Introduction
Colorectal cancer (CRC) is one of the leading causes
of cancer-associated mortalities in developed countries (1). In the past two decades, the morbidity
and mortality of CRC have risen rapidly in Chinese population
(2). A systemic therapy involving
neoadjuvant therapy, surgical resection and postoperative therapy
provides an opportunity for long-term survival. However, the
majority of patients with metastatic CRC (mCRC) experience a poor
survival rate (3,4). A growing body of research has
demonstrated the positive effect of molecularly targeted treatment
on the survival rate of patients with mCRC, especially with the use
of monoclonal antibodies against epidermal growth factor receptor
(EGFR) (5,6).
The tumorigenesis of CRC is a multistep process
through the accumulation of genetic alterations. KRAS mutations are
considered to be an early event in tumorigenesis (7). Proteins expressed by KRAS and
BRAF genes are involved in the Ras/Raf/mitogen-activated
protein kinase-extracellular-signal-regulated kinase kinase
(MEK)/extracellular-signal-regulated kinase signaling pathway,
which is a downstream pathway of EGFR. Mutations in KRAS and
BRAF genes lead to the persistent activation of this pathway
and accelerate the proliferation of tumor cells (8). It has been widely accepted that
KRAS mutations predict the poor efficacy of anti-EGFR
therapy in patients with mCRC (9).
However, whether KRAS mutations are correlated with
decreased survival in patients with mCRC remains controversial.
Previous studies have indicated that KRAS mutations present
statistically significant reductions in overall survival (OS) and
disease-free survival (DFS) (10–12).
Nevertheless, evidence of the association between KRAS
mutations and poor OS in patients with mCRC is obtained primarily
in Western countries and certain Asian countries; few data about
prognostic significance of KRAS mutations in mCRC are
available in Chinese patients (10–12).
Approximately 90% of KRAS mutations are
located in codon 12 and 13 (13).
Certain studies have demonstrated the diversity of biological
characteristics in CRC with distinct KRAS mutational sites
(14,15). An in vitro study revealed that
tumor cells with codon 12 mutations possess an increased ability
for cell transformation compared with those with codon 13 mutations
(16). However, in other studies,
codon 13 mutations are considered to be more relevant to the poorer
outcome than codon 12 mutations (17,18).
Compared with codon 12 mutations, codon 13 mutation (G13D) exhibits
increased efficacy of cetuximab treatment (15).
The results from previous studies did not reach a
consensus and very few studies were performed to analyze
KRAS mutation subtypes in Chinese patients with mCRC. Thus,
the present study aimed to identify the frequency of KRAS
and BRAF gene mutations in Chinese patients with mCRC, and
investigate the prognostic value of distinct codon-specific
KRAS mutations and their associations with
clinicopathological characteristics.
Materials and methods
Study population
Based on the database of Sir Run Run Shaw Hospital
(Hangzhou, China), a total of 580 patients were searched with
histologically confirmed CRC and imaging confirmed metastasis
between January 2010 and June 2016, among which 135 patients were
tested for mutations in KRAS gene and 128 in BRAF
gene. Characteristics of sex, age, body mass index (BMI), location
of primary tumor, metastatic sites and the time to metastasis were
collected. There were 135 patients, including 85 males and 50
females with the age range, 28–82. Survival analysis was performed
in 101 patients between January 2010 and September 2015 who
received curative resection for primary tumor. Adjuvant therapy
included 5-fluorouracil folinic acid in combination with irinotecan
(FOLFIRI) or oxaliplatin (FOLFOX); capecitabine in combination with
oxaliplatin (XELOX) or capecitabine alone; with or without targeted
drug (cetuximab or bevacizumab). A total of 30 patients received
neoadjuvant therapy prior to surgery. All the therapies were based
on the corresponding National Comprehensive Cancer Network
guideline. The present study was approved by the Institutional
Ethics Committee of Sir Run Run Shaw Hospital and informed consent
was obtained from each participant.
DNA preparation and quantitative
polymerase chain reaction
Tissue samples were fixed in 10% formalin at ambient
temperature for 6 h. DNA was extracted from formalin-fixed
paraffin-embedded samples of primary lesions or biopsy specimens.
