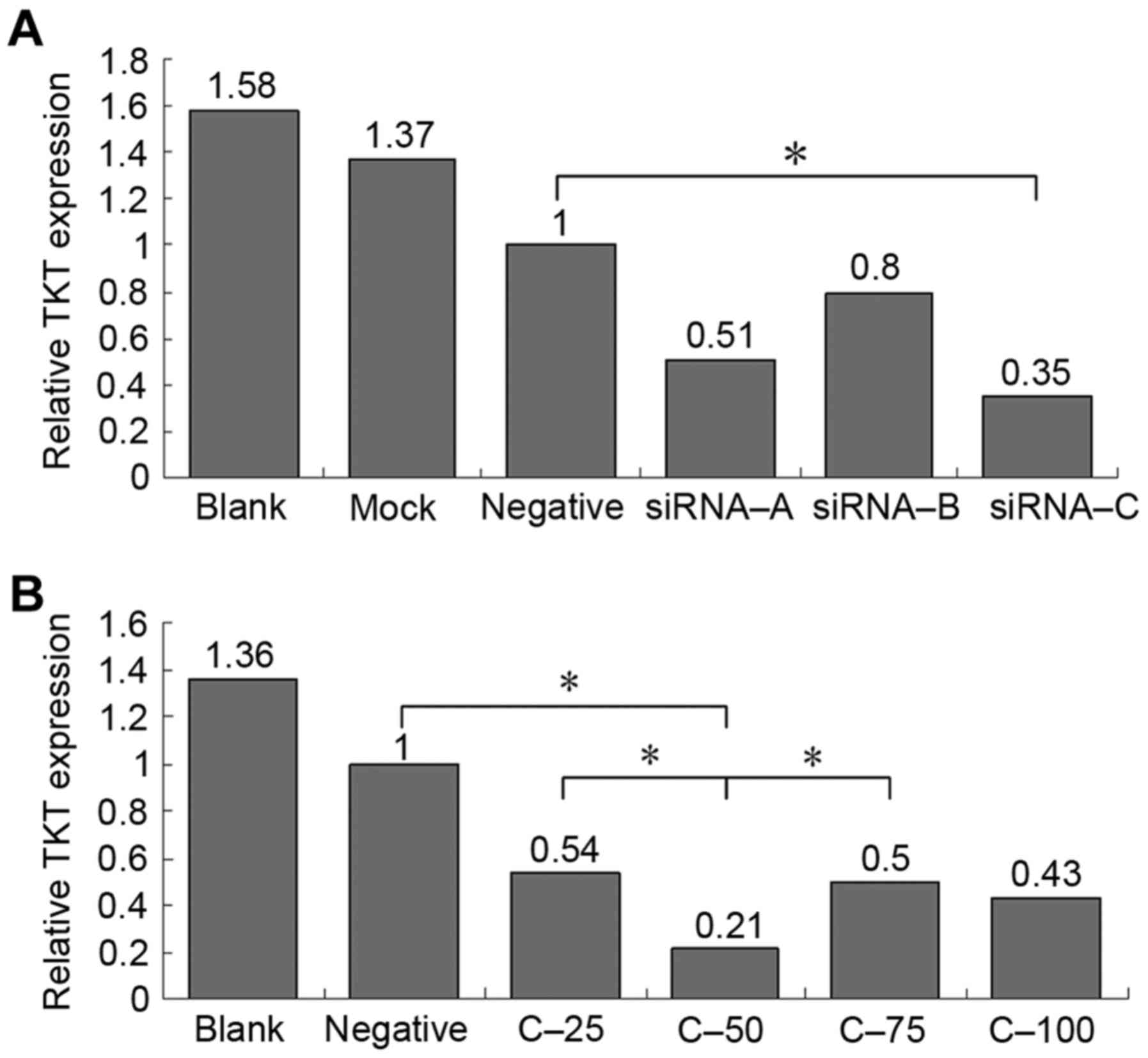
Fig. 2A presents the
TKT expression levels at 48 h of cell transfected with the
three TKT-specific siRNAs or negative control siRNA.
