Introduction
Gastric cancer (GC) is a stomach cancer with a
particularly high incidence in China, and in 2015 was the second
leading cause of cancer-associated mortality in China (1,2). It has
been estimated that in 2015 there were 498,000 GC-associated
mortalities in China in 2015 (1). Not
only is GC prevalent in China, patients also experience poor
survival rates (with the age-standardized 5-year relative survival
rate being <30%) (3). It is now
known that GC is a disease that results from environmental and
genetic factors and that Helicobacter pylori infection is
the most notable risk factor, with alcohol consumption also
potentially promoting the progression of GC (4–6). The link
between alcohol and GC has been determined, however, the underlying
molecular mechanism remains elusive (7,8). Thus, the
function of ethanol metabolism genes and signaling pathways in GC
require further investigation.
Previous studies have reported that alcohol
dehydrogenase (ADH) genes are associated with upper aerodigestive
types of cancer, and the genetic variation of the ADH cluster
increases the risk of cancer for alcohol drinkers (9,10).
Furthermore, ADH isoenzymes additionally exhibit diagnostic value
and prognostic prediction value in multiple different types of
cancer (11–13). Total ADH isoenzyme activity is
significantly increased in cancer tissues compared with healthy
organs, and ADH isoenzymes possess diagnostic value for patients
with GC (14,15). However, to the best of our knowledge,
the prognostic value of ADH isoenzymes in GC have yet to be
reported. The aim of the present study was to identify the
prognostic value of ADH genes in patients with GC.
Kaplan-Meier plotter (KM-plotter) (16) is an online survival analysis tool used
to rapidly assess the effect of genes on the prognosis of four
cancer types (breast, lung, ovarian and GC) using genome-wide
microarrays from the Gene Expression Omnibus (GEO) and Cancer
Biomedical Informatics Grid and The Cancer Genome Atlas (TCGA)
(17–20). A previous study by Li et al
(21) assessed the prognostic
functions of ADH 1 family member A1 isoenzyme (another alcohol
metabolism-associated isoenzyme) in GC using the KM-plotter tool,
and identified that aldehyde dehydrogenase (ALDH) family member A3
and ALDH family member L1 were potential prognostic biomarkers and
therapeutic targets for GC. In the present study, the KM-plotter
tool was used to further investigate the prognostic prediction
values of ADH genes in patients with GC.
Materials and methods
Bioinformatic analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) enrichment of ADH genes were analyzed using the
Database for Annotation, Visualization and Integrated Discovery
(david.ncifcrf.gov/home.jsp; accessed
February 20, 2017) v.6.8 (22).
Gene-gene and protein-protein interaction (PPI) networks were
constructed using the gene multiple association network integration
algorithm (GeneMANIA; genemania.org;
accessed February 20, 2017) (23,24) and
and the Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING v.10.0; string.embl.de; accessed February 20,
2017) (25,26), respectively.
Data sources
KM-plotter datasets were obtained from GEO and TCGA
(16). There were datasets available
on the websites for four different types of cancer, including,
breast, lung, ovarian and gastric cancer (16–20). In
the present study, KM-plotter (kmplot.com/analysis/index.php?p=service&cancer=gastric;
accessed February 20, 2017) was used to identify the distinct
prognostic values of ADH genes in GC (20). The clinical variables of GC, including
Tumor-Node-Metastasis stage (Seventh edition) (27), tumor differentiation, Lauren
classification (28), human epidermal
growth factor receptor 2 (HER2) status and treatment were available
on the websites. As the GSE52254 dataset was markedly different
with regards to characteristics, compared with other GC datasets in
the KM-plotter, it was excluded from the present study and the
remaining five datasets were included for further investigation
(GSE142210, GSE15459, GSE22377, GSE29272 and GSE51105) (20). Overall, the mRNA datasets of 593
patients with GC, with corresponding clinical data, were included
in the survival analysis and a total of 7 ADH genes were available
on the KM-plotter website (kmplot.com/analysis/index.php?p=service&cancer=gastric;
accessed February 20, 2017). The mRNA expression of the ADH genes
in GC tumor and adjacent non-tumor tissues were used from the
GSE29272 dataset (29), which
included the expression data of 134 pairs of GC tumor and adjacent
non-tumor tissue. The mRNA co-expression heat map of ADH genes was
constructed from the mRNA expression of GC tumor tissues from the
GSE29272 dataset. Microarray data were normalized using the RMA
method, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM723464).
