Introduction
The morbidity associated with gastric carcinoma (GC)
has declined in recent decades; however, GC remains the fourth most
common carcinoma and second highest cause of cancer-associated
mortality worldwide (1). In 2012,
there werean estimated 951,600 new cases and 723,100 GC-associated
mortalities (1). Despite progression
in diagnosis and treatment methods, the prognosis for patients with
GC remains poor. Due toinconspicuous symptoms in the early stages,
the vast majority of patients with GC are already in the advanced
stages at the time of first diagnosis, resulting in a poor
prognosis (2,3). Therefore, the early diagnosis and
treatment of GC are of critical importance for improving the
clinical outcome.
Mena (also referred to as ENAH-enabled homolog) is a
member of the Ena/vasodilator-stimulated phosphoprotein (VASP)
family of actin-binding proteins, which function in diverse types
of cell (4,5). Ena/VASP proteins are key regulatory
molecules that control the cell shape, movement and actin
organization on cadherin adhesion contacts, which are frequently
affected following malignant transformation (4,6). Mena is a
key mediator of cytoskeletal arrangement (7). It regulates cell movement by protecting
actin filaments from capping proteins during polymerization
(8). An upregulated Mena expression
level was previously reported in mouse and rat invasive breast
carcinoma (9), as well as in human
breast cancer cell lines and tissues (10). Similarly, Mena expression was observed
to beupregulated in human hepatocellular carcinoma (11), colorectal carcinoma (12), cervix precursor lesions (13) and pancreatic tumor cell lines as well
as in primary and metastatic pancreatic tumors (14); in normal tissue, Mena expression level
was reported at low or non-detectable levels (11). However, the clinical significance of
Mena in GC remains indistinct. The present study investigated the
expression level of Mena in GC to reveal its clinicopathological
significance.
Materials and methods
Patients and tissue samples
The present study was performed with 106 GC
paraffin-embedded tissue samples collected during resection from
the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou,
China) between January 2001 and December 2004 for
immunohistochemical (IHC) analysis. The median age of the patients
was 54 years old (range, 29–72 years) and the median tumor size was
6.0 cm (range, 0.8–15.0 cm); the group included 67 male and 39
female patients. From these 106 patients, 32 samples of adjacent
non-cancerous tissues were additionally collected as control
samples. All patients were pathologically diagnosed with gastric
adenocarcinoma. None of the patients had received any type of
neoadjuvant therapy and all underwent a radical excision. The
clinical information for these samples is summarized in Table I. The date of patient surgery was
defined as the initial event of survival analysis, and the date of
patient mortality or the censoring of the patient at the last
follow-up date was defined as the end time. The interval was
defined as the overall survival time for patients.
 | Table I.Association between Mena expression
level and clinicopathological characteristics. |
Table I.
Association between Mena expression
level and clinicopathological characteristics.
|
|
| Mena expression
status |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Total | Negative (%) | Positive (%) | P-value |
|---|
| Total | 106 | 50 | 56 |
|
| Gender |
|
|
| 0.573 |
|
Male | 67 (63.2) | 33 (49.3) | 34 (50.7) |
|
|
Female | 39 (36.8) | 17 (43.6) | 22 (56.4) |
|
| Age (years) |
|
|
| 0.065 |
|
≥60 | 46 (43.4) | 17 (37.0) | 29 (63.0) |
|
| <60 | 60 (56.6) | 33 (55.0) | 27 (45.0) |
|
| T stage |
|
|
| 0.033 |
| 1 | 10 (9.4) | 8 (80) | 2 (20) |
|
| 2 | 10 (9.4) | 7 (70) | 3 (30) |
|
| 3 | 84 (79.2) | 34 (40.5) | 50 (59.5) |
|
| 4a | 2 (1.9) | 1 (50) | 1 (50) |
|
| N stage |
|
|
| 0.313 |
| 0 | 21 (19.8) | 13 (61.9) | 8 (38.1) |
|
| 1 | 38 (35.8) | 17 (44.7) | 21 (55.3) |
|
| 3 | 47 (44.3) | 20 (42.6) | 27 (57.4) |
|
| M stage |
|
|
| 0.813 |
| 0 | 99 (93.4) | 47 (47.5) | 52 (52.3) |
|
| 1 | 7 (6.6) | 3 (42.9) | 4 (57.1) |
|
| TNM stage |
|
|
| <0.001 |
| I | 13 (12.3) | 12 (92.3) | 1 (7.7) |
|
| II | 18 (17.0) | 14 (77.8) | 4 (22.2) |
|
|
III | 68 (64.2) | 23 (33.8) | 45 (66.2) |
|
| IV | 7 (6.6) | 1 (14.3) | 6 (85.7) |
|
| Tumor size
(cm) |
|
|
| 0.419 |
| ≥5 | 74 (69.8) | 33 (44.6) | 41 (55.4) |
|
|
<5 | 32 (30.2) | 17 (53.1) | 15 (46.9) |
|
| Grade |
|
|
| 0.570 |
| 1 | 4 (3.8) | 3 (75) | 1 (25) |
|
| 2 | 25 (23.6) | 11 (44.0) | 14 (56.0) |
|
| 3 | 76 (71.7) | 36 (47.4) | 40 (52.6) |
|
| 4 | 1 (9) | 0 (0) | 1 (100) |
|
| Infiltration |
|
|
| 0.742 |
| 0 | 101 (95.3) | 48 (47.5) | 53 (52.5) |
|
| 1 | 5 (4.7) | 2 (40.0) | 3 (60.0) |
|
In addition, 10 paired GC and adjacent normal
tissues (the adjacent normal tissue was defined as at least 5cm
from the tumor edge) were collected from the Third Affiliated
Hospital of Sun Yat-sen University between June 2013 and February
2015 for reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) analysis. The group included 7 male and 3 female
patients, and the median age of the patients was 51 years old
(range, 32–69 years). Tissues were collected immediately after
surgery.