Compared with negative control group, the knockdown efficiency of
siRNA-C was the highest (TKT expression level, 0.35 relative
to the negative control group). Fig.
2B presents the TKT expression levels at 72 h in cells
transfected with various concentrations of siRNA-C. The results
demonstrated that a concentration of 50 nmol/l achieved the
greatest efficiency (TKT expression level, 0.21 relative to
the negative control group), and this was in agreement with the
flow cytometric analysis.
TKT knockdown decreases cell viability
and induces cell cycle arrest
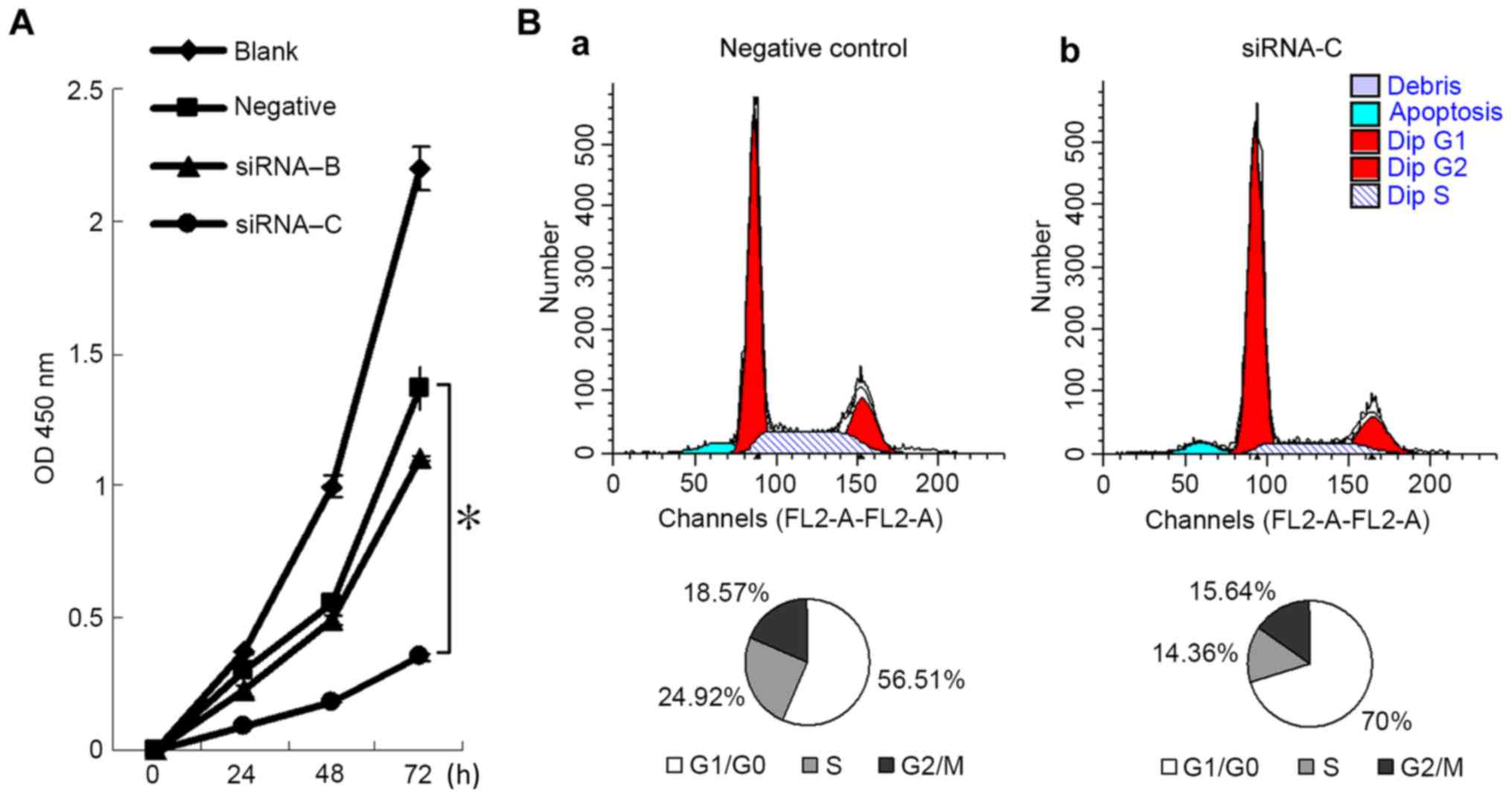
Fig. 3A presents the
cell viability of A549 cells following a 72-h transfection. After
24 h, the viability of cells transfected with siRNA-C was
significantly reduced compared with the negative control group [OD
values (mean ± standard deviation) of siRNA-C group vs. negative
control group were 0.2984±0.0371 vs. 0.0952±0.0063 (P<0.0001)
for 24 h; 0.5582±0.0090 vs. 0.1817±0.0037 (P<0.0001) for 48 h;
1.3663±0.0833 vs. 0.3519±0.0175 (P<0.0001)] for 72 h. This
indicated that downregulation of the TKT gene decreased the
viability of A549 cells.