Statistical analysis
Survival analysis in the KM-plotter tool used the
Kaplan-Meier method with log-rank test, whereas the comparison of
ADH genes mRNA expression between tumor and adjacent non-tumor used
paired Student's t-tests. Hazard ratios (HR) and 95% confidence
intervals (CI) were used to assess the relative risk of GC
survival. Pearson's correlation coefficient was used to assess the
co-expression correlation at the mRNA expression level, and a
co-expression heat map was constructed using the corrplot package
in R 3.3.0 platform (30). Scatter
plots were plotted using GraphPad Prism 6.0 (GraphPad Software,
Inc., La Jolla, CA, USA). Statistical analyses were performed using
SPSS v.20.0 software (IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Bioinformatic analysis
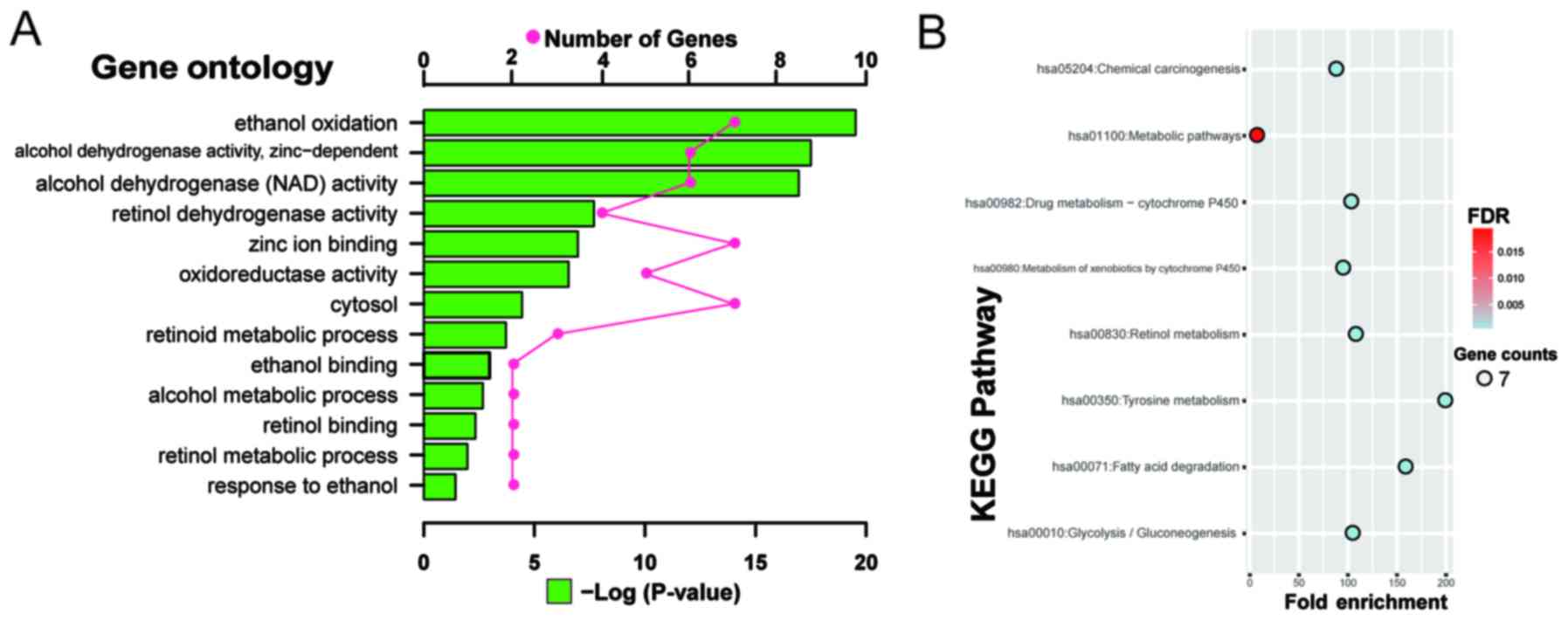
GO analysis suggested that ADH genes [ADH 1A (class
I), α polypeptide (ADH1A), ADH 1B (class I), β polypeptide
(ADH1B), ADH 1C (class I), γ polypeptide (ADH1C), ADH
4 (class II), π polypeptide (ADH4), ADH 5 (class III), χ
polypeptide (ADH5), ADH 6 (class V) (ADH6) and ADH 7
(class IV), µ or σ polypeptide (ADH7)] were involved in
ethanol oxidation, ADH activity, oxidoreductase activity, and
alcohol and retinol metabolic processes (Fig. 1A). Whereas KEGG analysis of ADH genes
indicated functions in tyrosine metabolism, retinol metabolism,
glycolysis and gluconeogenesis, fatty acid degradation, drug
metabolism via cytochrome P450 enzymes, metabolism of xenobiotics
by cytochrome P450 enzymes and chemical carcinogenesis (Fig. 1B). GO and KEGG function enrichment
analyses indicate that the ADH genes function in alcohol and toxic
chemical metabolism, and participate in tumorigenesis.
