Introduction
AXL is a member of the TYRO3-AXL-MER family of
receptor tyrosine kinases, which includes two other members: TYRO3
and MER. AXL is activated by its ligand, growth arrest-specific 6,
which in turn triggers several downstream signaling pathways
depending on the cell type, primarily phosphoinositide 3-kinase
(PI3K)/protein kinase B (AKT) and mitogen-activated protein
kinase/extracellular-signal-regulated kinase, nuclear factor-κB
(NF-κB) and signal transducer and activator of transcription 3
(STAT3) signaling pathways. It exerts a wide array of functions
including cell survival, proliferation, migration and adhesion
(1). AXL has also been implicated in
the pathophysiology of a number of types of cancer, including
breast, gastric, prostate, ovarian and lung (2–4). AXL
expression promotes cancer cell survival, angiogenesis, metastasis
and drug resistance (3,5).
AXL is significantly associated with
epithelial-mesenchymal transition (EMT) in several tumors. RNA
sequencing data of 643 cancer cell lines demonstrated that AXL
expression is markedly associated with a mesenchymal phenotype
(6). AXL expression is increased in
mesenchymal cells compared with epithelial cells in non-small cell
lung cancer (NSCLC) cell lines (7).
Furthermore, AXL downregulation in A549 and H460 mesenchymal cells
leads to enhanced expression of epithelial cadherin (E-cadherin)
and decreased expression of vimentin and neural cadherin
(N-cadherin), which are features of mesenchymal-epithelial
transition (7,8), suggesting that AXL is involved in
maintaining EMT. AXL has also been identified to regulate EMT in
other types of cancer, including breast (9,10),
prostate (11), ovarian (12) and pancreatic (13,14).
China is among the countries at highest risk of
esophageal squamous cell carcinoma (ESCC) (15). ESCC is the fourth most common cancer
in China, accounting for ~13% of all cancer cases in 2015 (16). ESCC is frequently associated with a
high risk of recurrence and high mortality rate and the 5-year
survival rate is <20% (17).
The function of AXL in ESCC has been demonstrated
only recently (18–20). AXL gene (19) and protein (19,20)
expression were upregulated in ESCC cells compared with normal
adjacent cells, and were identified to be associated with tumor
progression, increased risk of mortality and distant metastasis
(20). AXL was also identified to be
consistently overexpressed in ESCC cell lines (19). Knockdown of AXL expression inhibited
cell proliferation, survival, migration and invasion in
vitro and in vivo, and those effects were mediated by
the ΑKΤ/NF-κB and ΑKΤ/glycogen synthase kinase-3β signaling
pathways (19). Overexpression of AXL
also mediated resistance to the PI3Kα inhibitor BYL719 by
activating signaling pathways in ESCC cells (18).
However, the function of AXL in EMT in ESCC cells is
not well-documented. To the best of our knowledge, the present
study provides the first evidence that AXL is a marker for the
mesenchymal phenotype in ESCC using online mRNA profile data.
Materials and methods
Gene expression data
GSE47404 (21),
GSE23400 (22) and GSE21293 (23) gene expression data were obtained from
the Gene Expression Omnibus database (GEO; www.ncbi.nlm.nih.gov/geo). Further information on the
sample size and the microarray platforms used for the creation of
these datasets is presented in Table
I. Processed expression data and information of corresponding
platforms were downloaded and used further. Expression values (mRNA
log2 intensity) for each gene were examined manually and
used for further study.
 | Table I.Gene expression data. |
Table I.
Gene expression data.