Genomic DNA was extracted with QIAamp® DNA FFPE tissue
kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's
instructions. AmoyDx® KRAS Mutation Detection kit and
BRAF V600 Mutations Detection kit (both from Amoy Diagnostics Co.,
Ltd., Xiamen, China) were used to detect KRAS and
BRAF status of each DNA sample according to the
manufacturer's instructions. The quantitative polymerase chain
reaction (qPCR) experiment was performed on Cobas z480 (Roche
Molecular Diagnostics, Pleasanton, CA, USA) under the following
three stages: one cycle at 95°C for 5 min, 15 cycles at 95°C for 25
sec and 64°C for 20 sec and 72°C for 20 sec, 26 cycles at 93°C for
25 sec and 60°C for 35 sec and 72°C for 20 sec. A result was
considered mutation-positive if the Cq value was <30 with a
classic S-curve (19). The
aforementioned tests were performed in the Molecular Diagnostics
Laboratory of Sir Run Run Shaw Hospital.
Statistical analysis
Data was analyzed with χ2 test or
Fisher's exact test to compare proportions. The Student's t-test
was used to compare two groups of continuous data. Data are
presented as mean values. The Kaplan-Meier method was performed for
survival analysis and log rank test was used to compare the
survival distributions. Furthermore, Cox's proportional hazards
regression model was chosen to identify the impact of factors on
OS. Hazard ratio (HR) was calculated with 95% confidence interval
(CI). P<0.05 was considered to indicate a statistically
significant difference. All statistical analysis was performed with
SPSS statistical software (version 21.0; IBM Corp., Armonk, NY,
USA). Survival curves were plotted in Graph Pad Prism (version 6.0;
GraphPad Software, Inc., La Jolla, CA, USA).
Results
KRAS mutation characteristics
Among the 135 patients with KRAS gene
detection data, mutations in exon 2 were identified in 45 (33.3%)
patients, of which 34 (68.9%) exhibited a single mutation in codon
12, and 11 (31.1%) exhibited a single mutation in codon 13. Codon
12 represented 5 mutational types, while codon 13 represented only
1 (G13D). The most frequently observed mutation was 35G>A
(G12D), followed by 35G>T (G12V), 37.8 and 26.7%, respectively.
The mutational types G12A, G12C and G12S comprised a small part of
the mutations in these 2 codons. Of the 135 patients, BRAF
codon 600 status was tested in 128 patients. Only 3 patients
exhibited a mutation of V600E (3/128, 2.3%) and none of them
possessed KRAS mutations simultaneously. Mutations are
summarized in Table I.
 | Table I.Frequency of KRAS and
BRAF mutations in patients with metastatic colorectal
cancer. |
Table I.
Frequency of KRAS and
BRAF mutations in patients with metastatic colorectal
cancer.
| Amino acid | Case (total) | (%) |
|---|
| KRAS | 45 (135) | 33.3 |
|
G12A | 2 |
|
|
G12D | 17 |
|
|
G12V | 12 |
|
|
G12C | 2 |
|
|
G12S | 1 |
|
|
G13D | 11 |
|
| BRAF |
|
|
|
V600E | 3 (128) | 2.3 |
Association between KRAS gene
mutations and clinicopathological features
The association between clinicopathological features
and KRAS codon 12, 13, and a 12/13 mutation status are
presented in Table II. KRAS
gene mutations and KRAS codon 12 mutations were
significantly more common in female patients, compared with male
patients (P<0.05). Compared with left-sided colon, right-sided
colon experienced a significantly increased number of KRAS
codon 13 mutations (P<0.05). No other significant associations
were identified.
 | Table II.Clinicopathological features
according to KRAS codon status in 135 patients with
metastatic colorectal cancer. |
Table II.