The clinicopathological classification and staging
were determined according to the American Joint Committee on Cancer
criteria (15). Written informed
consent was obtained from all patients prior to enrollment in the
present study. The present study was approved by the Institutional
Research Ethics Committee of the Third Affiliated Hospital of Sun
Yat-sen University.
RT-qPCR analysis
Total RNA samples were extracted from 10 paired GC
and adjacent normal tissues using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. Extracted RNA was
pretreated with RNase-free DNase. For cDNA synthesis, 2 µg RNA from
each sample was used, according to the RevertAid™ First Strand cDNA
Synthesis kit instructions (K1622; Thermo Fisher Scientific,
Inc.).
For the PCR amplification of Mena cDNA, SYBR-Green
2X master mixture (170-8882AP; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used in a total volume of 20 µl, according
to the manufacturer's instructions, an initial amplification step
using Mena-specific primers was performed with a denaturation at
95°C for 10 min, which was followed by 28 denaturation cycles at
95°C for 60 sec, primer annealing at 58°C for 30 sec and primer
extension at 72°C for 30 sec. Upon completion of the cycling steps,
a final extension at 72°C for 5 min was performed prior to the
storage of the reaction mixture at 4°C. The primer sequences were
as follows: Mena sense, 5′-GTGCCATTCCTAAAGGGTTGA-3′ and antisense,
5′-GCTGCCAAAGTTGAGACCATAC-3′; GAPDH sense,
5′-TGTTGCCATCAATGACCCC-3′ and antisense, 5′-CTCCACGACGTACTCAGC-3′.
The primers were designed with Primer Express version 2.0 software
(Applied Biosystems; Thermo Fisher Scientific, Inc.).
GAPDH was used as an internal control, the relative
expression level of Mena was calculated using the 2−ΔΔCT
method (16); all experiments were
performed in triplicate.
IHC analysis
IHC staining was performed to investigate the
alteration to protein expression levels in 106 human GC tissues and
32 paired adjacent non-cancerous tissues. Briefly, 4-µm-thick
paraffin sections of the tissue were deparaffinized with xylene and
rehydrated in a descending alcohol series. Antigenic retrieval was
performed by submerging the slides in EDTA antigenic retrieval
buffer and microwave heating for 3 min at 650 W and thentwice more
at 350 W for 3 min. To quench endogenous peroxidase activity, the
slides were treated with 3% hydrogen peroxide in methanol and then
incubated with 1% bovine serum albumin (Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) at room temperature for 60 min to block
nonspecific binding. Subsequently, tissue sections were incubated
with a rabbit polyclonal anti-Mena antibody (dilution, 1:100; BD
Biosciences, Franklin Lakes, NJ, USA, catalog number: 5111-1) at
4°C overnight. Normal goat serum (Santa Cruz Biotechnology, Inc.)
was used at 4°C overnight as a negative control. The tissue
sections were incubated with a biotinylated anti-rabbit secondary
antibody (no dilution; Santa Cruz Biotechnology, Inc.; catalog
number: sc-2040) at room temperature for 30 min following 3 washes
in PBS, followed by further incubation with a
streptavidin-horseradish peroxidase complex (dilution, 1:1500;
Abcam, Cambridge, UK; catalog number: ab7403) at room temperature
for 30 min. Slides were immersed in 3-amino-9-ethyl carbazole room
temperature for 3 min and then counterstained with 10% Mayer's
hematoxylin at room temperature for 30 sec. Finally, they were
dehydrated and mounted with Crystal Mount.
Slides were imaged at magnification ×20 (0.5×0.5
µm2 pixel resolution) using a WSI instrument (ScanScope
CS, Aperio, Vista, CA, USA) fitted with a 20×/0.75 Plan Apo
objective lens (Olympus, Center Valley, PA, USA). For the
evaluation of immunostaining, the degree of immunostaining was
viewed and scored independently by two pathologists, who were
blinded to the histopathological characteristics and patient
information for the samples. The mean value of the scores provided
by the two independent pathologists was used for the comparative
evaluation of Mena expression.
The intensity of Mena staining was graded according
to the following criteria: 0, no staining; 1, weak staining (light
yellow); 2, moderate staining (yellow brown); and 3, strong
staining (brown). The percentage of stained tumor cells was scored
as follows: 0, no positive tumor cells; 1, 1–25% positive tumor
cells; 2, 26–50% positive tumor cells; 3, 51–75% positive tumor
cells; and 4, >75% positive tumor cells.