Gene and protein interaction networks suggested that
the ADH genes exist in a complex network associated with each
other, with the exception of ADH1C which was not recognized
by GeneMANIA and STRING. Gene-gene interaction networks revealed
that the ADH genes co-express with each other (Fig. 2A), whereas the PPI network data
revealed that ADH isoenzymes were directly or indirectly associated
with each other (Fig. 2B).
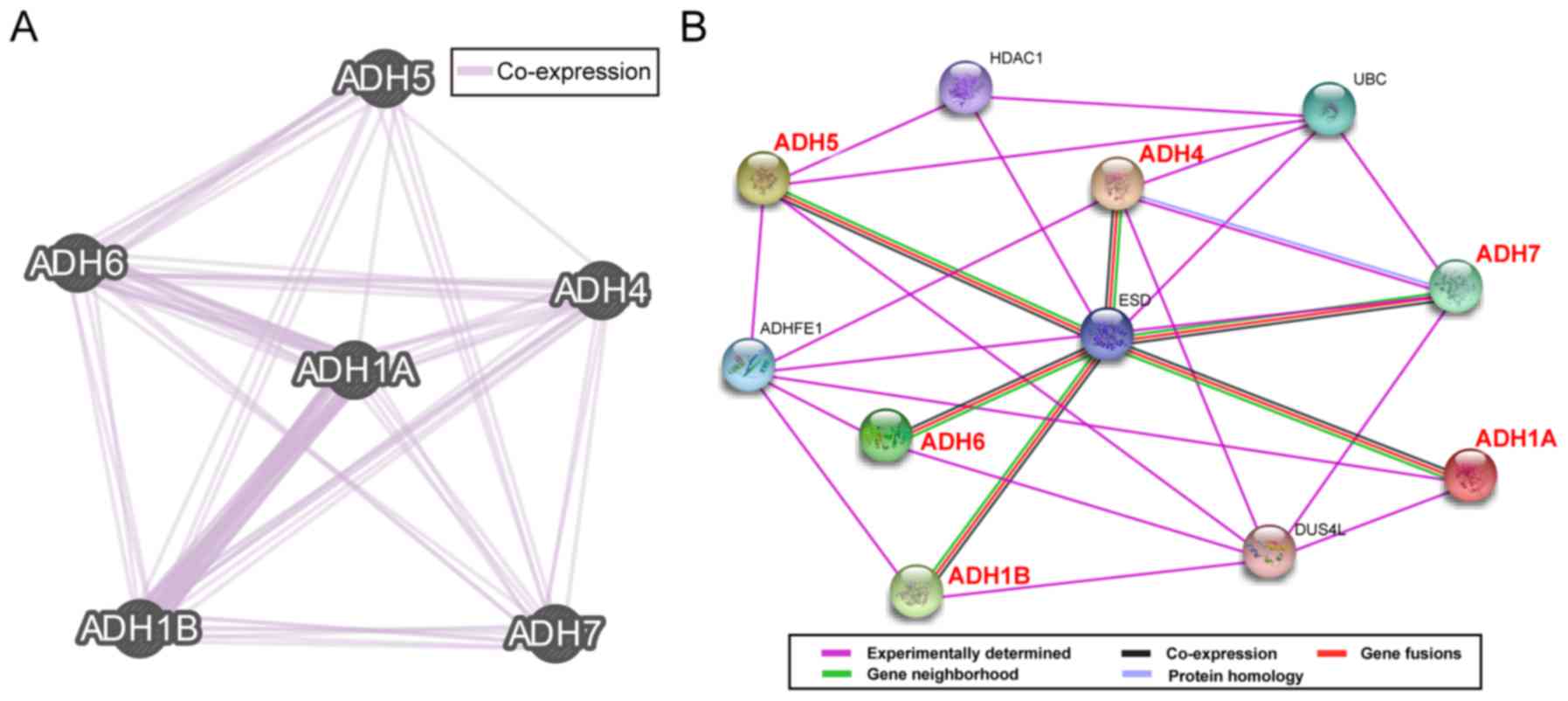 | Figure 2.Gene and protein interaction networks
of ADH genes. (A) Gene multiple association network integration
algorithm and (B) protein-protein interaction networks. ADH5,