Clinicopathological features
according to KRAS codon status in 135 patients with
metastatic colorectal cancer.
|
|
| KRAS codon 12 | KRAS codon 13 | KRAS codon 12 and
13 |
|---|
|
|
|
|
|
|
|---|
| Parameter | No. | Wt | Mutation (n,
%) | P-value | Wt | Mutation (n,
%) | P-value | Wt | Mutation (n,
%) | P-value |
|---|
| Sex |
|
|
| 0.03a,b |
|
|
>0.09c |
|
| 0.04a,b |
|
Male | 85 | 69 | 16 (18.8) |
| 78 | 7 (8.2) |
| 62 | 23 (27.1) |
|
|
Female | 50 | 32 | 18 (36.0) |
| 46 | 4 (8.0) |
| 28 | 22 (44.0) |
|
| Age
(years)d | 58.2 | 57.7 | 59.8 | 0.34e | 58.7 | 53.3 | 0.79e | 58.3 | 58.1 | 0.65e |
| Body-mass
indexd | 22.9 | 23.1 | 22.5 | 0.30e | 23.0 | 22.3 | 0.82e | 23.2 | 22.4 | 0.26e |
| Location |
|
|
| 0.44b |
|
| 0.04c |
|
| 0.51b |
|
Right-sided | 43 | 34 | 9 (20.9) |
| 36 | 7 (16.3) |
| 27 | 16 (37.2) |
|
|
Left-sided | 92 | 67 | 25 (27.2) |
| 88 | 4 (4.3) |
| 63 | 29 (31.5) |
|
| Metastasis |
|
|
| 0.81b |
|
| 0.74b |
|
| 1.00b |
|
Hepatic | 42 | 32 | 10 (23.8) |
| 38 | 4 (9.5) |
| 28 | 14 (33.3) |
|
|
Extrahepatic | 93 | 69 | 24 (25.8) |
| 86 | 7 (7.5) |
| 62 | 31 (33.3) |
|
| Synchronous
metastasis |
|
|
| 0.71b |
|
| 0.12b |
|
| 0.58b |
|
Yes | 100 | 74 | 26 (26.0) |
| 94 | 6 (6.0) |
| 68 | 32 (32.0) |
|
| No | 35 | 27 | 8 (22.9) |
| 30 | 5 (14.3) |
| 22 | 13 (37.1) |
|
Prognostic value of KRAS codon 12
mutation
Among the 135 patients, 101 patients that received
curative or palliative resection for CRC were included for survival
analysis. In a Kaplan-Meier survival analysis of the KRAS
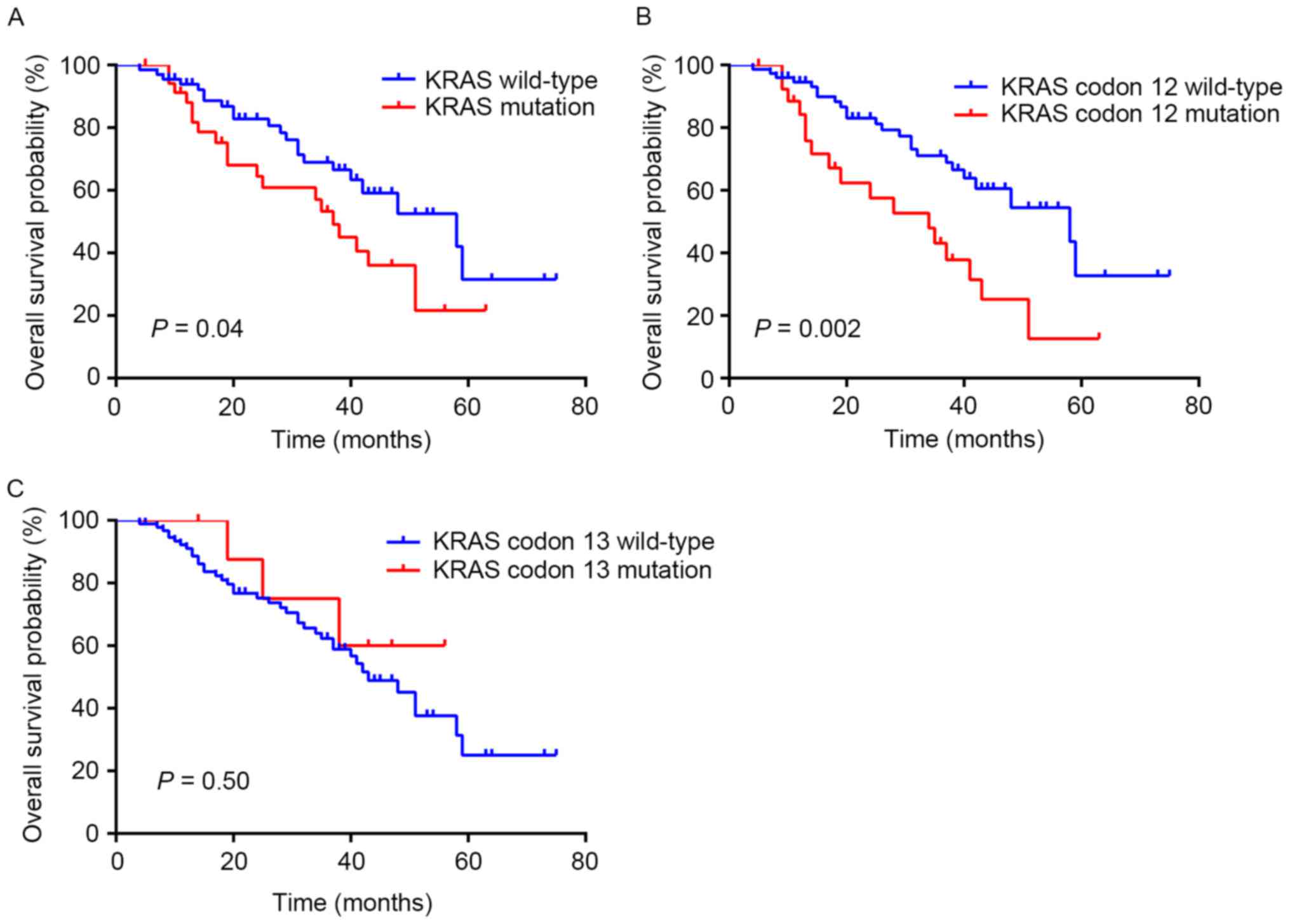
gene status in the 101 patients, KRAS mutations were highly