The staining score was evaluated as the product of
the proportion of positive tumor cells and the staining intensity
score. The expression level of Mena was defined as follows: ‘−’
(score 0, negative), ‘+’ (score 1–4, weakly positive), ‘++’ (score
5–8, positive) and ‘+++’ (score 9–12, strongly positive). Optimal
cut-off values for Mena expression were selected based on the
analysis of overall survival (OS) data with the log-rank test. A
staining index score of ≥4 was used to define tumors with high Mena
expression level whereas <4 indicated a low Mena expression
level.
Statistical analysis
All statistical analyses were performed using SPSS
20.0 software (IBM Corp., Armonk, NY, USA). The difference in Mena
expression levels between GC tissues and adjacent non-cancer
tissues were analyzed using the χ2 test. Survival curves
were plotted using the Kaplan-Meier method and compared using the
log-rank test. The association between Mena expression level and
other clinicopathological characteristics was analyzed usingthe
χ2 and Fisher's exact tests. Bivariate correlations
between the clinicopathological characteristics were determined
using Spearman's rank correlation coefficients. Clinicopathological
characteristics used to predict the prognosis in clinical practice
were evaluated by univariate and multivariate Cox regression
analyses. The selected type of Cox model for the univariate
analysis was the ‘enter’ method, and for the multivariate analysis,
the ‘forward’ method. P<0.05 was considered to indicate a
statistically significant difference.
Results
Mena is overexpressed in GC
tissues
To determine whether Mena expression isupregulated
in human GC, RT-qPCR was performed on 10 paired GC and adjacent
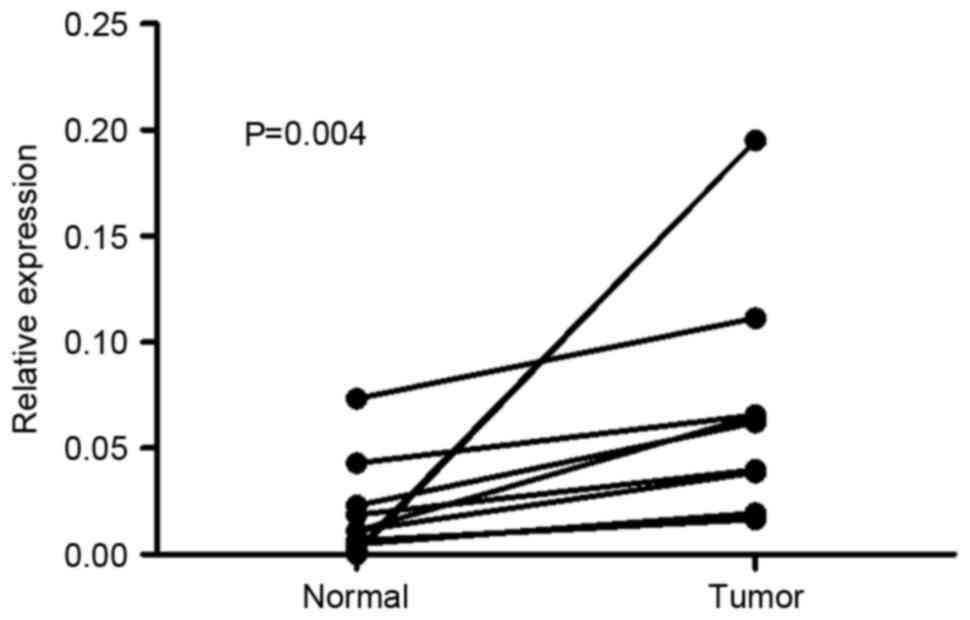
normal tissues. As presented in Fig.