| Dataset accession
no. | Description | Platform |
|---|
| GSE47404 | mRNA profiles of 71
primary tumor tissues from patients with ESCC from Japan | Whole Human Genome
4X44K Agilent G4112F microarray |
| GSE23400 | mRNA profiles of 51
pairs of primary tumor tissues from patients with ESCC and adjacent
normal tissues from China (only mRNA expression profiles from
patients with ESCC were used in this study) | Human Genome U133
Set microarray |
| GSE21293 | mRNA profiles of
invasive and non-invasive genetically engineered human esophageal
cells with hTERT and p53 mutations and ERFG overexpression | Human Genome
U133Plus 2.0 microarray |
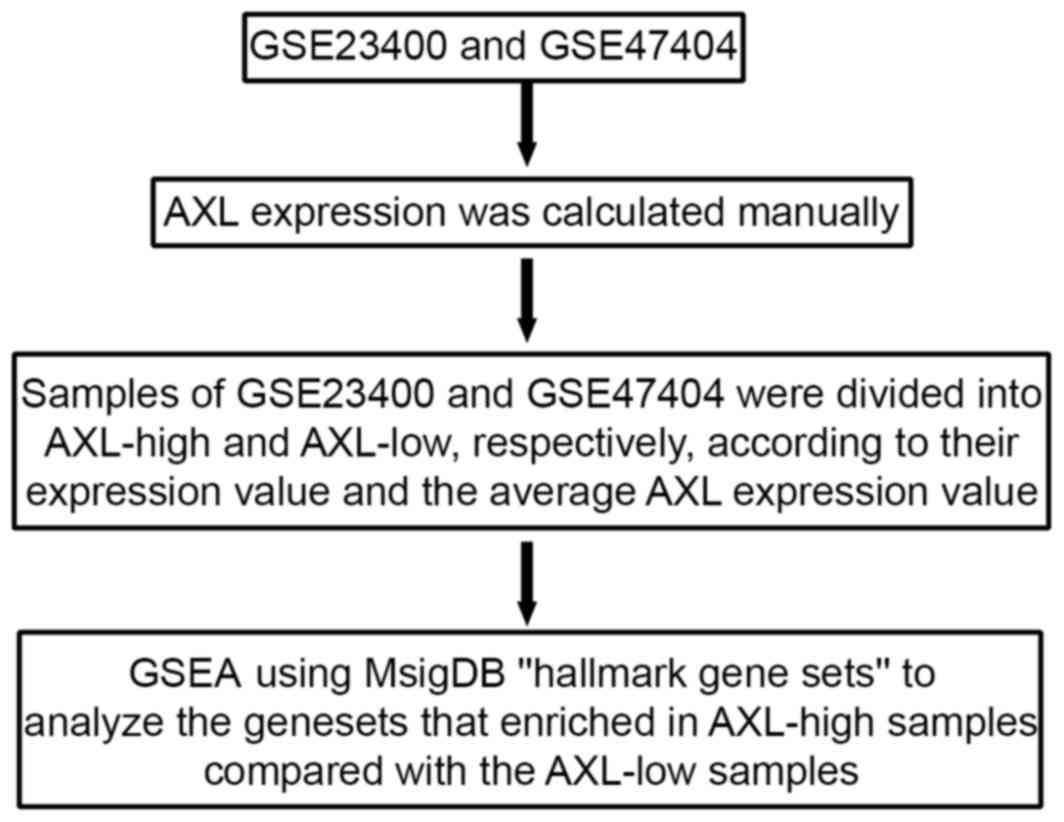
Gene Set Enrichment Analysis
(GSEA)
GSEA is a computational software that determines
whether predefined set of genes exhibit statistically significant
concordant differences between two biological states (http://software.broadinstitute.org/gsea/index.jsp)
(24). AXL expression was determined
on the basis of GSE47404 and GSE23400 datasets. If two probe sets
correspond to AXL, AXL expression is calculated as the average of
the value of the two probe sets. Following determination of the
mean expression of AXL, the samples were designated AXL-high ESCC
(samples with an above-average AXL value) or AXL-low ESCC (samples
with a below-average AXL value). AXL-low samples served as the
control samples. This collection of gene sets summarized and
represented specific well-defined biological states or processes,
and was generated by a computational methodology based on
identifying gene set overlaps and retaining genes that display
coordinate expression across several datasets (25). All the parameters were set to their
default values. A false discovery rate (FDR) value <0.25 was
considered to indicate significant enrichment. The procedure
followed for the GSEA is presented in Fig. 1.
Association of AXL expression and
cancer cell invasive ability in GSE21293
GSE21293 displayed the mRNA profiles of 34
genetically engineered human esophageal cells with different
invasive abilities (23). To
determine whether AXL is associated with the invasive ability of
ESCC cells, AXL expression in the samples was examined and the mean
expression value was calculated, and samples were divided into two
groups according to their AXL expression value as aforementioned:
AXL-high and AXL-low. AXL-low samples acted as the control samples.
The samples were also divided into two groups according to their
invasive ability: Invasive and non-invasive. Fisher's exact test
was used to examine their association. Furthermore, AXL expression
in the invasive and non-invasive groups was also compared using
Mann-Whitney U test.
Statistical analysis
Data were analyzed using SPSS (version 13.0; SPSS,
Inc., Chicago, IL, USA). Spearman's correlation was used to analyze
the association between AXL mRNA and mRNA of mesenchymal markers.
Fisher's Exact Test was applied to determine the correlation
between AXL expression and invasion ability of genetically
engineered esophageal cells. Mann-Whitney U test was used to assess
the expression difference of AXL mRNA in invasive and non-invasive
esophageal cells. P<0.05 was considered to indicate a
statistically significant difference.