Fig. 3B presents the
cell cycle distribution of the negative control and siRNA-C groups.
The proportions of cells in G1/G0 phase in
the negative control and siRNA-C groups were 56.51±2.0 and 70±2.5%,
respectively (P=0.002). The proportions of cells in G2/M
and S phases were 43.49±2.0 and 30±2.5% (P=0.002), respectively.
This suggests that the downregulation of TKT may arrest the
cell cycle of A549 cells in G1/G0 phase.
TKT knockdown decreases cell
migration
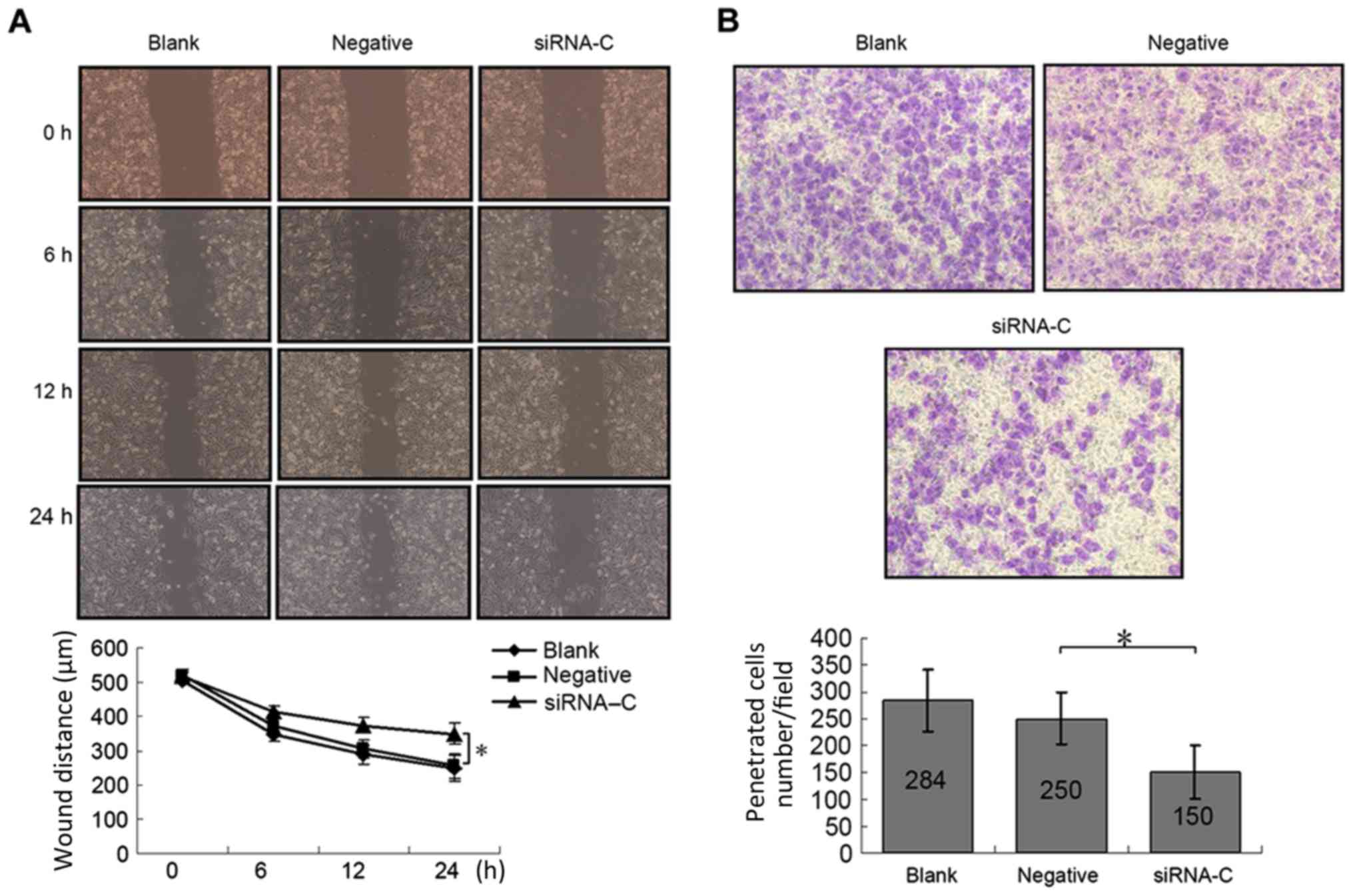
Compared with the negative control group, the wound
healing ability of cells in the siRNA-C group decreased markedly
(Fig. 4A); following a 24-h healing
period, the wound sizes of the negative control vs. siRNA-C groups
were 254.71±34.96 vs. 349.12±37.43 µm (P=0.0001). In the Transwell
migration assay (Fig. 4B), the number
of cells that migrated through the membrane was significantly
decreased in the siRNA-C group compared with the negative control
group (150±49.0/field vs. 250±47.8/field, respectively;
P<0.0001). This indicated that downregulation of TKT may
inhibit cell migration in A549 lung cancer cells.
TKT knockdown decreases the mRNA