1, the expression level of Mena mRNA was higher in all 10 GC
tissue samples compared with in adjacent normal tissues, with the
difference in expression level ranging from 1.5 to 84-fold. In IHC
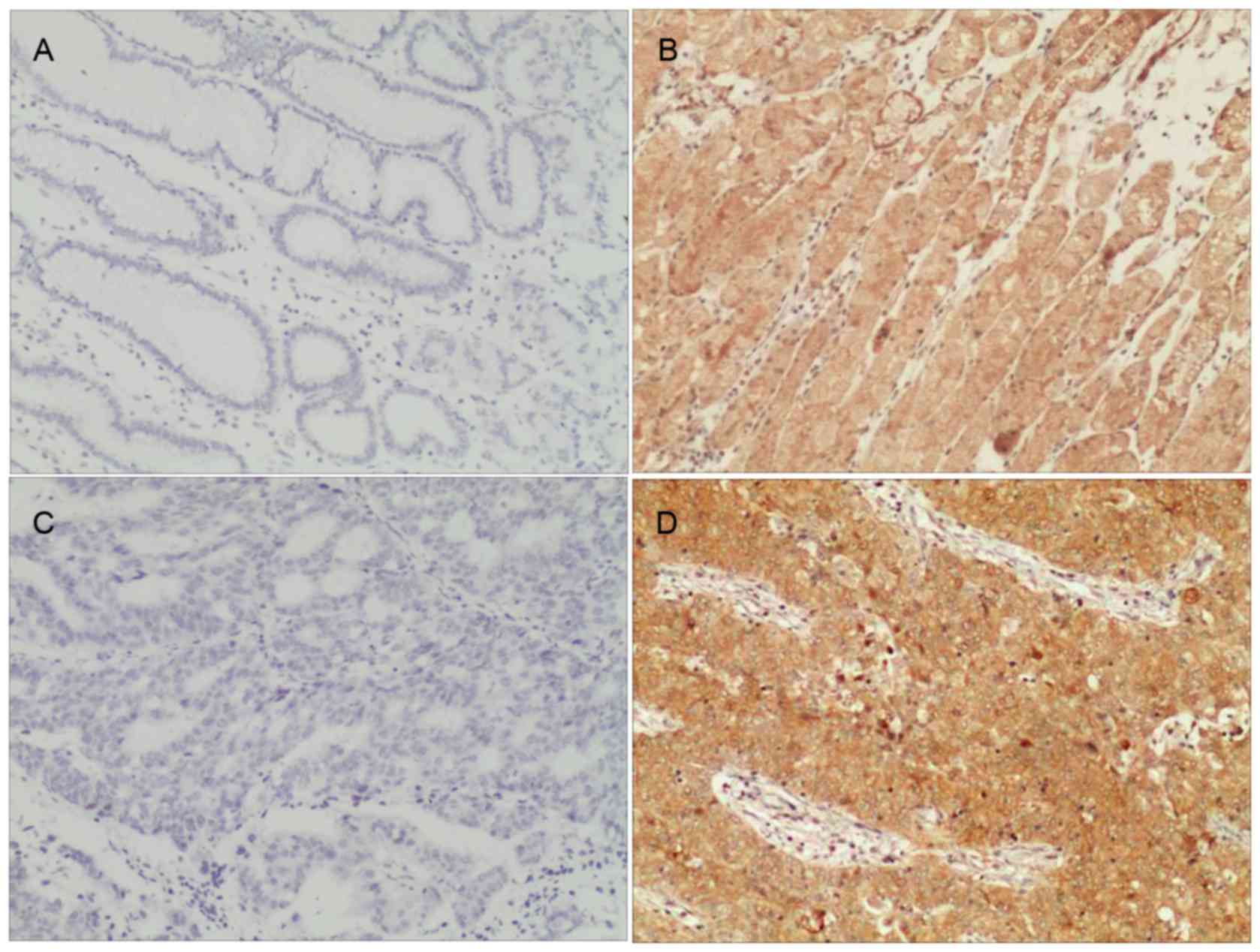
results, the high expression of Mena was observed in 52.83%
(56/106) of the patients with GC, whereas weak or no staining was
observed in 47.17% of the tumor samples (Table I). In the adjacent non-tumor tissues,
Mena protein staining was largely weak or absent; there was a 6.25%
(2/32) positive expressionrate detected. The difference between
these two groups was statistically significant
(χ2=18.910; P<0.001). As presented in Fig. 2, Mena staining occurred predominantly
in the cytoplasm.
Mena overexpression is associated with
GC clinical characteristics
To better understand the potential role of Mena in
the development and progression of GC, the association of Mena
expression level with other clinicopathological indexes in 106
paraffin-embedded archived GC tissues, including 10 stage I tumors,
10 stage II tumors, 84 stage III tumors and 2 stage IVa tumors, was
investigated.
As summarized in Table
I, there were no significant associations between Mena
expression level and the gender, age, node (N) or metastasis (M)
stage, tumor size, grade and the infiltration of adjacent organs in
the patients; however, the expression level of Mena was
significantly associated with the tumor (T; P=0.033) and TNM stages
(P<0.001).
Association between Mena expression
level and overall patient survival time
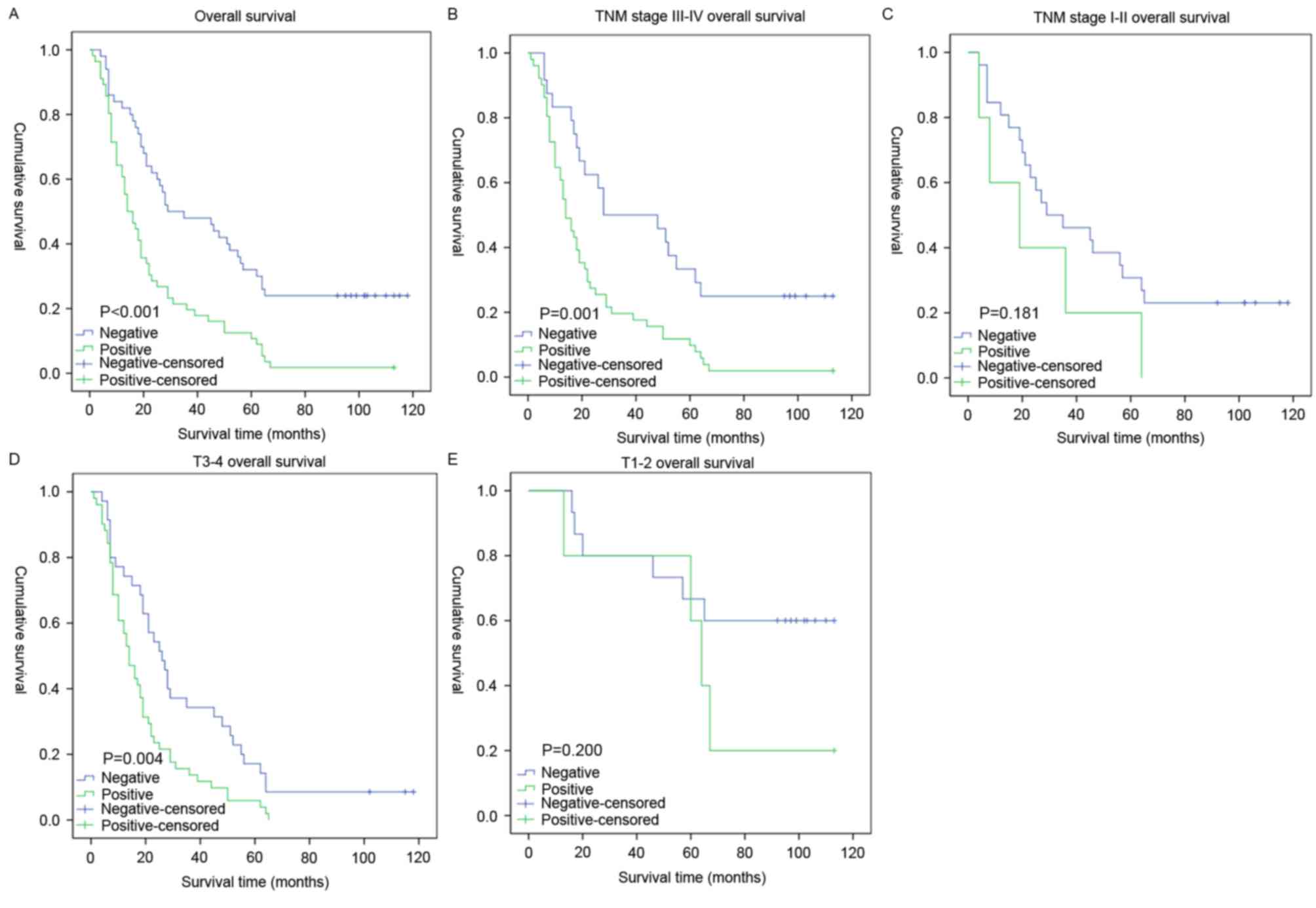
Survival analysis revealed a clear negative
association between the expression level of Mena protein and the OS
time of patients with GC (P<0.001; Fig. 3A). In addition, Cox regression
analysis revealed that Mena expression level, T stage and N stage
were independent prognostic factors for OS time (Table II).
 | Table II.Cox-regression analysis of various
prognostic parameters in patients. |
Table II.
Cox-regression analysis of various
prognostic parameters in patients.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| N stage |