Results
AXL mRNA expression is associated with
certain mesenchymal markers
To investigate the function of AXL in EMT, the
association of AXL with several EMT markers, including E-cadherin,
vimentin, fibronectin, snail family transcriptional repressor 1
(SNAI1), snail family transcriptional repressor 2 (SNAI2), twist
family basic helix-loop-helix transcription factor 1 (TWIST1), zinc
finger E-box-binding homeobox 1 (ZEB1) and N-cadherin, was examined
(Fig. 2). It was revealed that AXL
mRNA expression in GSE47404 and GSE23400 datasets is associated
with several EMT markers, including vimentin
(P=1.00×10−6 for GSE47404 and 1.00×10−6 for
GSE23400; Fig. 2B and J), fibronectin
(P=2.22×10−4 for GSE47404 and 1.00×10−6 for
GSE23400; Fig. 2D and L), TWIST1
(P=0.01 for GSE47404 and 1.90×10−4 for GSE23400;
Fig. 2G and O) and ZEB1 (P=0.04 for
GSE47404 and 1.00×10−6 for GSE23400; Fig. 2H and P). However, no association
between AXL expression and E-cadherin (Fig. 2A and I) or SNAI2 (Fig. 2F and N) was identified in either
dataset. A significant association was identified between AXL and
SNAI1 in GSE47404 (P=2.03×10−3; Fig. 2E), but not in GSE23400 (P=0.67;
Fig. 2M). Additionally, the
association between AXL and N-cadherin was significant in GSE23400
(P=6.59×10−3; Fig. 2K),
but not in GSE47404 (P=0.74; Fig.
2C). These preliminary results suggest that AXL may be involved
in EMT in ESCC.
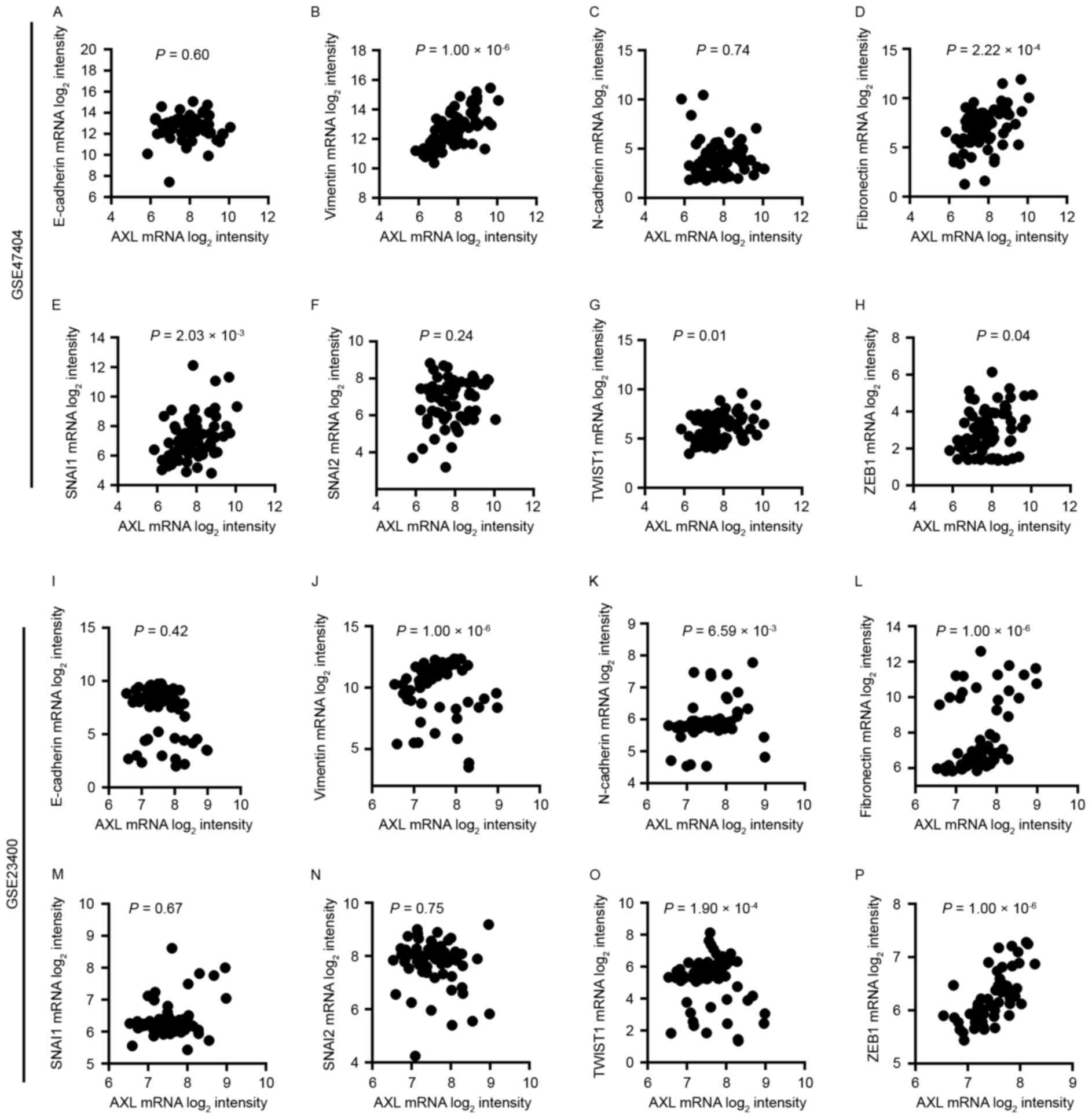 | Figure 2.Association between the expression of
AXL and (A) E-cadherin, (B) vimentin, (C) N-cadherin, (D)
fibronectin, (E) SNAI1, (F) SNAI2, (G) TWIST1 and (H) ZEB1 in the
GSE47404 dataset. Association between the expression of AXL and (I)
E-cadherin, (J) vimentin, (K) N-cadherin, (L) fibronectin, (M)
SNAI1, (N) SNAI2, (O) TWIST1 and (P) ZEB1 in the GSE23400 dataset.