associated with a poorer survival (log-rank P=0.04; median
survival, 37 months in the KRAS mutant vs. 58 months in the
KRAS wild-type; Fig. 1A). In
particular, patients with KRAS codon 12 wild-type
experienced a median survival of 58 months, which was significantly
increased, compared with patients with KRAS codon 12
mutation whose median survival was 34 months (log-rank P=0.002;
Fig. 1B). The survival analysis
indicated no difference between patients with and without
KRAS codon 13 mutation (P=0.50; Fig. 1C).
Analysis of prognostic risk
factors
Age, sex, the location of primary tumor and
metastasis, the time of metastasis, the use of target drug and
neoadjuvant therapy, and the KRAS gene status were analyzed
with the Cox regression model (Table
III). With the exception of KRAS codon 12 status, none
of these factors exhibited a predictive value for poor prognosis.
The patients with KRAS codon 12 mutations presented a
significant decrease in overall survival (HR=2.528, 95%
CI=1.369–4.668, P=0.003). In comparison, KRAS codon 13
mutants demonstrated no significant effect on survival (HR=0.657,
95% CI=0.202–2.135, P=0.49). Management of targeted and neoadjuvant
therapy was not associated with risk of mortality (P=0.85; P=0.78,
respectively). Further analysis revealed that rather than
c.35G>C (G12A) or c.35G>T (G12C), c.35G>A (G12D) and
c.35G>T (G12V) were associated with a significantly decreased OS
compared with KRAS wild-type (HR=2.313, 95% CI=1.069–5.004,
P=0.03; HR=2.621, 95% CI=1.057–6.497, P=0.04, respectively;
Table IV).
 | Table III.Univariate analysis of overall
survival for 101 patients. |
Table III.