| <0.001 |
| 0.002 |
| 0 | Reference |
| Reference |
|
| 1 | 4.022
(1.955–8.274) |
| 1.718
(0.784–3.766) |
|
| 3 | 7.015
(3.421–14.386) |
| 3.273
(1.533–6.988) |
|
| Age |
| 0.001 |
|
|
|
≥60 | Reference |
|
|
|
|
<60 | 0.481
(0.319–0.727) |
|
|
|
| Tumor size
(cm) |
| 0.001 |
|
|
|
<5 | Reference |
|
|
|
| ≥5 | 0.439
(0.272–0.709) |
|
|
|
| Mena expression
status |
| <0.001 |
| 0.010 |
|
Negative | Reference |
|
|
|
|
Positive | 0.433
(0.284–0.661) |
|
0.463(0.296–0.724) |
|
| T stage |
|
0.001 |
| 0.005 |
| 1 | Reference |
| Reference |
|
| 2 | 17.539
(2.207–139.398) |
| 9.680
(1.142–82.080) |
|
| 3 | 36.233
(4.970–264.173) |
| 16.096
(1.974–133.049) |
|
| 4 | 16.855
(1.516–188.064) |
| 2.845
(0.217–37.316) |
|
The prognostic significance of Mena expression
status in selective subgroups stratified by the T stage and TNM
stage was analyzed. For patients with late-stage tumors (stage
III–IVa), the expression level of Mena was strongly associated with
the OS duration (Fig. 3B; P=0.001),
which was not the case for patients with early-stage tumors (stages
I–II; Fig. 3C; P=0.181). Similarly,
when it was evaluated according to T stage, the effect on outcome
associated with the expression level of Mena was significant only
in the T3-4 subgroup (Fig. 3D;
P=0.004), and not in the T1-2 subgroup (Fig. 3E, P=0.200).
Discussion
GC is the fourth most common type of cancer and the
second leading cause for cancer-associated mortality worldwide,
although it exhibits a decreasing trend of incidence (1,17). There
has been significant clinical progress in the early diagnosis and
treatment of GC during recent decades; however, it is usually
diagnosed at a late stage, resulting in a high treatment cost and
decreasing the rate of successful curative surgery (18). The 5-year OS rate for GC is closely
associated with the tumor stage. Patients diagnosed at stage I
exhibit a 5-year OS rate of >90%, whereas patients diagnosed at
stage IV exhibit a 5-year OS rate of <5% (19). Therefore, there is currently a great
clinical demand for early diagnosis and treatment, which are
pivotal for improving the outcome of GC.
