Introduction
In 2013, gastric cancer (GC) had become the third
most common cause of cancer-associated mortality globally (1). It is estimated that 951,600 new cases
and 723,100 GC-associated mortalities were recorded worldwide in
2012 (2,3). Multistep processes and molecular markers
have been confirmed to be involved in the tumorigenesis and
invasiveness of GC (4). A combination
of surgery and chemotherapy has increased the survival time of
patients with GC (5,6). However, a significant number of patients
still suffer from relapse due to the resistance of tumor cells to
chemotherapeutic agents (5).
Therefore, understanding the molecular pathways underlying GC
carcinogenesis and progression will assist in improving diagnosis,
therapy, and prevention of this disease.
ADP-ribosylation factor 6 (Arf6), a member of the
Arf family, has emerged as a critical regulator of membrane
traffic, cell polarity and cytoskeletal organization (6,7). There is
increasing evidence to confirm that Arf6 is associated with cancer
development (8–10). Arf6 is overexpressed in various types
of human cancer, including breast cancer, lung adenocarcinoma,
clear cell renal cell carcinoma and head and neck squamous cell
carcinoma (11–13). In glioblastoma cells, early studies
have demonstrated that Arf6 is required for epidermal growth factor
(EGF)-induced cell proliferation (14). Arf6 has also been implicated in other
cellular processes associated with tumorigenesis, including the
epithelial-mesenchymal transition (EMT), migration and invasion
(12,15,16).
Furthermore, activation of Arf6 is associated with drug resistance
in the breast cancer cell lines MDA-MB-231, MDA-MB-453 and Hs578T
(17–19). Arf6 has also been demonstrated to
mediate EGF-induced EMT in GC cells (20). The EMT phenotype in cancers is
associated with migration, invasion and drug resistance (21,22).
However, the involvement of Arf6 in the growth, migration, invasion
and drug resistance of GC cells remains to be fully elucidated.
The present study was designed to explore the
function of Arf6 in the proliferation, migration, invasion and drug
resistance in GC cells. Our results demonstrate that Arf6
contributes to the proliferation, migration and invasion of GC
cells. Knockdown of Arf6 was demonstrated to increase the
sensitivity of GC cells to 5-fluorouracil (5-FU) via inhibition of
the extracellular signal-regulated kinase 1/2 (ERK1/2) signaling
pathway. Therefore, Arf6 may be a potential target for GC
therapy.
Materials and methods
Cell culture
The human GC cell line SGC-7901 was obtained from
the Type Culture Collection of the Institute of Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; high glucose) supplemented with 10%
(v/v) fetal bovine serum (FBS) (both from Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) and antibiotics (100 U/ml
streptomycin and 100 µg/ml penicillin; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a humidified incubator at
37°C with 5% CO2.
Small interfering RNA (siRNA)
transfection
siRNA sequences were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China) according to the
Arf6 gene sequence [GenBank (23);
accession no. NM 001663.3]. The sequences for Arf6 were as follows:
siArf6-1, 5′-GUGGCAAAUAAUGAGUAAUTT−3′; siArf6-2,
5′-GCGACCACUAUGAUAAUAUTT-3′; and siArf6-3,
5′-GACGCCAUAAUCCUCAUCUTT−3′. The control siRNA (siCtr) sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. SGC-7901 cells were seeded onto 6-well
plates at a density of 105 cells/well and were incubated
for 24 h. Cells were then transfected with control siRNA or Arf6
siRNA with Lipofectamine 2000 (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. The total siRNA
concentration in each well was 100 pmol and untransfected controls
were used to demonstrate that there was no significant difference
between untransfected controls and cells transfected with control
siRNA. Arf6 silencing was confirmed by assessing them mRNA and
protein expression levels in the SGC-7901 cells 48 h after
transfection, as follows.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. cDNA was synthesized using an RNA
to cDNA EcoDry™ Premix (Oligo dT) kit (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's instructions. The following gene-specific primers
were used in the present study: Arf6 forward,
5′-CAAGGTCTCATCTTCGTAGTG−3′ and reverse,
5′-CATGTGAGCCCCTCATAGAG-3′; GAPDH forward,
5′-TGAACGGGAAGCTCACTGG-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
PCR analysis was performed using Takara Ex Taq® DNA
polymerase (Takara Biotechnology Co., Ltd.) under the following
thermocycling conditions: 95°C for 3 min, followed by 28 cycles of
95°C for 30 sec, 57°C (Arf6) or 55°C (GAPDH) for 30 sec, and 72°C
for 1 min, with a final extension at 72°C for 6 min. The PCR
products were electrophoresed on 1% agarose gels and the bands were
visualized by UV fluorescence following staining with ethidium
bromide (2.5 g/ml) for 15 min. Data were analyzed by densitometry
using Tanon Gel Image System software, version 4.0 (Tanon Science
and Technology Co., Ltd., Shanghai, China).
Western blot analysis
Cells were lysed for 5 min on ice in
radioimmunoprecipitation assay buffer (cat. no. 89900; Thermo
Fisher Scientific, Inc.) containing 1% phenylmethanesulfonyl
fluoride (Beyotime Institute of Biotechnology, Haimen, China).
Lysate was then sonicated and centrifuged at 12,000 × g for 10 min
at 4°C. The protein concentration was quantified using a
bicinchoninic acid assay (Beyotime Institute of Biotechnology).