Results were assessed by Spearman's correlation. E-cadherin,
epithelial cadherin; N-cadherin, neural cadherin; SNAI, snail
family transcriptional repressor; TWIST1, twist family basic
helix-loop-helix transcription factor 1; ZEB1, zinc finger
E-box-binding homeobox 1. |
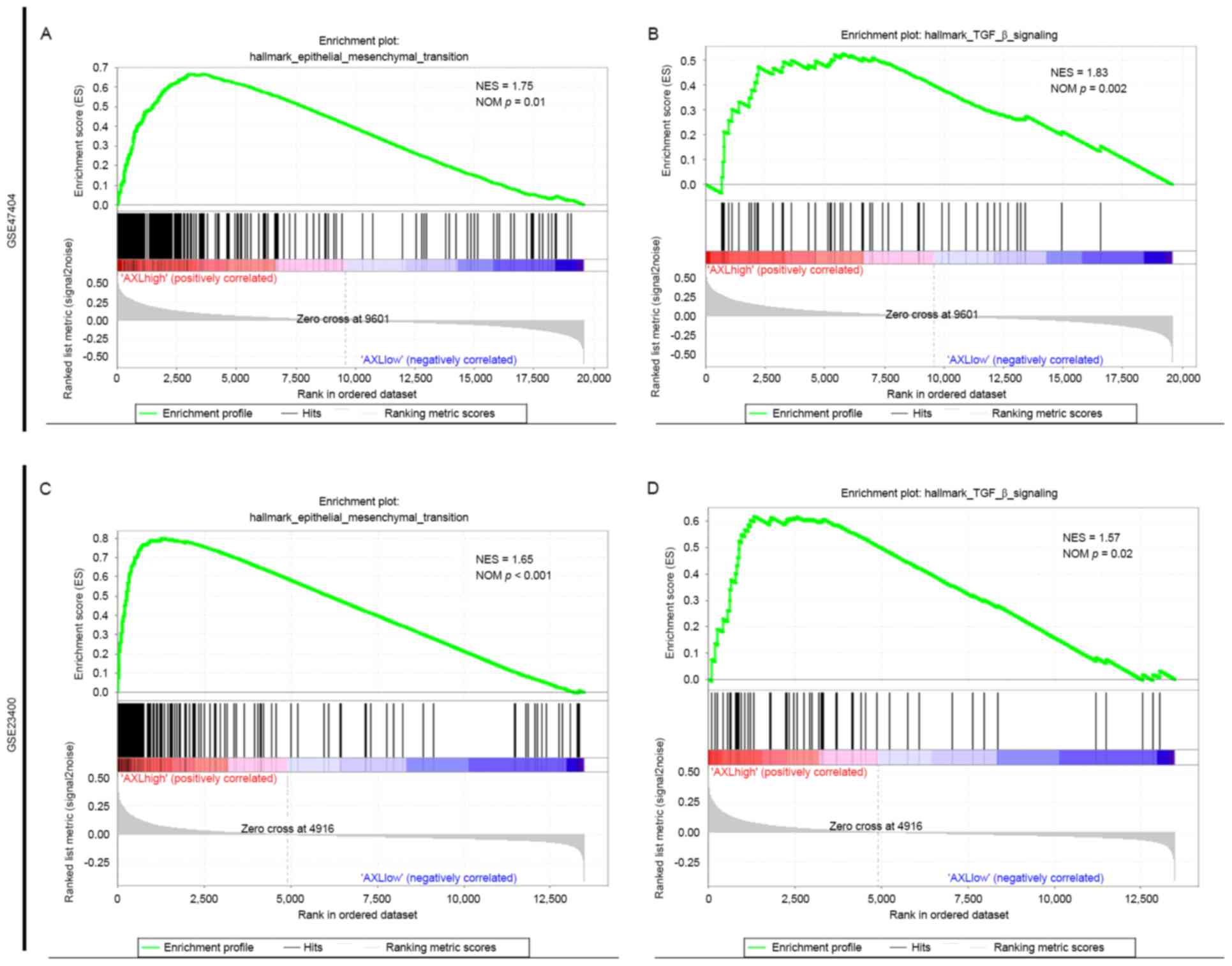
EMT gene set is enriched in the ESCC
group with high AXL expression
To further determine the function of AXL in EMT in
ESCC, GSEA was performed to examine whether the AXL-high and the
AXL-low expression groups display a different EMT state in ESCC.