Classical serum tumor markers, including
carcinoembryonic antigen and carbohydrate antigen 19-9 have
definite implications for GC diagnosis and monitoring, but the lack
of specificity and sensitivity impaired their function (20). In recent years, there have been
multiple novel tissue-based biomarkers for GC identified, including
vascular endothelial growth factor receptor 2 (21), excision repair cross-complementation
group 1 (22), human epidermal growth
factor receptor-2 (23,24), Bcl-2 and Ki-67 (25). However, most of these molecular
markers are not conventionally used in the clinical setting as they
do not accurately and efficiently predict the clinical outcome or
curative effect. Novel tumor molecular markers are thus required to
improve the detection, diagnosis and prognosis of GC.
Human ortholog of murine Mena, a member of the
Ena/VASP protein family that includes Mena, VASP and Evl in
mammals, is a key actin polymerization regulatory protein involved
in the assembly and dynamics of cytoplasmic actin networks
(26). The Ena/VASP family is an
important regulator of actin cytoskeleton dynamics involved in cell
motility. Additionally, alterations to the cellular actin network
serve an important role in malignant transformation and tumor
progression. Members of the Ena/VASP family that are localized at
the tips of protruding filopodia and lamellipodia and adhesion foci
function in the control of cell movement, shape and adhesion, which
are important biological processesin the development of metastatic
potential (27). Located on
chromosome 1, the Mena gene encodes the 570-residue Mena protein
and alternative splicing-derived isoforms (28). As a member of the Ena/VASP family,
Mena regulates membrane protrusion and cell movement in various
types of cells and contexts by influencing the geometry and
assembly of actin filament networks through the binding of G-actin
and F-actin (26,29–32).
Mena enhances tumor cell migration toward epidermal
growth factor (EGF) in part by interfering with the activity of the
inhibitory capping proteins and increasing actin filament
elongation, promoting actin polymerization (26,33,34). The
anti-capping activity of Mena is proposed to amplify the barbed end
output of the cofilin and Arp2/3 complex pathways, particularly in
response to EGF, which is important in the metastatic potential of
mammary tumors (26,29,35). Di
Modugno et al (28) revealed
that Mena is overexpressed in 75% of primary mammary carcinomas;
consistent with this observation, high expression levels of Mena in
breast cancer patients have been associated with poor prognosis
(28,36). Similarly, in precancerous lesions of
the cervix and colon, the expression of Mena was upregulated with
progressive transformation (13,37). It
was also detected in pancreatic carcinoma cell lines and in primary
and metastatic pancreatic tumor tissues (28,36,38). Mena
maintains the stability of invadopodia, actin-rich protrusions that
contain proteases, increasing the matrix degradation activity of
tumor cells. Mena activity potentiates EGF-induced tumor cell
invasion and membrane protrusion. These previous studies
demonstrate that the overexpression of Mena in cancer enables the
invasion and metastasis of tumor cells in response to otherwise
benign EGF stimulus levels, increasingthe responsiveness to
macrophage signaling (26).
The present study presented novel evidence that the
upregulation of Mena was associated with poor clinical outcomes in
patients with GC, particularly for those with late-stage disease.
It was clearly demonstrated that in GC tissues, the expression of
Mena at the mRNA and protein levels was markedly higher compared
with in the adjacent normal tissues. Therefore, Mena may be a
biomarker for GC, which may aid precise diagnoses. However, at
present, the precise functions of Mena in human cancer remain
unclear. The overexpression of Mena in GC may reflect the aberrant
regulation of actin dynamics. However, understanding of the precise
mechanism underlying Mena in GC requires further investigation.
The present study additional lyinvestigated the
association between Mena expression level and other clinical
features of patients with GC. There was a significant association
between Mena expression level and the T and TNM stages, which
revealed that Mena may be used as an independent biomarker for the
recognition of a subpopulation of GC patients with more aggressive
disease. However, the associations between Mena and the gender,
age, N stage, M stage, tumor size, grade, and infiltration in
patients with GC were not significant.
Previous studies have reported the prognostic value
of Mena in human cancer. For example, numerous studies have
observed that high expression levels of Mena areassociated with a
poor prognosis in patients with breast cancer (28,36).
However, to the best of our knowledge, the prognostic value of Mena
in GC has not previously been explored. In the present study,
patients with high Mena expression levels had a 1.79% cumulative
10-year survival rate, which was significantly lower than patients
in the low Mena expression group (24.0%). Multivariate analysis
revealed that the expression level of Mena may be an independent
prognostic factor for OS time in GC patients (Table II). Of note, a sub-group analysis
demonstrated that patients with high Mena expressionand poor
clinical out comes also demonstrated the features of late TNM and T
stages.