expression levels of other metabolism-associated genes
As the key enzyme of the non-oxidative pathway, a
decreased expression level of TKT can influence the
expression levels of other key metabolic enzymes. The knockdown of
TKT induced the downregulation of G6PDH, TAL,
PRPS1, SORD and HK1 mRNA in A549 cells
(Fig. 5).
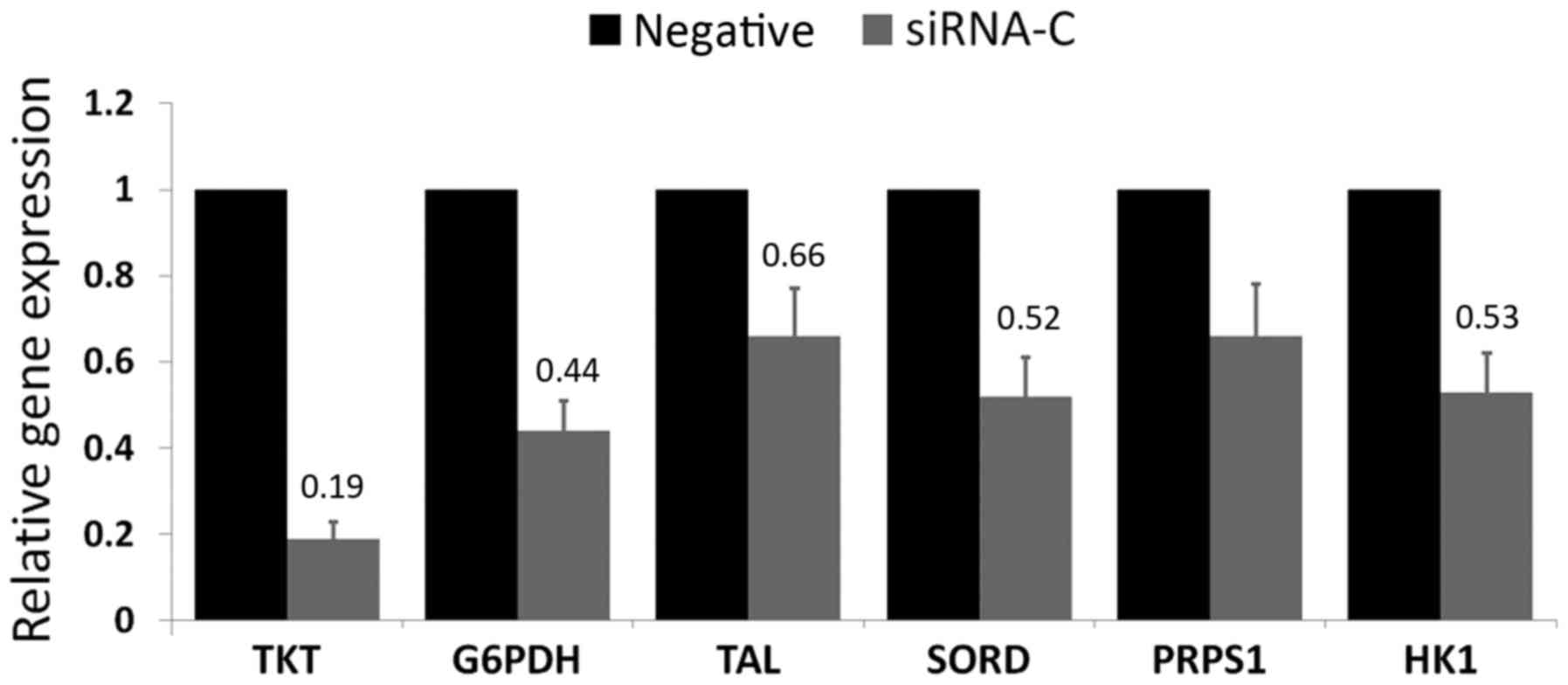 | Figure 5.mRNA expression levels of TKT
and other key metabolic enzymes determined by reverse
transcription-quantitative polymerase chain reaction. Relative to
the negative control group (set as 1.0), the expression levels of
TKT, G6PDH, TAL, SORD, PRPS1 and
HK1 were 0.19, 0.44, 0.66, 0.52, 0.66 and 0.53,
respectively. Results are presented as the mean ± standard
deviation. Each sample was performed in triplicate. TKT,
transketolase; TAL, transaldolase; G6PDH, glucose-6-phosphate
dehydrogenase; PRPS1, phosphoribosyl pyrophosphate synthetase 1;
SORD, sorbitol dehydrogenase; HK1, hexokinase 1. |
Detection of TKT family genes
The human TKT gene family includes
TKT, TKTL1 and TKTL2 (9). In the present study, the mRNA expression
levels of TKTL1 and TKTL2 were detected in untreated
A549 lung cancer cells using RT-qPCR. The results revealed that the
Cq values of TKT, TKTL1-1, TKTL1-2, TKTL1-3 and TKTL2 were
20.73±1.35, 31.97±1.71, 33.42±1.33, 34.71±0.79, respectively. This
indicated that the expression levels of TKTL1 and
TKTL2 mRNAs are low or absent in these cells.
Discussion
The non-oxidative pathway mediated by TKT exploits
the intermediates of glycolysis to synthesize ribose phosphate
reversibly (6). A previous study
confirmed that the non-oxidative pathway serves a vital role in
malignant cell proliferation and metabolism (10). High activity and expression levels of
TKT have been identified in multiple tumor types (11). Furthermore, enhanced TKT activity may
be a biomarker for tumor relapse and metastasis, indicating a poor
prognosis (12).
The results of the present study indicated that the
downregulation of TKT and decrease of ribose reduced A549
cell proliferation rate markedly, and arrested cell cycle at the
G1/G0 phase. Targeting metabolic enzymes has
great potential as an anticancer therapy. As a coenzyme of TKT,
thiamine (vitamin B1) may be a target for tumor therapy, and its
analogues, oxythiamine and pyrithiamine, have been demonstrated to
inhibit tumor growth significantly (4,13).