alcohol dehydrogenase 5 (class III), χ polypeptide; ADH6, alcohol
dehydrogenase 6 (class V); ADH4, alcohol dehydrogenase 4 (class
II), π polypeptide; ADH1A, alcohol dehydrogenase 1A (class I), α
polypeptide; ADH1B, alcohol dehydrogenase 1B (class I), β
polypeptide; ADH7, alcohol dehydrogenase 7 (class IV), µ or σ
polypeptide; HDAC1, histone deacetylase 1; UBC, ubiquitin C; ESD,
esterase D; ADHFE1, alcohol dehydrogenase, iron containing 1;
DUS4L, dihydrouridine synthase 4 like. |
Survival analysis
By searching the GC database of the KM-plotter tool,
only the GSE29272 dataset was revealed to have an mRNA dataset of
GC tumor and adjacent non-tumor tissues. Consequently, only the
GSE29272 dataset was used in order to compare GC tumor and adjacent
non-tumor tissues. Due to missing data for ADH4 in the
GSE29272 dataset, only six ADH genes were included in the
comparison and co-expression heat map construction. ADH1A
(P<0.001), ADH1B (P<0.001), ADH1C (P<0.001)
and ADH7 (P<0.001) exhibited significantly decreased
expression in GC tumor tissue compared with non-tumor tissues,
whereas ADH5 was significantly upregulated (P=0.004) and
ADH6 demonstrated no significant difference (P=0.343) in
tumor tissues compared with non-tumor tissues (Fig. 3A). Furthermore, all the ADH genes