In conclusion, to the best of our knowledge, this is
the first study to investigate Mena expression level and its
clinicopathological and prognostic significance in GC. The results
of the present study suggested that Mena was upregulated in GC
tissues and associated with the T and TNM stages. Multivariate
analysis revealed that Mena may be an independent molecular marker
for the prediction of GC prognosis and survival. Therefore,
detecting the Mena protein expression level may aid the
stratification of patients as a novel therapeutic strategy and
establish a rational treatment selection criterion for patients
with GC. Further, in-depth study will berequired to investigate the
molecular mechanism underlying Mena involvement in the development
and progression of GC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81672661 and
81502268) and grants from the Guangdong Province Natural Science
Foundation (grant nos. 2015A030310126 and 2015A030313182).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim BS, Cho SW, Min SK and Lee BH:
Differences in prognostic factors between early and advanced
gastric cancer. Hepatogastroenterology. 58:1032–1040.
2011.PubMed/NCBI
|
|
3
|
Saragoni L: Upgrading the definition of
early gastric cancer: Better staging means more appropriate
treatment. Cancer Biol Med. 12:355–361. 2015.PubMed/NCBI
|
|
4
|
Krause M, Dent EW, Bear JE, Loureiro JJ
and Gertler FB: Ena/VASP proteins: Regulators of the actin
cytoskeleton and cell migration. Annu Rev Cell Dev Biol.
19:541–564. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang D, Zhang X, Huang S, Yuan H, Li J and
Wang Y: Mena-GRASP65 interaction couples actin polymerization to
Golgi ribbon linking. Mol Biol Cell. 27:137–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen XJ, Squarr AJ, Stephan R, Chen B,
Higgins TE, Barry DJ, Martin MC, Rosen MK, Bogdan S and Way M:
Ena/VASP proteins cooperate with the WAVE complex to regulate the
actin cytoskeleton. Dev Cell. 30:569–584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi K and Suzuki K: WAVE2, N-WASP,
and Mena facilitate cell invasion via phosphatidylinositol
3-kinase-dependent local accumulation of actin filaments. J Cell
Biochem. 112:3421–3429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barzik M, Kotova TI, Higgs HN, Hazelwood
L, Hanein D, Gertler FB and Schafer DA: Ena/VASP proteins enhance
actin polymerization in the presence of barbed end capping
proteins. J Biol Chem. 280:28653–28662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Wyckoff JB, Goswami S, Wang Y,
Sidani M, Segall JE and Condeelis JS: Coordinated regulation of
pathways for enhanced cell motility and chemotaxis is conserved in
rat and mouse mammary tumors. Cancer Res. 67:3505–3511. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Du JW, Xu KY, Fang LY and Qi XL: Clinical
significance of Mena and Her-2 expression in breast cancer. Eur J
Gynaecol Oncol. 33:455–458. 2012.PubMed/NCBI
|
|
11
|
Hu K, Wang J, Yao Z, Liu B, Lin Y, Liu L
and Xu L: Expression of cytoskeleton regulatory protein Mena in
human hepatocellular carcinoma and its prognostic significance. Med
Oncol. 31:9392014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toyoda A, Kawana H, Azuhata K, Yu J, Omata
A, Kishi H, Higashi M and Harigaya K: Aberrant expression of human
ortholog of mammalian enabled (hMena) in human colorectal
carcinomas: Implications for its role in tumor progression. Int J
Oncol. 34:53–60. 2009.PubMed/NCBI
|
|
13
|
Gurzu S, Jung I, Prantner I, Chira L and
Ember I: The immunohistochemical aspects of protein Mena in
cervical lesions. Rom J Morphol Embryol. 50:213–216.
2009.PubMed/NCBI
|
|
14
|
Pino MS, Balsamo M, Di Modugno F,
Mottolese M, Alessio M, Melucci E, Milella M, McConkey DJ,
Philippar U, Gertler FB, et al: Human Mena+11a isoform serves as a
marker of epithelial phenotype and sensitivity to epidermal growth
factor receptor inhibition in human pancreatic cancer cell lines.
Clin Cancer Res. 14:4943–4950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rutkowski P, Wozniak A, Debiec-Rychter M,
Kąkol M, Dziewirski W, Zdzienicki M, Ptaszynski K, Jurkowska M,
Limon J and Siedlecki JA: Clinical utility of the new American
Joint Committee on Cancer staging system for gastrointestinal
stromal tumors: Current overall survival after primary tumor
resection. Cancer. 117:4916–4924. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arocho A, Chen B, Ladanyi M and Pan Q:
Validation of the 2-DeltaDeltaCt calculation as an alternate method
of data analysis for quantitative PCR of BCR-ABL P210 transcripts.
Diagn Mol Pathol. 15:56–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hamashima C: Current issues and future
perspectives of gastric cancer screening. World J Gastroenterol.
20:13767–13774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matsuda T, Ajiki W, Marugame T, Ioka A,
Tsukuma H and Sobue T; Research Group of Population-Based Cancer
Registries of Japan, : Population-based survival of cancer patients
diagnosed between 1993 and 1999 in Japan: A chronological and
international comparative study. Jpn J Clin Oncol. 41:40–51. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He CZ, Zhang KH, Li Q, Liu XH, Hong Y and
Lv NH: Combined use of AFP, CEA, CA125 and CAl9-9 improves the
sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol.