Univariate analysis of overall
survival for 101 patients.
| Variable | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Sex |
| 0.17 |
|
Male | Ref. |
|
|
Female | 1.547
(0.832–2.875) |
|
| Age (years) | 1.012
(0.985–1.039) | 0.39 |
| Location |
| 0.41 |
|
Right-sided | Ref. |
|
|
Left-sided | 0.758
(0.393–1.462) |
|
| Metastasis |
| 0.64 |
|
Hepatic | Ref. |
|
|
Extrahepatic | 1.194
(0.569–2.507) |
|
| Synchronous
metastasis |
| 0.16 |
|
Yes | Ref. |
|
| No | 0.633
(0.336–1.195) |
|
| Targeted drug |
| 0.85 |
| No | Ref. |
|
|
Yes | 0.937
(0.479–1.834) |
|
| Neoadjuvant
therapy |
| 0.78 |
| No | Ref. |
|
|
Yes | 1.101
(0.569–2.129) |
|
| KRAS status |
| 0.04a |
| Wt | Ref. |
|
|
Mutant | 1.884
(1.026–3.462) |
|
| Codon 12
status |
| <0.01 |
| Wt | Ref. |
|
|
Mutant | 2.528
(1.369–4.668) |
|
| Codon 13
status |
| 0.49 |
| Wt | Ref. |
|
|
Mutant | 0.657
(0.202–2.135) |
|
 | Table IV.Analysis of overall survival
according to KRAS codon 12 mutations by Cox regression
analysis. |
Table IV.
Analysis of overall survival
according to KRAS codon 12 mutations by Cox regression
analysis.
| Nucleotide | Amino acid | Total number | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Wild-type |
| 74 | Ref. |
|
| c.35G>A | G12D | 13 | 2.313
(1.069–5.004) | 0.03a |
| c.35G>T | G12V | 10 | 2.621
(1.057–6.497) | 0.04a |
| c.35G>C | G12A | 2 | 3.308
(0.432–25.358) | 0.25 |
| c.34G>T | G12C | 2 | 3.991
(0.928–17.165) | 0.06a |
Discussion
In the present study, the frequencies of KRAS
and BRAF gene mutations were determined in Chinese patients
with mCRC. The association between KRAS mutations with
clinicopathological features was investigated. KRAS codon 12
mutations, especially G12D and G12V were revealed to exhibit
predictive value for poor overall survival. To the best of our
knowledge, studies concerning KRAS mutations have primarily
been conducted only in the Western population (10,11). The
impact of KRAS mutation, especially its different mutational
sites, on the survival of Chinese population was uncertain and
controversial.
The frequency of KRAS mutations (33.3%) and
BRAF mutations (2.3%) in the present study were in
accordance with a previous retrospective observational study that
also involved Chinese patients with mCRC (34.8 and 3.4%,
respectively) (20). However, the
KRAS and BRAF mutation rate was slightly higher in
the Western population (37.3 and 6%) (10). Since the morbidity and mortality of
CRC in China were different from that in Western countries, the
KRAS and BRAF mutation rates in different regions
required further exploration (21).
The majority of KRAS mutations have been
revealed to occur in codon 12 and 13 (3). The detection of associations between
KRAS mutations and clinicopathological features had been
performed widely; however, the data was lacking when codon 12 and
13 were considered separately (14,22,23). In
the present study, compared with male patients, KRAS codon
12 mutations were more common in female patients compared with
codon 13 mutations. Notably, while KRAS mutations (including
codon 12 and 13) were not associated with the location of primary
tumor, codon 13 mutation (G13D) itself was likely to occur in the
tumors arising from right-sided colon. Recently, it was accepted
that patients with right-sided colon cancer experienced an inferior
OS rate (24). Additionally,
increasing evidence suggests that mutations in KRAS codon 13
predict a poor outcome (12,18). In the present study, G13D mutation was
associated with right-sided colon cancer; however, exhibited no
prognostic significance, which may be explained by the small sample
size. As for patterns of recurrence, Margonis et al
(18) suggested that patients with
KRAS codon 13 mutations, rather than codon 12 mutations
possessed an increased risk of extrahepatic recurrence and
lung-specific recurrence. Nevertheless, the results of the present
study identified no difference in the patterns of tumor progression
(including metastatic sites and time to recurrence) according to
codon 12 or 13 mutation status.