demonstrated a positive association with each other (Fig. 3B).
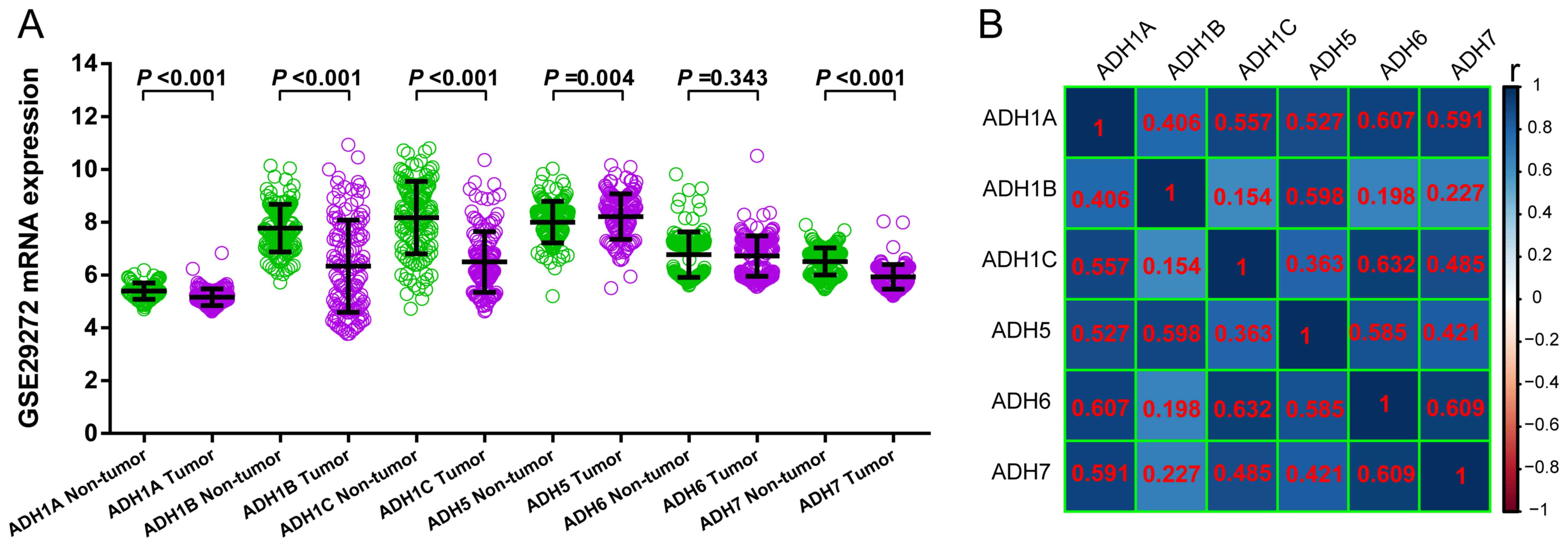 | Figure 3.Comparison of the expression of ADH
genes between GC tumor and adjacent non-tumor tissue and ADH gene
co-expression analysis. (A) Scatter plot of the expression of ADH
genes between tumor and adjacent non-tumor tissue in patients with
GC. (B) Co-expression heat map of ADH genes (the numbers shown in
red are the r-values of pearson correlation coefficient). GC,
gastric cancer; ADH1A, alcohol dehydrogenase 1A (class I), α
polypeptide; ADH1B, alcohol dehydrogenase 1B (class I), β
polypeptide; ADH1C, alcohol dehydrogenase 1C (class I), γ