Proteins from each sample (20 µg) were subjected to 10%
SDS-polyacrylamide gel electrophoresis and transferred to
polyvinylidene difluoride membranes. The membranes were then
blocked with 5% skim milk at room temperature for 1 h and incubated
overnight with mouse anti-Arf6 (1:500 dilution, cat. no. sc-7971;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit
anti-ERK1/2 (1:1,000 dilution, cat. no. 4695; Cell Signaling
Technology, Inc., Danvers, MA, USA), rabbit anti-phosphorylated
ERK1/2 (pERK1/2) (1:1,000 dilution, cat. no. 4377; Cell Signaling
Technology, Inc.), and mouse anti-GAPDH (1:2,000 dilution, cat. no.
sc-47724; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Following washing with TBST 3 times, the membranes were incubated
with goat anti-rabbit IgG (1:3,000 dilution, cat. no. sc-2004;
Santa Cruz Biotechnology, Inc.) or goat anti-mouse IgG (1:3,000
dilution, cat. no. sc-2005; Santa Cruz Biotechnology, Inc.)
horseradish peroxidase-conjugated secondary antibodies for 1 h at
37°C, and visualized using enhanced chemiluminescence detection
reagents (Thermo Fisher Scientific, Inc.) and were exposed to
chemiluminescent film (Thermo Fisher Scientific, Inc.). Data were
analyzed using ImageJ software (version 1.6; National Institutes of
Health, Bethesda, MD, USA) and were normalized to GAPDH
expression.
Cell proliferation assay
Cell proliferation was measured using Cell Counting
Kit-8 (CCK-8) according to the manufacturer's instructions
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Briefly, control
and transfected cells were seeded at a density of 3×103
cells/well in 96-well plates and treated with 0, 1, 10, 25, 50, 100
µg/ml 5-FU for 48 h at 37°C or 10 µM ERK1/2 inhibitor U0126 (both
from Sigma-Aldrich; Merck KGaA) for 12 h prior to being treated
with 20 µg/ml 5-FU for 48 h at 37°C. CCK-8 (10 µl) was added to
each well and incubated for an additional 4 h at 37°C. Optical
density (OD) was measured using a microplate reader (Omega Bio-Tek,
Inc., Norcross, GA, USA) at 450 nm. Each time-point was repeated in
three wells, and the experiment was independently performed three
times.
Colony formation assay
Control and transfected cells were seeded at a
density of 5×102 cells/well in 6-well plates, and
cultured in DMEM in an environment with 5% CO2 at 37°C
for 14 days to allow colonies to form. The plates were stained with