The GSEA of the present study utilized ‘Hallmark gene sets
[h.all.v5.2.symbols.gmt (Hallmarks)]’ as the gene sets to be
analyzed.
The results revealed that in GSE47404, the ESCC
group with high AXL expression levels displayed a more marked
mesenchymal state compared with the ESCC group with low AXL
expression levels (Fig. 3A, and
Table II). Furthermore, transforming
growth factor-β signaling, which has a predominant function in EMT
in cancer (26,27), is also enriched in the AXL-high
expression group (Fig. 3B and
Table II). Analyze on GSE23400 also
showed similar pattern (Fig. 3C and D
and Table II). STAT3 signaling,
which is reported to serve a function in EMT in ESCC (28), is also highly enriched in the AXL-high
expression group in the two datasets (Table II).
 | Table II.Gene Set Enrichment Analysis on
GSE47404 and GSE23400 with hallmark gene sets. |
Table II.
Gene Set Enrichment Analysis on
GSE47404 and GSE23400 with hallmark gene sets.
| Dataset accession
no. | Gene set name | NES | NOM P-value | FDR Q-value |
|---|
| GSE47404 |
HALLMARK_ALLOGRAFT_REJECTION | 1.87 | <0.001 | 0.15 |
| GSE47404 |
HALLMARK_NOTCH_SIGNALING | 1.85 | <0.001 |
9.59×10−2 |
| GSE47404 |
HALLMARK_COMPLEMENT | 1.84 | <0.001 | 6.73×10–2 |
| GSE47404 |
HALLMARK_COAGULATION | 1.84 | <0.001 |
5.04×10−2 |
| GSE47404 |
HALLMARK_TGF_BETA_SIGNALING | 1.83 | 1.95×10-3 |
4.38×10−2 |
| GSE47404 |
HALLMARK_IL2_STAT5_SIGNALING | 1.82 | <0.001 | 3.95×10-2 |
| GSE47404 |
HALLMARK_ANGIOGENESIS | 1.79 |
7.39×10−3 | 4.98×10-2 |
| GSE47404 |
HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 1.75 |
1.13×10−2 | 4.67×10-2 |
| GSE47404 |
HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.73 |
5.93×10−3 | 4.73×10-2 |
| GSE47404 |
HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.73 |
1.79×10−2 | 4.58×10-2 |
| GSE47404 |
HALLMARK_INFLAMMATORY_RESPONSE | 1.71 |
1.38×10−2 | 4.83×10-2 |
| GSE47404 |
HALLMARK_UV_RESPONSE_DN | 1.64 |
1.24×10−2 | 7.29×10-2 |
| GSE47404 |
HALLMARK_KRAS_SIGNALING_UP | 1.63 |
5.78×10−3 | 7.14×10-2 |
| GSE47404 |
HALLMARK_APOPTOSIS | 1.62 |
3.12×10−2 | 6.76×10-2 |
| GSE47404 |
HALLMARK_INTERFERON_ALPHA_RESPONSE | 1.62 |
3.43×10−2 | 6.54×10-2 |
| GSE47404 |
HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.53 |
8.74×10−2 | 0.10 |
| GSE47404 |
HALLMARK_HYPOXIA | 1.38 | 0.11 | 0.20 |
| GSE23400 |