The distribution of mutations in codon 12 and 13
varied in distinct studies. G12D in the present study was the most
prominent subtype which was congruent with data from Caucasian and
other Chinese mCRC (25–27). It had been reported that different
subtypes of KRAS mutations may confer variable tumor biology
(28,29). A previous study involving surgically
resected lung adenocarcinoma revealed that G12C mutation was
associated with poorer outcome compared with other subtypes of
mutations (28). Another study in
Chinese patients with CRC demonstrated that KRAS codon 13
mutations, in particular G13D, were associated with significantly
decreased OS rates (12). However, in
the present study, KRAS codon 13 mutations had no effect on
OS, but mutations in KRAS codon 12 were predictive for poor
prognosis. This result was consistent with the evidence from in
vitro studies, in which codon 12 mutations may increase
aggressiveness due to increased transforming capacity and decreased
levels of apoptosis (16,30). Previously, meta-analysis also revealed
the predictive value that codon 12 mutations possessed by pooling
the data from five randomized trials researching patients with mCRC
in Western countries (10). In that
meta-analysis, G12C was associated with inferior OS compared with
KRAS wild-type; however, the results of the present study
demonstrated that G12D and G12V were associated with decreased OS
times. The prognostic value of codon and amino acid specific
KRAS mutations has been discussed on several cancer types.
For example, Bournet et al (31) reported that KRAS G12D mutant
was an independent prognostic factor for unresectable pancreatic
cancer. It had been proposed that in non-small cell lung cancer,
the G12D mutation subtype was associated with the activation of the
phosphoinositide 3-kinase/AKT serine/threonine kinase (AKT) and MEK
signaling whereas mutation G12V or G12C preferred to activate Raf
and Ral, and decreased growth factor-dependent AKT activation
(32,33). These findings led to the present study
in which the association between signaling pathways and the role of
KRAS mutation subtypes on prognosis of patients with CRC
were investigated, particularly in patients with mCRC from
different regions.
BRAF V600E mutation has also been widely
identified in CRC (34). It is
considered to be associated with poor clinical outcome and may have
confounding effect with KRAS mutations (35,36).
BRAF V600E mutation was not included in the further analysis
due to its extremely low mutation rate (2.3%).
There were several limitations to the present study,
for example it was a retrospective study with a small sample size.
Microsatellite instability and other family members of RAS
gene that serve a function in CRC were not included. The
therapeutic regimens varied between the patients, which may have
resulted in the heterogeneity. However, no significant difference
was observed in the number of patients receiving neoadjuvant
therapy and targeted therapy between the groups of codon 12
mutation, and codon 12 wild-type (P>0.05; data not shown).
Additionally, the Cox regression model identified that whether
receiving these two therapies or not was not a factor affecting the
survival.
In conclusion, the results of the present study
indicate that codon 12 mutations may predict poor OS in Chinese
patients with mCRC, and further investigation demonstrated that
G12D and G12V served as an indicator of poor prognosis in this
specific population. In future clinical studies of CRC, the
significance of gene mutation subtypes should be brought into
consideration.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81272681). The
AmoyDx® KRAS Mutation Detection kit, BRAF V600 Mutations
Detection kit and corresponding primers are patented (patent no. ZL
2009 1 0111501.6).
References
|
1
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ng SC and Wong SH: Colorectal cancer
screening in Asia. Br Med Bull. 105:29–42. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung JJ, Lau JY, Young GP, Sano Y, Chiu
HM, Byeon JS, Yeoh KG, Goh KL, Sollano J, Rerknimitr R, et al: Asia
Pacific consensus recommendations for colorectal cancer screening.
Gut. 57:1166–1176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shao YC, Chang YY, Lin JK, Lin CC, Wang
HS, Yang SH, Jiang JK, Lan YT, Lin TC, Li AF, et al: Neoadjuvant
chemotherapy can improve outcome of colorectal cancer patients with
unresectable metastasis. Int J Colorectal Dis. 28:1359–1365. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohhara Y, Fukuda N, Takeuchi S, Honma R,
Shimizu Y, Kinoshita I and Dosaka-Akita H: Role of targeted therapy
in metastatic colorectal cancer. World J Gastrointest Oncol.