0.5% (w/v) crystal violet in 70% ethanol for 20 min at room
temperature, and colonies were counted under a light microscope
(TS100; Nikon Corporation, Tokyo, Japan). The experiment was
independently performed three times.
In vitro migration and invasion
assays
For the Transwell migration assay, untransfected and
transfected SGC-7901 cells in the exponential growth phase were
trypsinized with 1X trypsin, washed twice with phosphate-buffered
saline, and suspended in DMEM without FBS. Cells (2×104
cells/well) were seeded into polycarbonate membrane inserts (8-µm
pore size) in 24-well Transwell cell culture dishes. Cells were
permitted to attach to the membrane for 30 min. The lower chamber
was filled with 600 µl DMEM with 10% FBS. Cells were permitted to
migrate for 24 h at 37°C. Following incubation, stationary cells
were removed from the upper surface of the membranes. The cells
that had migrated to the lower surface were fixed with 4%
paraformaldehyde at room temperature for 15 min and were stained
with 0.1% crystal violet for 15 min at the same temperature. The
cells that had migrated through the membrane were manually counted
at ×200 magnification from 5 fields/filter using a light microscope
(TS100; Nikon Corporation). The invasion assay was performed using
Matrigel-coated chambers from the BioCoat Matrigel Invasion Chamber
kit (BD Biosciences, Franklin Lakes, NJ, USA) using the same method
as aforementioned for the migration assay.
Statistical analysis
Data were statistically analyzed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Data were presented as the
mean ± SD of three independent experiments. Differences between
experimental groups were analyzed using one way analysis of
variance (ANOVA). The Student-Newman-Keuls test was used as a post
hoc test following ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
Knockdown of Arf6 in SGC-7901 cells by
siRNA
To investigate the function of Arf6 in GC cells, our
group identified and validated three independent and
non-overlapping siRNA sequences to deplete endogenous Arf6
expression in SGC-7901 cells. RT-PCR and western blot analysis were
used to evaluate the ability of different Arf6 siRNAs to silence
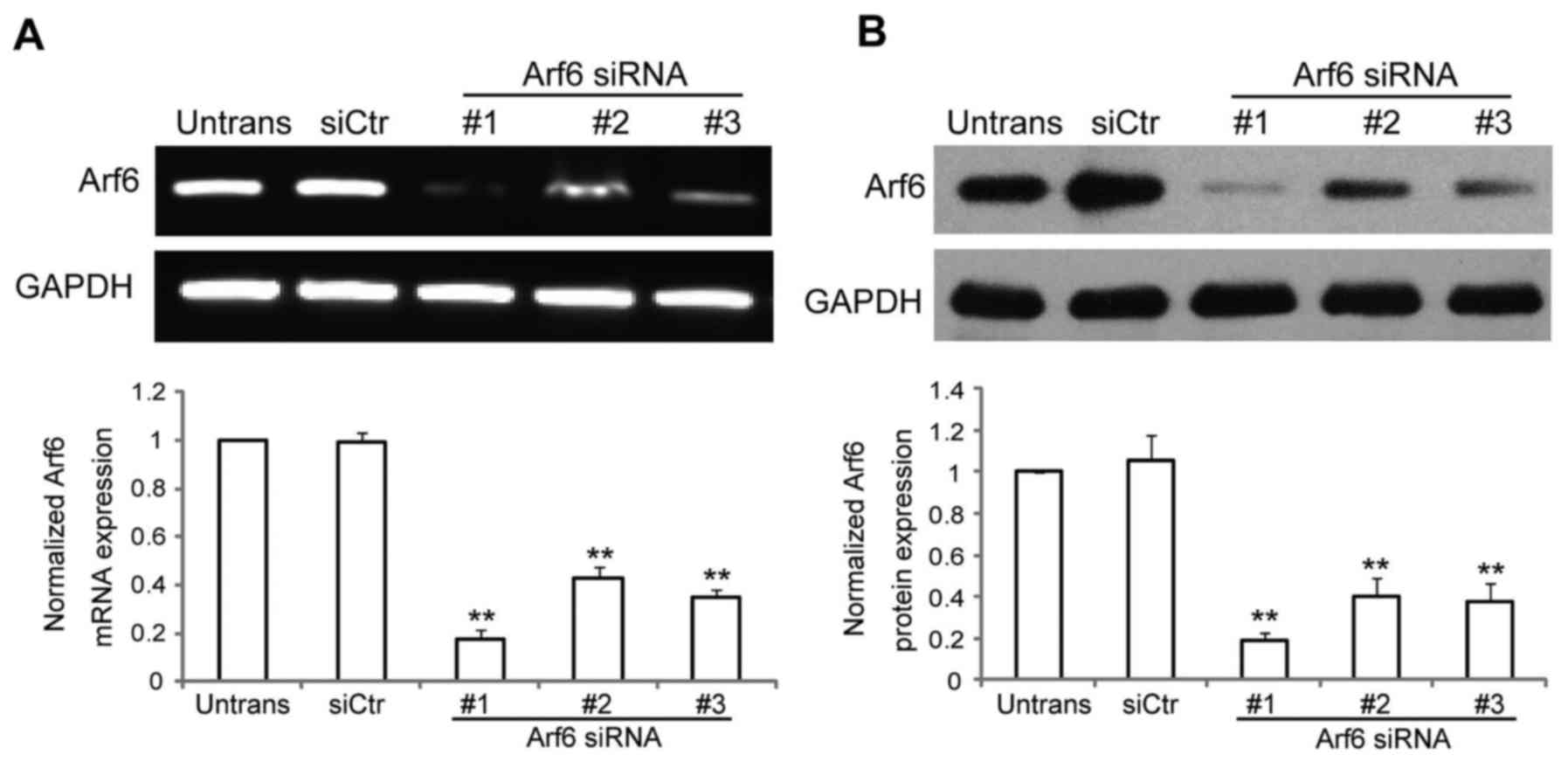
Arf6 expression in vitro. As presented in Fig. 1A and B, Arf6 expression was
significantly reduced in cells transfected with specific siRNAs