HALLMARK_COAGULATION | 1.71 | 5.86×10-3 |
7.27×10−2 |
| GSE23400 |
HALLMARK_KRAS_SIGNALING_UP | 1.71 | <0.001 | 3.64×10-2 |
| GSE23400 |
HALLMARK_UV_RESPONSE_DN | 1.66 |
1.91×10−3 | 4.74×10-2 |
| GSE23400 |
HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 1.65 | <0.001 |
4.51×10−2 |
| GSE23400 |
HALLMARK_APICAL_JUNCTION | 1.58 | 4.10×10-3 |
8.88×10−2 |
| GSE23400 |
HALLMARK_TGF_BETA_SIGNALING | 1.57 | 1.74×10-2 |
7.69×10−2 |
| GSE23400 |
HALLMARK_ANGIOGENESIS | 1.55 | 2.10×10-2 |
8.26×10−2 |
| GSE23400 |
HALLMARK_MYOGENESIS | 1.54 | 3.97×10-2 |
7.94×10−2 |
| GSE23400 |
HALLMARK_ALLOGRAFT_REJECTION | 1.54 | 4.45×10-2 |
7.36×10−2 |
| GSE23400 |
HALLMARK_COMPLEMENT | 1.49 | 3.75×10-2 | 0.10 |
| GSE23400 |
HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.47 |
6.22×10−2 | 0.11 |
| GSE23400 |
HALLMARK_IL2_STAT5_SIGNALING | 1.45 | <0.001 | 0.11 |
| GSE23400 |
HALLMARK_APOPTOSIS | 1.45 | 9.77×10-3 | 0.11 |
| GSE23400 |
HALLMARK_INFLAMMATORY_RESPONSE | 1.41 |
7.68×10−2 | 0.13 |
| GSE2340 |
HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.36 | 0.01 | 0.17 |
| GSE23400 |
HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.31 | 0.18 | 0.23 |
| GSE23400 |
HALLMARK_PROTEIN_SECRETION | 1.31 | 0.12 | 0.21 |
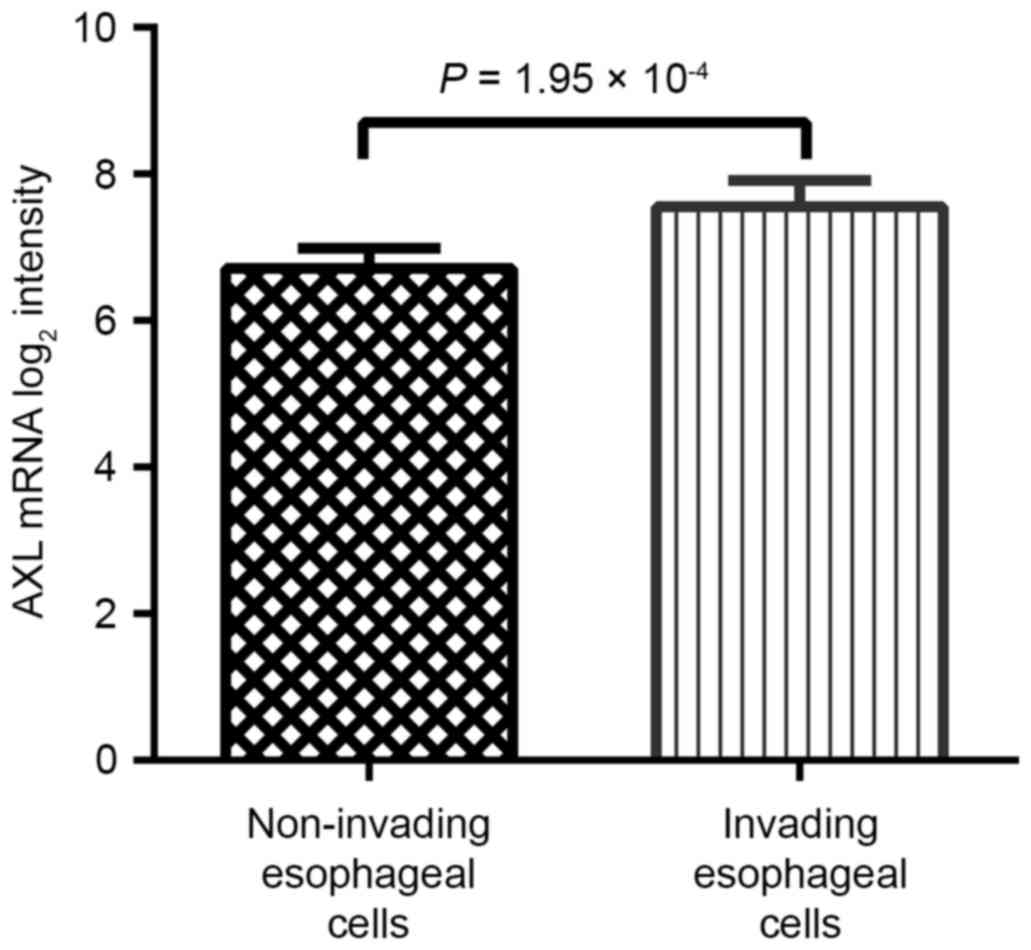
AXL expression is associated with
increased invasive ability of ESCC cells
A prominent feature of EMT is to confer enhanced
invasive ability on cancer cells. Therefore, the present study
assessed the association between AXL expression and the invasive
ability of ESCC on the basis of the GSE21293 dataset (23). This dataset includes normal esophageal
cells, which are genetically engineered with human telomerase
reverse transcriptase, epidermal growth factor receptor
overexpression and p53 mutations, in order to immortalize them and
display differential invasion. According to their ability to
invade, these cells were divided into invasive and non-invasive
groups. As presented in Table III,
the AXL-high expression group displayed increased ability of
invasion (P=2.61×10−4). AXL expression in non-invasive
and invasive groups was also compared and it was revealed that the
invasive group expressed significantly increased AXL levels
(P=1.95×10−4; Fig. 4).
 | Table III.Association between AXL expression
and invasive ability of genetically engineered esophageal
cells. |
Table III.