8:642–655. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graham DM, Coyle VM, Kennedy RD and Wilson
RH: Molecular subtypes and personalized therapy in metastatic
colorectal cancer. Curr Colorectal Cancer Rep. 12:141–150. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McCubrey JA, Steelman LS, Abrams SL, Lee
JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA,
D'Assoro AB, et al: Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT
pathways in malignant transformation and drug resistance. Adv
Enzyme Regul. 46:249–279. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Cutsem E, Köhne CH, Láng I, Folprecht
G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D,
Tejpar S, et al: Cetuximab plus irinotecan, fluorouracil, and
leucovorin as first-line treatment for metastatic colorectal
cancer: Updated analysis of overall survival according to tumor
KRAS and BRAF mutation status. J Clin Oncol. 29:2011–2019. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Modest DP, Ricard I, Heinemann V,
Hegewisch-Becker S, Schmiegel W, Porschen R, Stintzing S, Graeven
U, Arnold D, von Weikersthal LF, et al: Outcome according to KRAS-,
NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled
analysis of five randomized trials in metastatic colorectal cancer
by the AIO colorectal cancer study group. Ann Oncol. 27:1746–1753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andreatos N, Ronnekleiv-Kelly S, Margonis
GA, Sasaki K, Gani F, Amini N, Wilson A and Pawlik TM: From bench
to bedside: Clinical implications of KRAS status in patients with
colorectal liver metastasis. Surg Oncol. 25:332–338. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Guo F, Shi X, Zhang L, Zhang A,
Jin H and He Y: BRAF V600E mutation and KRAS codon 13 mutations
predict poor survival in Chinese colorectal cancer patients. BMC
Cancer. 14:8022014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Neumann J, Zeindl-Eberhart E, Kirchner T
and Jung A: Frequency and type of KRAS mutations in routine
diagnostic analysis of metastatic colorectal cancer. Pathol Res
Pract. 205:858–862. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Z, Chen Y, Wang D, Wang G, He L and Suo
J: Detection of KRAS mutations and their associations with
clinicopathological features and survival in Chinese colorectal
cancer patients. J Int Med Res. 40:1589–1598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Roock W, Jonker DJ, Di Nicolantonio F,
Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M,
Piessevaux H, et al: Association of KRAS p.G13D mutation with
outcome in patients with chemotherapy-refractory metastatic
colorectal cancer treated with cetuximab. JAMA. 304:1812–1820.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guerrero S, Casanova I, Farré L, Mazo A,
Capellà G and Mangues R: K-ras codon 12 mutation induces higher
level of resistance to apoptosis and predisposition to
anchorage-independent growth than codon 13 mutation or
proto-oncogene overexpression. Cancer Res. 60:6750–6756.
2000.PubMed/NCBI
|
|
17
|
Samowitz WS, Curtin K, Schaffer D,
Robertson M, Leppert M and Slattery ML: Relationship of Ki-ras
mutations in colon cancers to tumor location, stage, and survival:
A population-based study. Cancer Epidemiol Biomarkers Prev.
9:1193–1197. 2000.PubMed/NCBI
|
|
18
|
Margonis GA, Kim Y, Sasaki K, Samaha M,
Amini N and Pawlik TM: Codon 13 KRAS mutation predicts patterns of
recurrence in patients undergoing hepatectomy for colorectal liver
metastases. Cancer. 122:2698–2707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang H, Zheng X, Ji T, Fu L, Bai D, Liao
Y, Zhang H, Ding Y and Zheng L: Comparative screening of K-ras
mutations in colorectal cancer and lung cancer patients using a
novel real-time PCR with ADx-K-ras Kit and Sanger DNA sequencing.
Cell Biochem Biophys. 62:415–420. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li ZZ, Wang F, Zhang ZC, Wang F, Zhao Q,
Zhang DS, Wang FH, Wang ZQ, Luo HY, He MM, et al: Mutation
profiling in chinese patients with metastatic colorectal cancer and
its correlation with clinicopathological features and anti-EGFR
treatment response. Oncotarget. 7:28356–28368. 2016.PubMed/NCBI
|
|
21
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Price TJ, Bruhn MA, Lee CK, Hardingham JE,
Townsend AR, Mann KP, Simes J, Weickhardt A, Wrin JW, Wilson K, et
al: Correlation of extended RAS and PIK3CA gene mutation status
with outcomes from the phase III AGITG MAX STUDY involving
capecitabine alone or in combination with bevacizumab plus or minus
mitomycin C in advanced colorectal cancer. Br J Cancer.