However, thiamine is also a coenzyme for pyruvate dehydrogenase and
ketoglutarate dehydrogenase, which are key enzymes of the Krebs
cycle. Furthermore, thiamine deficiency may affect
thiamine-dependent enzymes dysfunction to various degrees and
transketolase is the most vulnerable, induced by thiamine
deficiency compared with other thiamine-dependent enzymes (pyruvate
dehydrogenase and ketoglutarate dehydrogenase) (14). Previous studies have revealed that
numerous natural compounds, including sugiol and epicatechin
gallate, may inhibit cancer cell growth via the disturbance of TKT
(15,16). In the present study, TKT-siRNA
inhibited A549 cell growth via G0/G1 cell
cycle arrest, in agreement with previous experiments, and decreased
the expression of G6PDH mRNA, which is the key enzyme of the
oxidative signaling pathway. Jung et al (16) examined intracellular glutathione
expression levels in TKT-knockdown cells, and revealed that
glutathione levels were decreased, which induced reactive oxygen
species production in DU145 cells. Thus, the presence of TKT was
necessary for the production of the antioxidant enzyme NADPH and
TKT knockdown increased reactive oxygen species generation,
which may induce inflammation-injury (16).
TKT is associated with numerous metabolic
signaling pathways, including the polyol pathway, which includes
SORD as its key enzyme (17).
SORD expression was revealed to be decreased in
TKT-knockdown cells in the present study. The polyol pathway
provides substantial fructose for entry into the non-oxidative
signaling pathway. When the non-oxidative pathway is inhibited,
metabolites accumulate, fructose demand is reduced and SORD
expression decreases. Fig. 6 presents
the pentose phosphate pathway and its associated metabolic
signaling pathways.
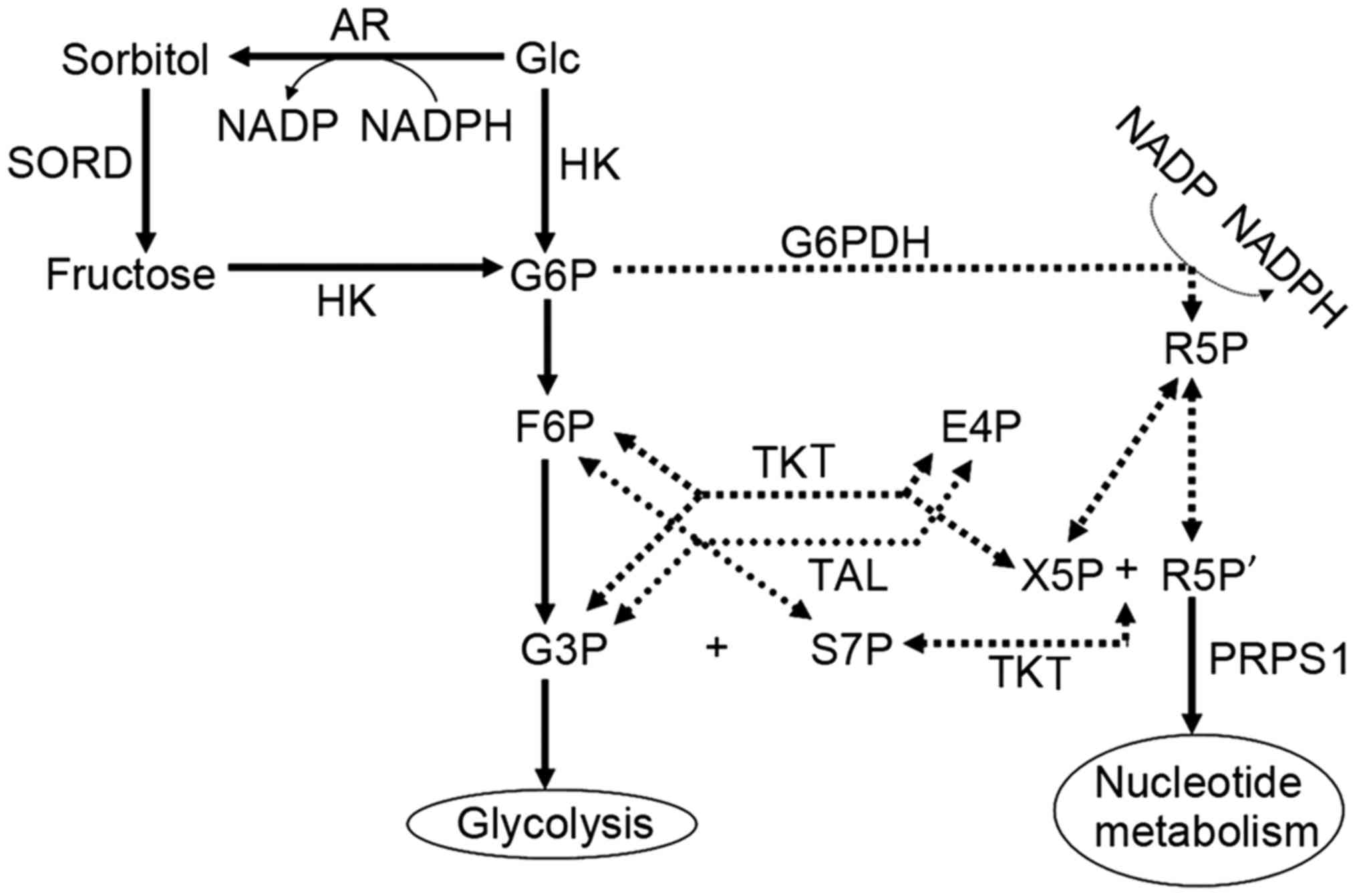 | Figure 6.Pentose phosphate pathway (dotted
lines) and associated metabolic pathways (solid lines). TKT,
transketolase; TAL, transaldolase; G6PDH, glucose-6-phosphate
dehydrogenase; HK, hexokinase; AR, aldose reductase; SORD, sorbitol