Association between AXL expression
and invasive ability of genetically engineered esophageal
cells.
|
| Invasive
ability |
|
|
|---|
|
|
|
|
|
|---|
| AXL expression | Non-invasive | Invasive | Total | P-value |
|---|
| AXL-low | 19 | 2 | 21 |
|
| AXL-high | 4 | 10 | 14 |
|
| Total | 23 | 12 |
| 2.61×10-4 |
Discussion
EMT is a fundamental process in body development,
and in the differentiation of multiple tissues and organs (29). EMT endows cancer cells with invasive
properties and with stem-cell like characteristics, prevents
drug-induced apoptosis and senescence, and contributes to
immunosuppression (29,30). The EMT state defined by the
E-cadherin/vimentin ratio has been demonstrated to be associated
with a poor 5-year survival rate of patients with ESCC (31).
AXL has been demonstrated to be an important factor
in certain tumors including NSCLC, breast cancer, prostate cancer,
myeloid leukemia and ovarian cancer (2,3,12,32). AXL
enhances tumor proliferation, promotes EMT and induces drug
resistance (2,3,32).
Clinical trials for melanoma, NSCLC and acute myeloid leukemia are
ongoing to determine the safety and efficiency of AXL inhibitors
(https://clinicaltrials.gov/ct2/results?cond=&term=BGB324&cntry1=&state1=&Search=Search).
The function of AXL in ESCC has only recently been
identified (18–20). However, its function in EMT in ESCC is
even less well-elucidated. The present study provides preliminary
evidence from three independent mRNA array studies indicating that
AXL is a marker for the mesenchymal state of ESCC. According to the
results of the present study, it was revealed that AXL expression
is associated with certain EMT markers, including vimentin,
fibronectin, TWIST1 and ZEB1. However, AXL was not associated with
other EMT markers, including E-cadherin. These results are
consistent with those in a study by Wilson et al (6) which demonstrated that AXL mRNA is
associated with vimentin mRNA expression in a group of 643 human
cancer cell lines. In contrast, Sato et al (33) indicated that AXL expression is
associated with E-cadherin, but not vimentin, in lung
adenocarcinomas. Additionally, the ESCC group with high AXL
expression was enriched with the EMT gene set as revealed by GSEA.
Another data cohort revealed that AXL is markedly associated with
the invasive ability of genetically engineered esophageal cells.
Collectively, the results of the present study suggest that AXL is
an EMT marker for ESCC. However, further studies, including in
vitro and in vivo cancer models, are required to confirm
those results. As mRNA and protein expression are not necessarily
associated (34), experiments
analyzing AXL protein levels in ESCC are required.
Acknowledgements
The present study is supported by a Shandong
Province Higher Educational Science and Technology Program (grant
no. J13LK14), Natural Science Foundation of Shandong province
(grant no. 2013ZRC03007), Jining Scientific and Technological
Project (grant nos. 2012jnjc02, 2014jnjc17 and 2015-57-71), Youth
Foundation of Jining Medical University (grant nos. JY2013KJ035 and
JY2015KJ023), The Research Innovation Program for College Graduates
of Shandong Province (grant no. SDYY14014) and Projects of
Education Planning Office of Shandong Province (grant no.
2013GG043). The authors thank Dr Liyan Guo (Associate Professor of
Epidemiology, Jining Medical University) for assistance with
statistical analysis.
References
|
1
|
Hafizi S and Dahlbäck B: Signalling and
functional diversity within the Axl subfamily of receptor tyrosine
kinases. Cytokine Growth Factor Rev. 17:295–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paccez JD, Vogelsang M, Parker MI and
Zerbini LF: The receptor tyrosine kinase Axl in cancer: Biological
functions and therapeutic implications. Int J Cancer.
134:1024–1033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Graham DK, DeRyckere D, Davies KD and Earp
HS: The TAM family: Phosphatidylserine sensing receptor tyrosine
kinases gone awry in cancer. Nat Rev Cancer. 14:769–785. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qu X, Liu J, Zhong X, Li X and Zhang Q:
Role of AXL expression in non-small cell lung cancer. Oncol Lett.
12:5085–5091. 2016.PubMed/NCBI
|
|
5
|
Zhang Z, Lee JC, Lin L, Olivas V, Au V,
LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al:
Activation of the AXL kinase causes resistance to EGFR-targeted
therapy in lung cancer. Nat Genet. 44:852–860. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilson C, Ye X, Pham T, Lin E, Chan S,
McNamara E, Neve RM, Belmont L, Koeppen H, Yauch RL, et al: AXL
inhibition sensitizes mesenchymal cancer cells to antimitotic
drugs. Cancer Res. 74:5878–5890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Byers LA, Diao L, Wang J, Saintigny P,
Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al: An
epithelial-mesenchymal transition gene signature predicts
resistance to EGFR and PI3K inhibitors and identifies Axl as a
therapeutic target for overcoming EGFR inhibitor resistance. Clin
Cancer Res. 19:279–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu F, Li J, Jang C, Wang J and Xiong J:
The role of Axl in drug resistance and epithelial-to-mesenchymal
transition of non-small cell lung carcinoma. Int J Clin Exp Pathol.