against Arf6. Arf6 mRNA expression was reduced by ~82% with
siArf6-1, ~57% with siArf6-2 and ~65% with siArf6-3, while protein
expression was reduced by ~81% with siArf6-1, ~60% with siArf6-2
and ~62% with siArf6-3, compared with cells transfected with siCtr.
Thus, siArf6-1 was selected as the most efficient and specific
sequence to silence the expression of Arf6 in the SGC-7901 cells,
and was used in all subsequent experiments.
Knockdown of Arf6 inhibits the
proliferation of SGC-7901 cells in vitro
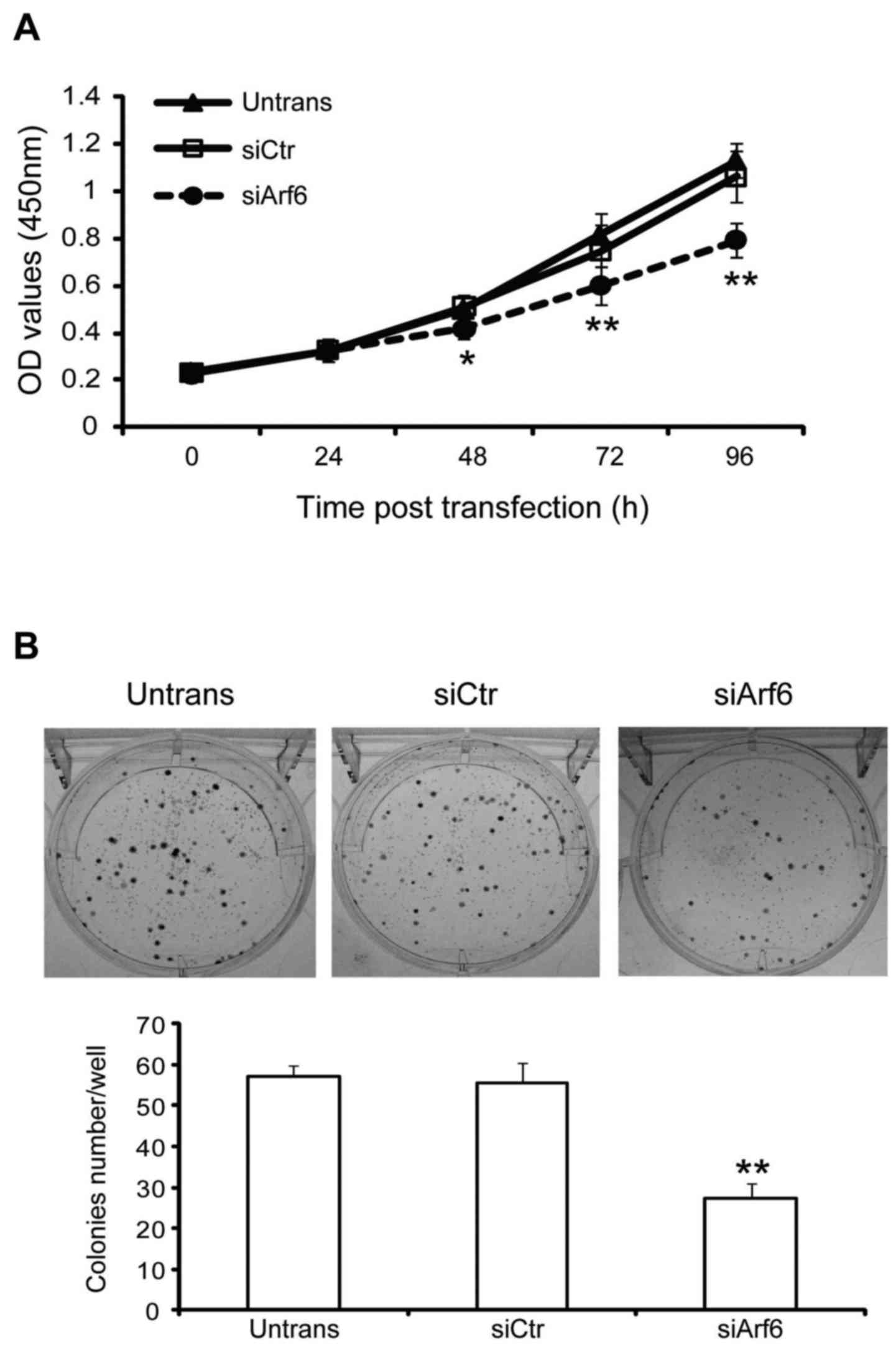
The effect of Arf6 knockdown on cell proliferation
and tumorigenesis were assessed using CCK-8 and colony formation
assays, respectively. As presented in Fig. 2A, Arf6 knockdown resulted in a
significant decrease in the proliferation of SGC-7901 cells at 48,
72, and 96 h. In addition, cells transfected with siArf6 formed
fewer and smaller colonies as compared with cells transfected with
siCtr (Fig. 2B). Taken together,
these data indicate that knockdown of Arf6 resulted in a
significant inhibitory effect on cell proliferation and colony
formation in SGC-7901 cells in vitro.
Knockdown of Arf6 inhibits the
migration and invasion of SGC-7901 cells in vitro
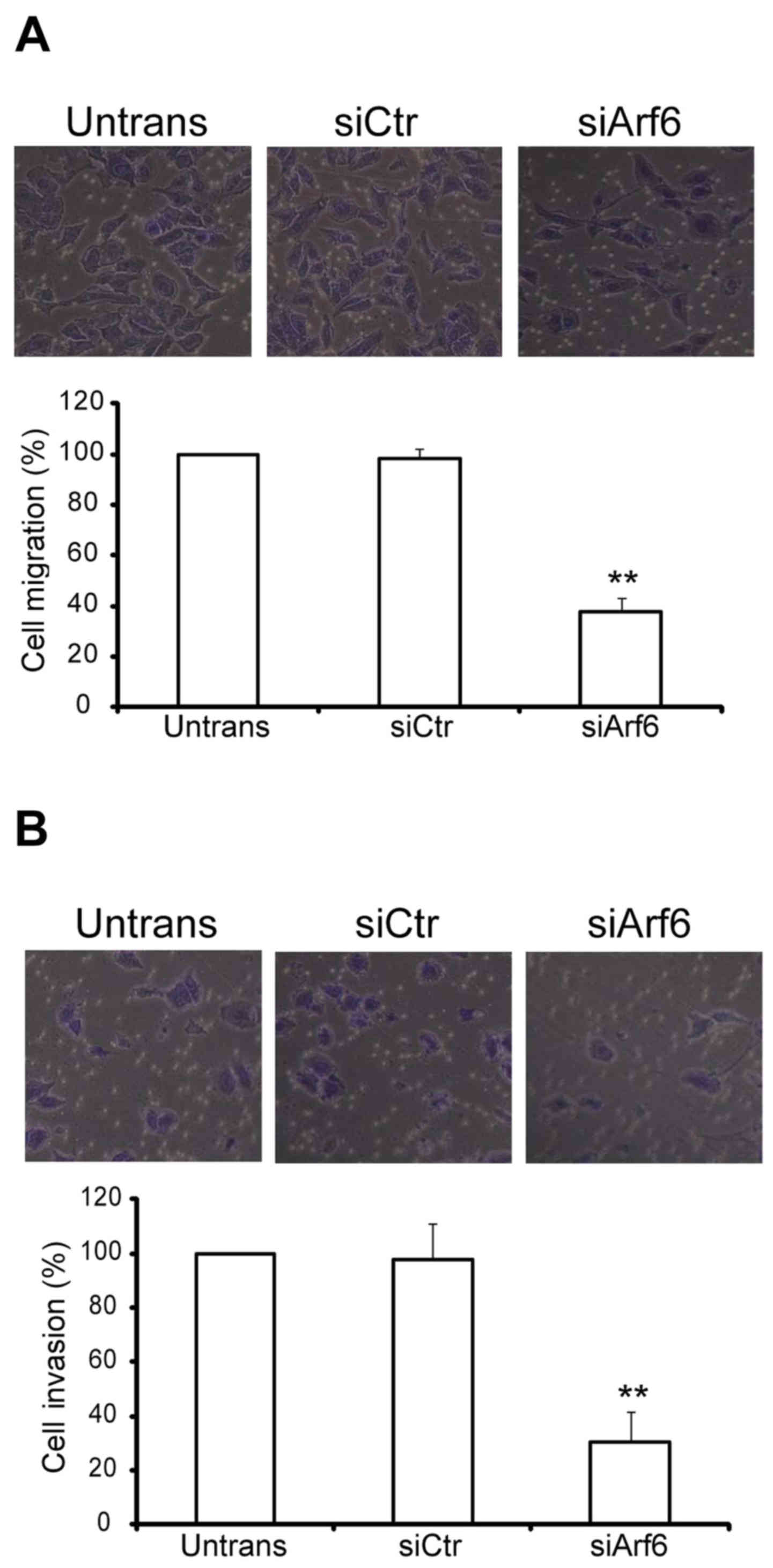
The in vitro migration and invasion assays
were designed to investigate the function of Arf6 in SGC-7901 cell
migratory and invasive processes. For the migration assay,
untransfected and transfected cells were seeded on Transwell
chambers with uncoated filters. In total, 100% of the untransfected
SGC-7901 cells were able to migrate to the filters in 24 h, while
the migratory percentage of siCtr-transfected cells was 98% and
that of siArf6-transfected cells was 38% (Fig. 3A). For the invasion assay,
untransfected and transfected cells were seeded on Transwell
chambers with Matrigel-coated filters. After 24 h of incubation,
the invasion of siArf6 cells was significantly reduced (Fig. 3B). Taken together, these results
indicated that silencing Arf6 reduces SGC-7901 cell migration and
invasion in vitro.
Knockdown of Arf6 decreases activation
of the ERK1/2 pathway
According to the results of previous studies, Arf6
regulates the activation of ERK1/2 (24–26).
Furthermore, activation of ERK1/2 has been demonstrated to increase
cell proliferation, migration and invasion in GC (27–29). Thus,