112:963–970. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tejpar S, Stintzing S, Ciardiello F,
Tabernero J, Van Cutsem E, Beier F, Esser R, Lenz HJ and Heinemann
V: Prognostic and predictive relevance of primary tumor location in
patients with RAS wild-type metastatic colorectal cancer:
Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA
Oncol. Oct 10–2016.(Epub ahead of print).
|
|
25
|
Li FH, Shen L, Li ZH, Luo HY, Qiu MZ,
Zhang HZ, Li YH and Xu RH: Impact of KRAS mutation and PTEN
expression on cetuximab-treated colorectal cancer. World J
Gastroenterol. 16:5881–5888. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao C, Zhou J, Yang Z, Huang Y, Wu X, Shen
H, Tang J and Chen Q: KRAS, BRAF and PIK3CA mutations and the loss
of PTEN expression in Chinese patients with colorectal cancer. PLoS
One. 7:e366532012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Frattini M, Saletti P, Romagnani E, Martin
V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F and
Mazzucchelli L: PTEN loss of expression predicts cetuximab efficacy
in metastatic colorectal cancer patients. Br J Cancer.
97:1139–1145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nadal E, Chen G, Prensner JR, Shiratsuchi
H, Sam C, Zhao L, Kalemkerian GP, Brenner D, Lin J, Reddy RM, et
al: KRAS-G12C mutation is associated with poor outcome in
surgically resected lung adenocarcinoma. J Thorac Oncol.
9:1513–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tejpar S, Celik I, Schlichting M,
Sartorius U, Bokemeyer C and Van Cutsem E: Association of KRAS G13D
tumor mutations with outcome in patients with metastatic colorectal
cancer treated with first-line chemotherapy with or without
cetuximab. J Clin Oncol. 30:3570–3577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ward RL, Todd AV, Santiago F, O'Connor T
and Hawkins NJ: Activation of the K-ras oncogene in colorectal
neoplasms is associated with decreased apoptosis. Cancer.
79:1106–1113. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bournet B, Muscari F, Buscail C, Assenat
E, Barthet M, Hammel P, Selves J, Guimbaud R, Cordelier P and
Buscail L: KRAS G12D mutation subtype is a prognostic factor for
advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol.
7:e1572016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ihle NT, Byers LA, Kim ES, Saintigny P,
Lee JJ, Blumenschein GR, Tsao A, Liu S, Larsen JE, Wang J, et al:
Effect of KRAS oncogene substitutions on protein behavior:
Implications for signaling and clinical outcome. J Natl Cancer
Inst. 104:228–239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guibert N, Ilie M, Long E, Hofman V,
Bouhlel L, Brest P, Mograbi B, Marquette CH, Didier A, Mazieres J
and Hofman P: KRAS mutations in lung adenocarcinoma: Molecular and
epidemiological characteristics, methods for detection, and
therapeutic strategy perspectives. Curr Mol Med. 15:418–432. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li WQ, Kawakami K, Ruszkiewicz A, Bennett
G, Moore J and Iacopetta B: BRAF mutations are associated with
distinctive clinical, pathological and molecular features of
colorectal cancer independently of microsatellite instability
status. Mol Cancer. 5:22006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yokota T, Ura T, Shibata N, Takahari D,
Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K and
Yatabe Y: BRAF mutation is a powerful prognostic factor in advanced
and recurrent colorectal cancer. Br J Cancer. 104:856–862. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Imamura Y, Morikawa T, Liao X, Lochhead P,
Kuchiba A, Yamauchi M, Qian ZR, Nishihara R, Meyerhardt JA, Haigis
KM, et al: Specific mutations in KRAS codons 12 and 13, and patient
prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer
Res. 18:4753–4763. 2012. View Article : Google Scholar : PubMed/NCBI
|