7:6653–6661. 2014.PubMed/NCBI
|
|
9
|
Asiedu MK, Beauchamp-Perez FD, Ingle JN,
Behrens MD, Radisky DC and Knutson KL: AXL induces
epithelial-to-mesenchymal transition and regulates the function of
breast cancer stem cells. Oncogene. 33:1316–1324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gjerdrum C, Tiron C, Høiby T, Stefansson
I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT,
et al: Axl is an essential epithelial-to-mesenchymal
transition-induced regulator of breast cancer metastasis and
patient survival. Proc Natl Acad Sci USA. 107:pp. 1124–1129. 2010;
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mishra A, Wang J, Shiozawa Y, McGee S, Kim
J, Jung Y, Joseph J, Berry JE, Havens A, Pienta KJ and Taichman RS:
Hypoxia stabilizes GAS6/Axl signaling in metastatic prostate
cancer. Mol Cancer Res. 10:703–712. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Antony J, Tan TZ, Kelly Z, Low J, Choolani
M, Recchi C, Gabra H, Thiery JP and Huang RY: The GAS6-AXL
signaling network is a mesenchymal (Mes) molecular subtype-specific
therapeutic target for ovarian cancer. Sci Signal. 9:ra972016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koorstra JB, Karikari CA, Feldmann G,
Bisht S, Rojas PL, Offerhaus GJ, Alvarez H and Maitra A: The Axl
receptor tyrosine kinase confers an adverse prognostic influence in
pancreatic cancer and represents a new therapeutic target. Cancer
Biol Ther. 8:618–626. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kirane A, Ludwig KF, Sorrelle N, Haaland
G, Sandal T, Ranaweera R, Toombs JE, Wang M, Dineen SP, Micklem D,
et al: Warfarin blocks gas6-Mediated Axl activation required for
pancreatic cancer epithelial plasticity and metastasis. Cancer Res.
75:3699–3705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Allum WH, Stenning SP, Bancewicz J, Clark
PI and Langley RE: Long-term results of a randomized trial of
surgery with or without preoperative chemotherapy in esophageal
cancer. J Clin Oncol. 27:5062–5067. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elkabets M, Pazarentzos E, Juric D, Sheng
Q, Pelossof RA, Brook S, Benzaken AO, Rodon J, Morse N, Yan JJ, et
al: AXL mediates resistance to PI3Kα inhibition by activating the
EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell
carcinomas. Cancer Cell. 27:533–546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paccez JD, Duncan K, Vava A, Correa RG,
Libermann TA, Parker MI and Zerbini LF: Inactivation of GSK3β and
activation of NF-kB pathway via Axl represents an important
mediator of tumorigenesis in esophageal squamous cell carcinoma.
Mol Biol Cell. 26:821–831. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsieh MS, Yang PW, Wong LF and Lee JM: The
AXL receptor tyrosine kinase is associated with adverse prognosis
and distant metastasis in esophageal squamous cell carcinoma.
Oncotarget. 7:36956–36970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sawada G, Niida A, Uchi R, Hirata H,
Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y,
et al: Genomic landscape of esophageal squamous cell carcinoma in a
japanese population. Gastroenterology. 150:1171–1182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su H, Hu N, Yang HH, Wang C, Takikita M,
Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al: Global
gene expression profiling and validation in esophageal squamous
cell carcinoma and its association with clinical phenotypes. Clin
Cancer Res. 17:2955–2966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Michaylira CZ, Wong GS, Miller CG,
Gutierrez CM, Nakagawa H, Hammond R, Klein-Szanto AJ, Lee JS, Kim
SB, Herlyn M, et al: Periostin, a cell adhesion molecule,
facilitates invasion in the tumor microenvironment and annotates a
novel tumor-invasive signature in esophageal cancer. Cancer Res.
70:5281–5292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:pp. 15545–15550. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui Y, Li YY, Li J, Zhang HY, Wang F, Bai
X and Li SS: STAT3 regulates hypoxia-induced epithelial mesenchymal
transition in oesophageal squamous cell cancer. Oncol Rep.
36:108–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Niwa Y, Yamada S, Koike M, Kanda M, Fujii
T, Nakayama G, Sugimoto H, Nomoto S, Fujiwara M and Kodera Y:
Epithelial to mesenchymal transition correlates with tumor budding
and predicts prognosis in esophageal squamous cell carcinoma. J
Surg Oncol. 110:764–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bivona TG and Okimoto RA: AXL receptor
tyrosine kinase as a therapeutic target in NSCLC. Lung Cancer
(Auckl). 6:27–34. 2015.PubMed/NCBI
|
|
33
|
Sato K, Suda K, Shimizu S, Sakai K,
Mizuuchi H, Tomizawa K, Takemoto T, Nishio K and Mitsudomi T:
Clinical, pathological and molecular features of lung
adenocarcinomas with AXL expression. PLoS One. 11:e01541862016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vogel C and Marcotte EM: Insights into the
regulation of protein abundance from proteomic and transcriptomic
analyses. Nat Rev Genet. 13:227–232. 2012.PubMed/NCBI
|