dehydrogenase; Glc, glucose; G6P, glucose 6-phosphate; F6P,
fructose 6-phosphate; R5P, ribose 5-phosphate; G3P, glyceraldehyde
3-phosphate; E4P, erythrose 4-phosphate; S7P, sedoheptulose
7-phosphate; X5P, xylulose 5-phosphate; R5P', ribulose 5-phosphate;
NADP, nicotinamide-adenine dinucleotide phosphate; NADPH, reduced
NADP; PRPS1, phosphoribosyl pyrophosphate synthetase 1. |
The human TKT gene family includes three
members: TKT, TKTL1 and TKTL2. TKTL1
was considered to be a pseudogene until Coy et al (18) identified its encoded protein and named
it in 2005; however, there are controversies over the results of
previous studies on TKTL1 (19,20), and
whether TKTL1 can be regarded as a potential tumor marker
requires further data for validation.
In conclusion, silencing TKT mRNA expression
by RNAi can inhibit A549 lung cancer cell proliferation and cell
migration, induce cell cycle arrest, and lead to the downregulation
of other metabolic enzyme mRNAs. TKT may be a potential
target for tumor therapy in the future. Further research
investigating the mechanism underlying oncotherapy mediated by RNAi
may provide a powerful tool for clinical application.
Acknowledgements
The present study was supported by a Basic Research
Key Project from the Shanghai Science and Technology Commission
(grant no. 12JC1402202) and the Key Project from the Shanghai
Health Technical Committee (grant no. 201440041).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
3
|
Patra KC and Hay N: The pentose phosphate
pathway and cancer. Trends Biochem Sci. 39:347–354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raïs B, Comin B, Puigjaner J, Brandes JL,
Creppy E, Saboureau D, Ennamany R, Lee WN, Boros LG and Cascante M:
Oxythiamine and dehydroepiandrosterone induce a G1 phase cycle
arrest in Ehrlich's tumor cells through inhibition of the pentose
cycle. FEBS Lett. 456:113–118. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salas E, Roy S, Marsh T, Rubin B and
Debnath J: Oxidative pentose phosphate pathway inhibition is a key
determinant of antimalarial induced cancer cell death. Oncogene.