the effect of Arf6 knockdown on the ERK1/2 pathway was investigated
in SGC-7901 cells. As presented in Fig.
4A, p-ERK1/2 levels were significantly reduced in the
Arf6-knocdown SGC-7901 cells, while total ERK1/2 expression was
comparable to that observed in the control cells. This association
between Arf6 and p-ERK1/2 expression suggested that Arf6 may be
involved in the regulation of the ERK1/2 signaling pathway.
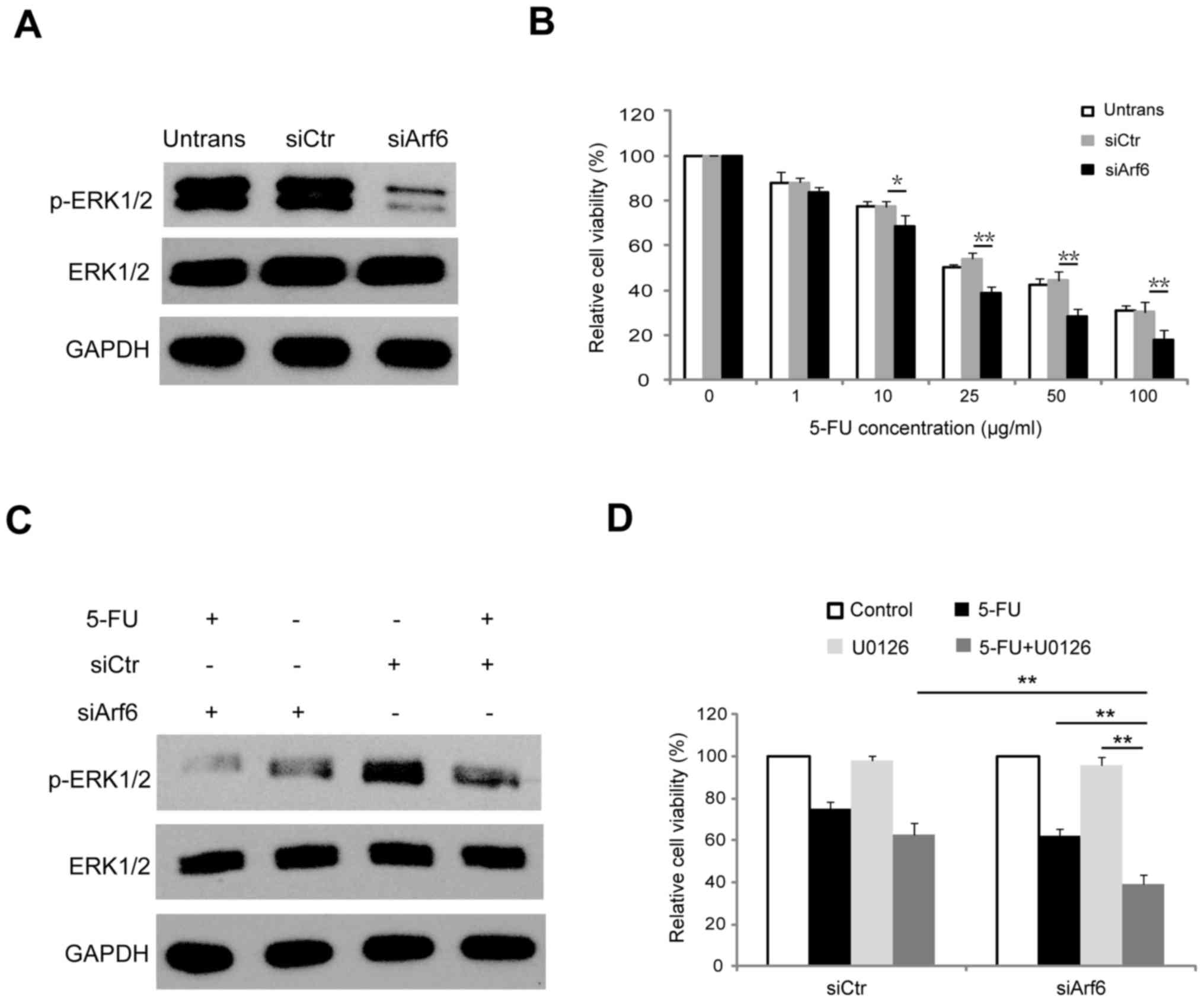 | Figure 4.Knockdown of Arf6 increases the
sensitivity of SGC-7901 cells to 5-FU through modulation of ERK1/2
signaling. (A) SGC-7901 cells were transfected with siCtr or
siArf6, and 48 h following transfection proteins were extracted and
subjected to western blot analysis. (B) Dose-dependent CCK-8 assay
was employed to investigate the effect of Arf6-knockdown on the
viability of SGC-7901 cells. (C) Western blot analysis of ERK1/2
and p-ERK1/2 in SGC-7901 cells transfected with siCtr or siArf6 and
treated with 20 µg/ml 5-FU for 48 h. (D) SGC-7901 cells transfected
with siCtr or siArf6 were treated with 10 µM U0126 for 12 h prior
to being treated with 5-FU, and cell viability was determined using
a CCK-8 assay. *P<0.05 and **P<0.01 vs. siCtr. Arf6,
ADP-ribosylation factor 6; 5-FU, 5-fluorouracil; siCtr, control
siRNA; siArf6, siRNA targeting Arf6; CCK-8, Cell Counting Kit-8;
ERK1/2, extracellular signal-regulated kinase 1/2; p,
phosphorylated; Untrans, untransfected. |
Knockdown of Arf6 enhances sensitivity
of SGC-7901 cells to 5-FU through modulating the ERK1/2 signaling
pathway
Previous studies have reported that Arf6 is involved
in drug resistance in a variety of types of cancer cell (17–19). The
effect of Arf6 knockdown on the sensitivity of SGC-7901 cells to
5-FU was further investigated. To determine the sensitivity of
cells to 5-FU, siArf6 or siCtr-transfected SGC-7901 cells were
exposed to different concentrations of 5-FU, ranging from 0 to 100
µg/ml, for 48 h. Cell viability was examined using a CCK-8 assay.
The cell survival rate appeared to show a dose-dependent decrease
in response to 5-FU treatment, and knockdown of Arf6 resulted in
increased sensitivity to 5-FU treatment in SGC-7901 cells (Fig. 4B). Next, the present study aimed to
determine whether the Arf6 knockdown-enhanced 5-FU sensitivity of
SGC-7901 cells was due to inactivation of the ERK1/2 signaling