13:872013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fuchs CS, Tomasek J, Yong CJ, Dumitru F,
Passalacqua R, Goswami C, Safran H, dos Santos LV, Aprile G, Ferry
DR, et al: Ramucirumab monotherapy for previously treated advanced
gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An
international, randomised, multicentre, placebo-controlled, phase 3
trial. Lancet. 383:31–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamada Y, Boku N, Nishina T, Yamaguchi K,
Denda T, Tsuji A, Hamamoto Y, Konishi K, Tsuji Y, Amagai K, et al:
Impact of excision repair cross-complementing gene 1 (ERCC1) on the
outcomes of patients with advanced gastric cancer: Correlative
study in Japan Clinical Oncology Group Trial JCOG9912. Ann Oncol.
24:2560–2565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tewari M, Kumar A, Mishra RR, Kumar M and
Shukla HS: HER2 expression in gastric and gastroesophageal cancer:
Report from a tertiary care hospital in North India. Indian J Surg.
77 Suppl 2:S447–S451. 2015. View Article : Google Scholar
|
|
24
|
Ye P, Zhang M, Fan S, Zhang T, Fu H, Su X,
Gavine PR, Liu Q and Yin X: Intra-tumoral heterogeneity of HER2,
FGFR2, cMET and ATM in gastric cancer: Optimizing personalized
healthcare through innovative pathological and statistical
analysis. PLoS One. 10:e01432072015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou Y, Li Y, Zheng J, Liu K and Zhang H:
Detecting of gastric cancer by Bcl-2 and Ki67. Int J Clin Exp
Pathol. 8:7287–7290. 2015.PubMed/NCBI
|
|
26
|
Philippar U, Roussos ET, Oser M, Yamaguchi
H, Kim HD, Giampieri S, Wang Y, Goswami S, Wyckoff JB,
Lauffenburger DA, et al: A Mena invasion isoform potentiates
EGF-induced carcinoma cell invasion and metastasis. Dev Cell.
15:813–828. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kwiatkowski AV, Gertler FB and Loureiro
JJ: Function and regulation of Ena/VASP proteins. Trends Cell Biol.
13:386–392. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Di Modugno F, Bronzi G, Scanlan MJ, Del
Bello D, Cascioli S, Venturo I, Botti C, Nicotra MR, Mottolese M,
Natali PG, et al: Human Mena protein, a serex-defined antigen
overexpressed in breast cancer eliciting both humoral and CD8+
T-cell immune response. Int J Cancer. 109:909–918. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gertler FB, Niebuhr K, Reinhard M, Wehland
J and Soriano P: Mena, a relative of VASP and drosophila enabled,
is implicated in the control of microfilament dynamics. Cell.
87:227–239. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bear JE, Loureiro JJ, Libova I, Fässler R,
Wehland J and Gertler FB: Negative regulation of fibroblast
motility by Ena/VASP proteins. Cell. 101:717–728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drees F and Gertler FB: Ena/VASP: Proteins
at the tip of the nervous system. Curr Opin Neurobiol. 18:53–59.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Neel NF, Barzik M, Raman D,
Sobolik-Delmaire T, Sai J, Ham AJ, Mernaugh RL, Gertler FB and
Richmond A: VASP is a CXCR2-interacting protein that regulates
CXCR2-mediated polarization and chemotaxis. J Cell Sci.
122:1882–1894. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Entenberg D, Wyckoff J, Gligorijevic B,
Roussos ET, Verkhusha VV, Pollard JW and Condeelis J: Setup and use
of a two-laser multiphoton microscope for multichannel intravital
fluorescence imaging. Nat Protoc. 6:1500–1520. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goswami S, Philippar U, Sun D, Patsialou
A, Avraham J, Wang W, Di Modugno F, Nistico P, Gertler FB and
Condeelis JS: Identification of invasion specific splice variants
of the cytoskeletal protein Mena present in mammary tumor cells
during invasion in vivo. Clin Exp Metastasis. 26:153–159. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Condeelis J, Singer RH and Segall JE: The
great escape: When cancer cells hijack the genes for chemotaxis and
motility. Annu Rev Cell Dev Biol. 21:695–718. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Di Modugno F, Mottolese M, Di Benedetto A,
Conidi A, Novelli F, Perracchio L, Venturo I, Botti C, Jager E,
Santoni A, et al: The cytoskeleton regulatory protein hMena (ENAH)
is overexpressed in human benign breast lesions with high risk of
transformation and human epidermal growth factor
receptor-2-positive/hormonal receptor-negative tumors. Clin Cancer
Res. 12:1470–1478. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gurzu S, Jung I, Prantner I, Ember I,
Pávai Z and Mezei T: The expression of cytoskeleton regulatory
protein Mena in colorectal lesions. Rom J Morphol Embryol.
49:345–349. 2008.PubMed/NCBI
|
|
38
|
Di Modugno F, DeMonte L, Balsamo M, Bronzi
G, Nicotra MR, Alessio M, Jager E, Condeelis JS, Santoni A, Natali
PG and Nisticò P: Molecular cloning of hMena (ENAH) and its splice
variant hMena+11a: Epidermal growth factor increases their
expression and stimulates hMena+11a phosphorylation in breast
cancer cell lines. Cancer Res. 67:2657–2665. 2007. View Article : Google Scholar : PubMed/NCBI
|