35:2913–2922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Williams JF, Arora KK and Longenecker JP:
The pentose pathway: A random harvest. Impediments which oppose
acceptance of the classical (F-type) pentose cycle for liver, some
neoplasms and photosynthetic tissue. The case for the L-type
pentose pathway. Int J Biochem. 19:749–817. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu H, Lan WX, Bo L, Niu C, Zhou JJ and Zhu
HL: Metabolic response of LLC xenografted mice to oxythiamine, as
measured by [1H] NMR spectroscopy. Genet Mol Res. 14:11043–11051.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boros LG, Torday JS, Lim S, Bassilian S,
Cascante M and Lee WN: Transforming growth factor beta2 promotes
glucose carbon incorporation into nucleic acid ribose through the
nonoxidative pentose cycle in lung epithelial carcinoma cells.
Cancer Res. 60:1183–1185. 2000.PubMed/NCBI
|
|
11
|
Lin CC, Chen LC, Tseng VS, Yan JJ, Lai WW,
Su WP, Lin CH, Huang CY and Su WC: Malignant pleural effusion cells
show aberrant glucose metabolism gene expression. Eur Respir J.
37:1453–1465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ricciardelli C, Lokman NA, Cheruvu S, Tan
IA, Ween MP, Pyragius CE, Ruszkiewicz A, Hoffmann P and Oehler MK:
Transketolase is upregulated in metastatic peritoneal implants and
promotes ovarian cancer cell proliferation. Clin Exp Metastasis.
32:441–455. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang CM, Liu YZ, Liao JW and Hu ML: The in
vitro and in vivo anti-metastatic efficacy of oxythiamine and the
possible mechanisms of action. Clin Exp Metastasis. 27:341–349.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pekovich SR, Martin PR and Singleton CK:
Thiamine deficiency decreases steady-state transketolase and
pyruvate dehydrogenase but not alpha-ketoglutarate dehydrogenase
mRNA levels in three human cell types. J Nutr. 128:683–687.
1998.PubMed/NCBI
|
|
15
|
Sánchez-Tena S, Alcarraz-Vizán G, Marín S,
Torres JL and Cascante M: Epicatechin gallate impairs colon cancer
cell metabolic productivity. J Agric Food Chem. 61:4310–4317. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jung SN, Shin DS, Kim HN, Jeon YJ, Yun J,
Lee YJ, Kang JS, Han DC and Kwon BM: Sugiol inhibits STAT3 activity
via regulation of transketolase and ROS-mediated ERK activation in
DU145 prostate carcinoma cells. Biochem Pharmacol. 97:38–50. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uzozie A, Nanni P, Staiano T, Grossmann J,
Barkow-Oesterreicher S, Shay JW, Tiwari A, Buffoli F, Laczko E and
Marra G: Sorbitol dehydrogenase overexpression and other aspects of
dysregulated protein expression in human precancerous colorectal
neoplasms: A quantitative proteomics study. Mol Cell Proteomics.
13:1198–1218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coy JF, Dressler D, Wilde J and Schubert
P: Mutations in the transketolase-like gene TKTL1: Clinical
implications for neurodegenerative diseases, diabetes and cancer.
Clin Lab. 51:257–273. 2005.PubMed/NCBI
|
|
19
|
Mayer A, Von Wallbrunn A and Vaupel P:
Glucose metabolism of malignant cells is not regulated by
transketolase-like (TKTL)-1. Int J Oncol. 37:265–271. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hartmannsberger D, Mack B, Eggert C,
Denzel S, Stepp H, Betz CS and Gires O: Transketolase-like protein
1 confers resistance to serum withdrawal in vitro. Cancer Lett.
300:20–29. 2011. View Article : Google Scholar : PubMed/NCBI
|