pathway. As presented in Fig. 4C,
5-FU suppressed the phosphorylation of ERK1/2 in SGC-7901 cells.
However, knockdown of Arf6 resulted in a smaller reduction of
p-ERK1/2 expression, while total ERK1/2 expression was unaffected.
The CCK-8 results revealed that U0126 (a specific MEK inhibitor)
effectively increased siArf6-mediated 5-FU sensitivity of SGC-7901
cells (Fig. 4D). Collectively, these
results indicated that knockdown of Arf6 enhanced the
chemosensitivity of SGC-7901 cells to 5-FU by suppressing ERK1/2
activity.
Discussion
Arf6 is a member of the Arf family which exhibits
pleiotropic biological functions (7,30–33). Arf6 has been reported to be
upregulated in multiple types of tumor, and has been demonstrated
to be involved in a number of biological processes, including
cancer cell growth, EMT, cell adhesion, migration, invasion,
angiogenesis, malignant transformation and resistance to
chemotherapy (18,24,34–36).
However, the biological functions of Arf6 in GC remain to be fully
elucidated.
To investigate the potential associations between
Arf6 expression and the biological features of GC cells, Arf6
expression was knocked down in GC cells using three siRNA
sequences, and siRNA targeting of Arf6 in SGC-7901 cells in
vitro resulted in efficient, specific inhibition of endogenous
Arf6 mRNA and protein. Further experiments demonstrated that
knockdown of Arf6 in SGC-7901 cells significantly inhibited the
migration and invasion of SGC-7901 cells in vitro. These
results indicated that Arf6 expression is associated with
pro-metastatic events in SGC-7901 cells. These data are consistent
with previous results in other tumor cell lines, including breast
cancer cells (37) and lung cancer
cells (13). Furthermore, Arf6 has
also been implicated in the modulation of cancer cell growth and
the tumorigenic phenotype of cancer cells in pancreatic and lung
cancer (10,35). The present study also demonstrated
that Arf6-knockdown SGC-7901 cells had reduced proliferation and a
reduced ability to form colonies. Taken together, these results
suggest that Arf6 expression is associated with migration,
invasion, proliferation and tumorigenicity in SGC-7901 cells.
Previous studies have demonstrated the presence of
an association between Arf6 and ERK1/2 signaling in several cancer
cell lines, and this association has been implicated in cancer
progression (20,24,25).
Furthermore, ERK1/2 signaling has been demonstrated to mediate cell
proliferation, migration and invasion in various types of tumor
cell, including GC cells (27–29). In
the present study, the effect of Arf6 knockdown on ERK1/2
activation was investigated in SGC-7901 cells. Phosphorylation of
ERK1/2 was markedly reduced in Arf6 siRNA-transfected cells
compared with the control cells, indicating that the migration,
invasion, proliferation and tumorigenicity of SGC-7901 cells are
regulated via the ERK1/2 pathway. However, the precise mechanisms
by which Arf6 knockdown inhibits tumor growth, migration and
invasion require further study.
Previous studies have demonstrated that Arf6 confers
resistance to multiple chemotherapy agents, including gemcitabine,
fluorouracil and temsirolimus (17–19).
However, whether Arf6 is involved in chemoresistance in GC cells
specifically remains unclear. In the present study, knockdown of
Arf6 was revealed to sensitize SGC-7901cells to 5-FU in
vitro, suggesting that Arf6 induces 5-FU resistance in GC
cells. Inhibition of the ERK1/2 pathway has been reported to
increase 5-FU efficacy in multiple cancer cell lines, including GC
cell lines. Furthermore, the results of the present study
demonstrated that Arf6 knockdown significantly decreased ERK1/2
signaling pathway activity. Thus, whether Arf6 regulates
chemosensitivity to 5-FU by modulating ERK1/2 in SGC-7901 cells was
investigated. The results revealed that the specific ERK1/2
inhibitor U0126 effectively increased Arf6 siRNA-mediated 5-FU
sensitivity. These results indicated that Arf6 may regulate
chemosensitivity to 5-FU through the ERK1/2 signaling pathway in
SGC-7901 cells.
In conclusion, the results of the present study
demonstrated that knockdown of Arf6 inhibits SGC-7901 cell
proliferation, migration and invasion, and increases the
sensitivity of SGC-7901 cells to 5-FU, with the increasing drug
sensitivity potentially associated with the inhibition of ERK1/2
signals. Understanding the mechanisms underlying these effects may
provide novel strategies for GC treatment. Combining Arf6 gene
therapy with traditional chemotherapy may be an effective anti-GC
strategy in the future.
References
|
1
|
Piazuelo MB and Correa P: Gastric cáncer:
Overview. Colomb Med (Cali). 44:192–201. 2013.PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qi X, Liu Y, Wang W, Cai D, Li W, Hui J,
Liu C, Zhao Y and Li G: Management of advanced gastric cancer: An
overview of major findings from meta-analysis. Oncotarget.
7:78180–78205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akhavan-Niaki H and Samadani AA: Molecular
insight in gastric cancer induction: An overview of cancer stemness
genes. Cell Biochem Biophys. 68:463–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Flotow H: The use of high-throughput
screening in identifying chemotherapeutic agents for gastric
cancer. Future Med Chem. 6:2103–2112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mukhamedova N, Hoang A, Cui HL, Carmichael
I, Fu Y, Bukrinsky M and Sviridov D: Small GTPase ARF6 regulates
endocytic pathway leading to degradation of ATP-binding cassette
transporter A1. Arterioscler Thromb Vasc Biol. 36:2292–2303. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu X, Zhou T, Chen L, Zheng S, Chen S,
Zhang D, Li G and Wang Z: Arf6 controls endocytosis and polarity
during asexual development of Magnaporthe oryzae. FEMS Microbiol
Lett. 363:fnw2482016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hashimoto S, Mikami S, Sugino H, Yoshikawa
A, Hashimoto A, Onodera Y, Furukawa S, Handa H, Oikawa T, Okada Y,
et al: Lysophosphatidic acid activates Arf6 to promote the
mesenchymal malignancy of renal cancer. Nat Commun. 7:106562016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hongu T, Yamauchi Y, Funakoshi Y, Katagiri
N, Ohbayashi N and Kanaho Y: Pathological functions of the small
GTPase Arf6 in cancer progression: Tumor angiogenesis and
metastasis. Small GTPases. 7:47–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang C, Qin Y, Zhang B, Ji S, Shi S, Xu
W, Liu J, Xiang J, Liang D, Hu Q, et al: ARF6, induced by mutant
Kras, promotes proliferation and Warburg effect in pancreatic
cancer. Cancer Lett. 388:303–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hashimoto S, Onodera Y, Hashimoto A,
Tanaka M, Hamaguchi M, Yamada A and Sabe H: Requirement for Arf6 in
breast cancer invasive activities. Proc Natl Acad Sci USA. 101:pp.
6647–6652. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morishige M, Hashimoto S, Ogawa E, Toda Y,
Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, et al:
GEP100 links epidermal growth factor receptor signalling to Arf6
activation to induce breast cancer invasion. Nat Cell Biol.
10:85–92. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oka S, Uramoto H, Shimokawa H, Yamada S
and Tanaka F: Epidermal growth factor receptor-GEP100-Arf6 axis
affects the prognosis of lung adenocarcinoma. Oncology. 86:263–270.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li M, Wang J, Ng SS, Chan CY, He ML, Yu F,
Lai L, Shi C, Chen Y, Yew DT, et al: Adenosine
diphosphate-ribosylation factor 6 is required for epidermal growth
factor-induced glioblastoma cell proliferation. Cancer.
115:4959–4972. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hashimoto A, Hashimoto S, Ando R, Noda K,
Ogawa E, Kotani H, Hirose M, Menju T, Morishige M, Manabe T, et al:
GEP100-Arf6-AMAP1-cortactin pathway frequently used in cancer
invasion is activated by VEGFR2 to promote angiogenesis. PLoS One.
6:e233592011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsumoto Y, Sakurai H, Kogashiwa Y,
Kimura T, Matsumoto Y, Shionome T, Asano M, Saito K and Kohno N:
Inhibition of epithelial-mesenchymal transition by cetuximab via
the EGFR-GEP100-Arf6-AMAP1 pathway in head and neck cancer. Head
Neck. 39:476–485. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dani N, Barbosa AJ, Del Rio A and Di
Girolamo M: ADP-ribosylated proteins as old and new drug targets
for anticancer therapy: The example of ARF6. Curr Pharm Des.
19:624–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hashimoto A, Hashimoto S, Sugino H,
Yoshikawa A, Onodera Y, Handa H, Oikawa T and Sabe H: ZEB1 induces
EPB41L5 in the cancer mesenchymal program that drives ARF6-based
invasion, metastasis and drug resistance. Oncogenesis. 5:e2592016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hashimoto A, Oikawa T, Hashimoto S, Sugino
H, Yoshikawa A, Otsuka Y, Handa H, Onodera Y, Nam JM, Oneyama C, et
al: P53- and mevalonate pathway-driven malignancies require Arf6
for metastasis and drug resistance. J Cell Biol. 213:81–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Du J, Zheng J, Liu J, Xu R, Shen
T, Zhu Y, Chang J, Wang H, Zhang Z, et al: EGF-reduced Wnt5a
transcription induces epithelial-mesenchymal transition via
Arf6-ERK signaling in gastric cancer cells. Oncotarget.
6:7244–7261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mitra A, Mishra L and Li S: EMT, CTCs and
CSCs in tumor relapse and drug-resistance. Oncotarget.
6:10697–10711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Clark K, Karsch-Mizrachi I, Lipman DJ,
Ostell J and Sayers EW: GenBank. Nucleic Acids Res. 44:D67–D72.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Z, Du J, Yang L, Zhu Y, Yang Y, Zheng
D, Someya A, Gu L and Lu X: GEP100/Arf6 is required for epidermal
growth factor-induced ERK/Rac1 signaling and cell migration in
human hepatoma HepG2 cells. PLoS One. 7:e387772012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu Z, Xu R, Liu J, Zhang Y, Du J, Li W,
Zhang W, Li Y, Zhu Y and Gu L: GEP100 regulates epidermal growth
factor-induced MDA-MB-231 breast cancer cell invasion through the
activation of Arf6/ERK/uPAR signaling pathway. Exp Cell Res.
319:1932–1941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davies JC, Tamaddon-Jahromi S, Jannoo R
and Kanamarlapudi V: Cytohesin 2/ARF6 regulates preadipocyte
migration through the activation of ERK1/2. Biochem Pharmacol.
92:651–660. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Akter H, Park M, Kwon OS, Song EJ, Park WS
and Kang MJ: Activation of matrix metalloproteinase-9 (MMP-9) by
neurotensin promotes cell invasion and migration through ERK
pathway in gastric cancer. Tumour Biol. 36:6053–6062. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li P, Jia YF, Ma XL, Zheng Y, Kong Y,
Zhang Y, Zong S, Chen ZT and Wang YS: DEC2 suppresses tumor
proliferation and metastasis by regulating ERK/NF-κB pathway in
gastric cancer. Am J Cancer Res. 6:1741–1757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Teng H, Huang Q and Chen L: Inhibition of
cell proliferation and triggering of apoptosis by agrimonolide
through MAP kinase (ERK and p38) pathways in human gastric cancer
AGS cells. Food Funct. 7:4605–4613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eva R, Crisp S, Marland JR, Norman JC,
Kanamarlapudi V, Ffrench-Constant C and Fawcett JW: ARF6 directs
axon transport and traffic of integrins and regulates axon growth
in adult DRG neurons. J Neurosci. 32:10352–10364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Torii T, Miyamoto Y, Yamamoto M, Ohbuchi
K, Tsumura H, Kawahara K, Tanoue A, Sakagami H and Yamauchi J: Arf6
mediates Schwann cell differentiation and myelination. Biochem
Biophys Res Commun. 465:450–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
George AA, Hayden S, Stanton GR and
Brockerhoff SE: Arf6 and the 5′phosphatase of Synaptojanin 1
regulate autophagy in cone photoreceptors. Inside Cell. 1:117–133.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Grossmann AH, Zhao H, Jenkins N, Zhu W,
Richards JR, Yoo JH, Winter JM, Rich B, Mleynek TM, Li DY and
Odelberg SJ: The small GTPase ARF6 regulates protein trafficking to
control cellular function during development and in disease. Small
GTPases. 1–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen PW, Jian X, Yoon HY and Randazzo PA:
ARAP2 signals through Arf6 and Rac1 to control focal adhesion
morphology. J Biol Chem. 288:5849–5860. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hongu T, Funakoshi Y, Fukuhara S, Suzuki
T, Sakimoto S, Takakura N, Ema M, Takahashi S, Itoh S, Kato M, et
al: Arf6 regulates tumour angiogenesis and growth through
HGF-induced endothelial β1 integrin recycling. Nat Commun.
6:79252015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bourmoum M, Charles R and Claing A: The
GTPase ARF6 controls ROS production to mediate angiotensin
II-induced vascular smooth muscle cell proliferation. PLoS One.
11:e01480972016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sabe H, Hashimoto S, Morishige M, Ogawa E,
Hashimoto A, Nam JM, Miura K, Yano H and Onodera Y: The
EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer
invasion and metastasis. Traffic. 10:982–993. 2009. View Article : Google Scholar : PubMed/NCBI
|