polypeptide; ADH5, alcohol dehydrogenase 5 (class III), χ
polypeptide; ADH6, alcohol dehydrogenase 6 (class V); ADH7, alcohol
dehydrogenase 7 (class IV), µ or σ polypeptide. |
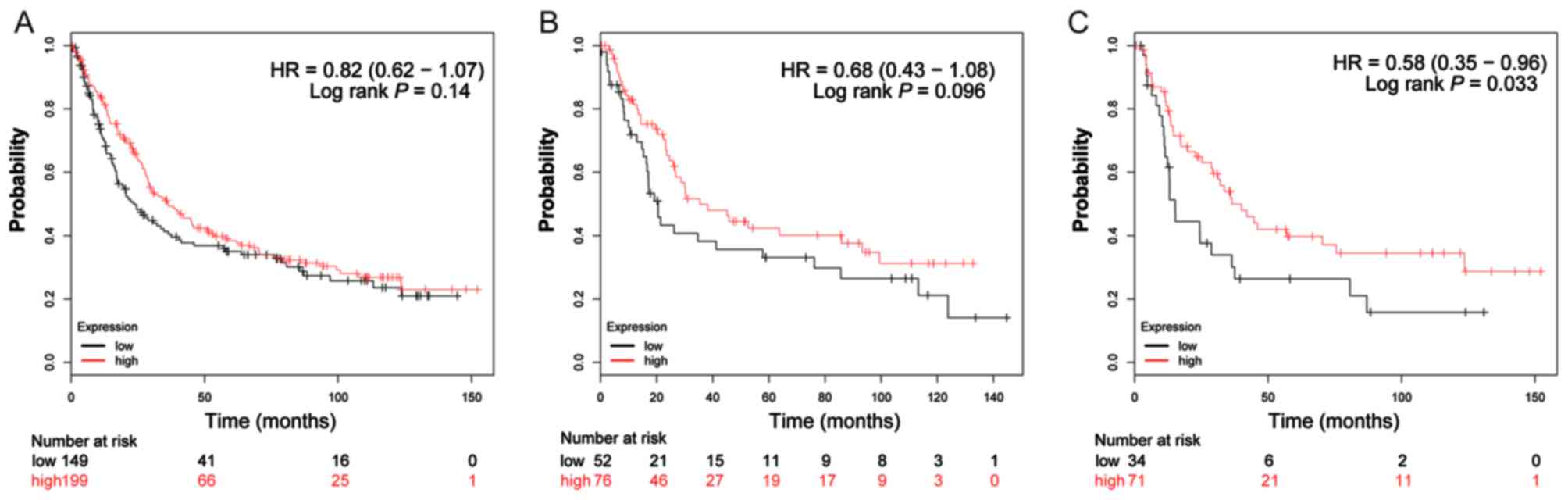
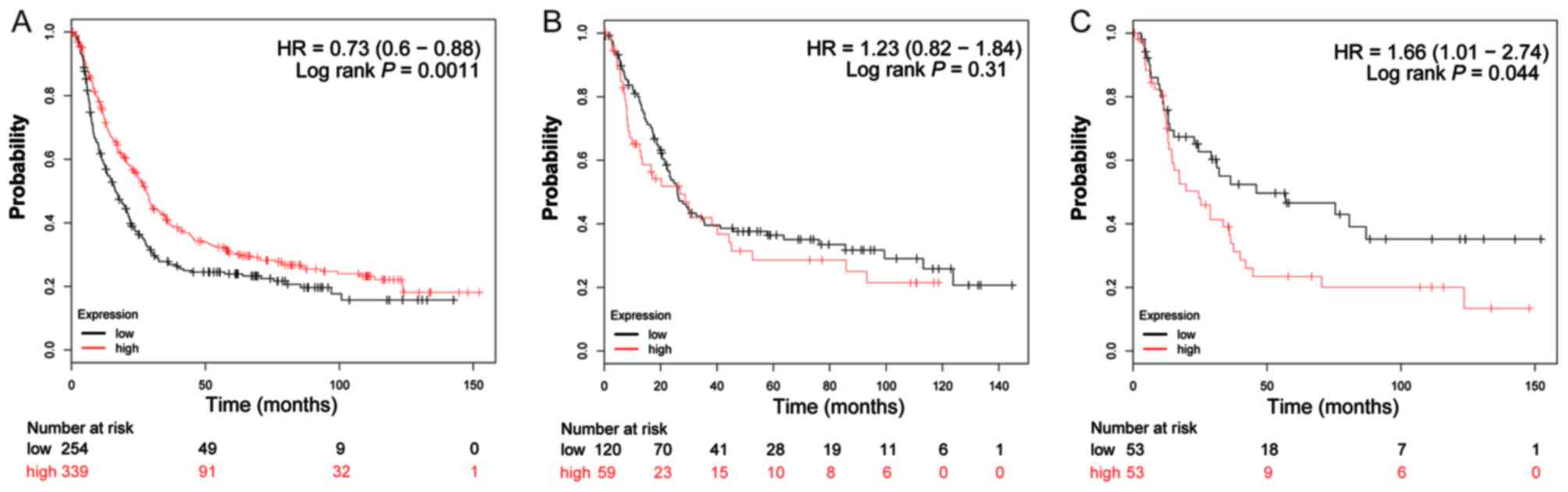
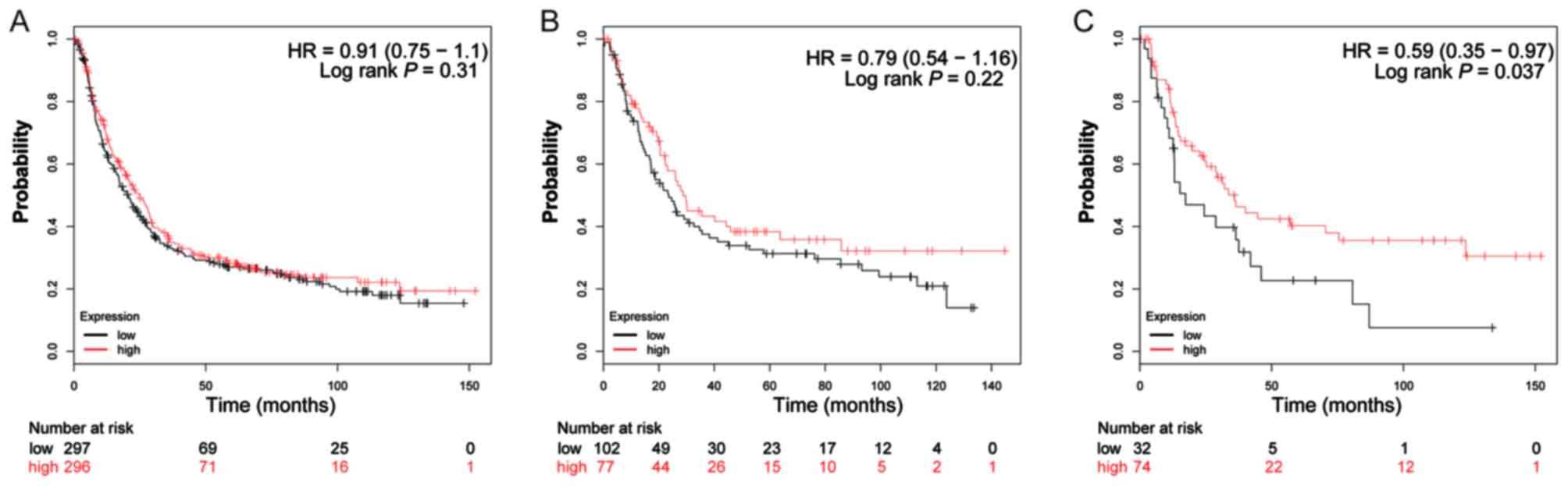
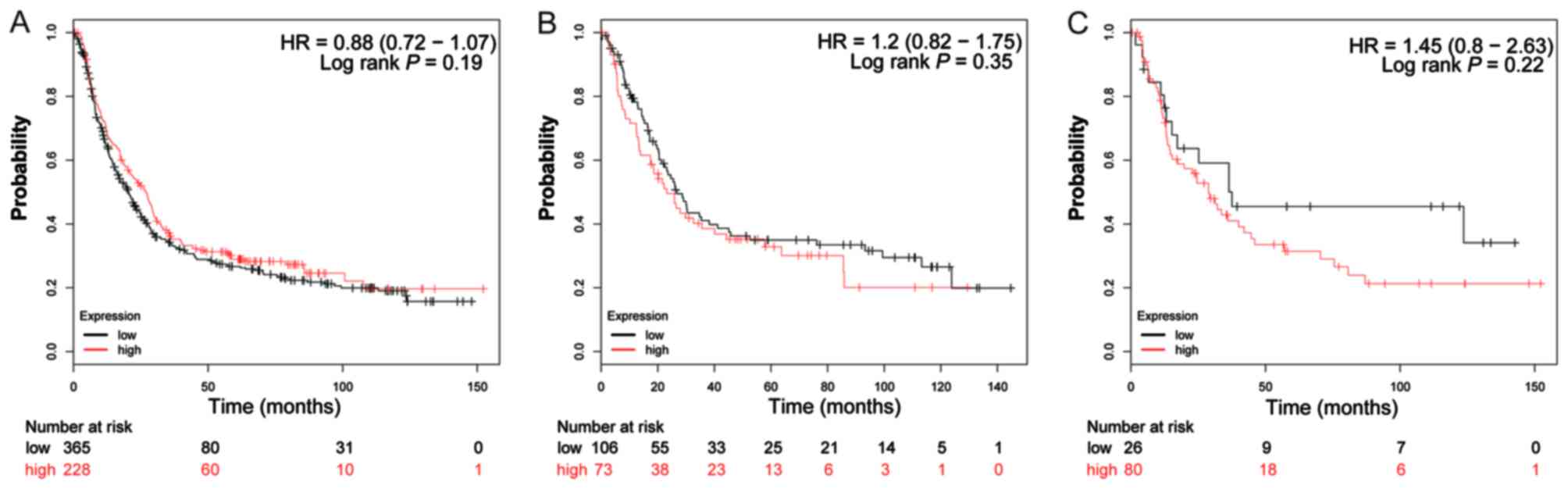
The valid gene Affymetrix ID of ADH genes in the
KM-plotter tool were 207820_at (ADH1A; Fig. 4A-C), 209613_at (ADH1B; Fig. 5A-C), 206262_at (ADH1C; Fig. 6A-C), 231678_at (ADH4; Fig. 7A-C), 208848_at (ADH5; Fig. 8A-C), 214261_at (ADH6; Fig. 9A-C) and 21055_at (ADH7;
Fig. 10A-C). Survival analysis
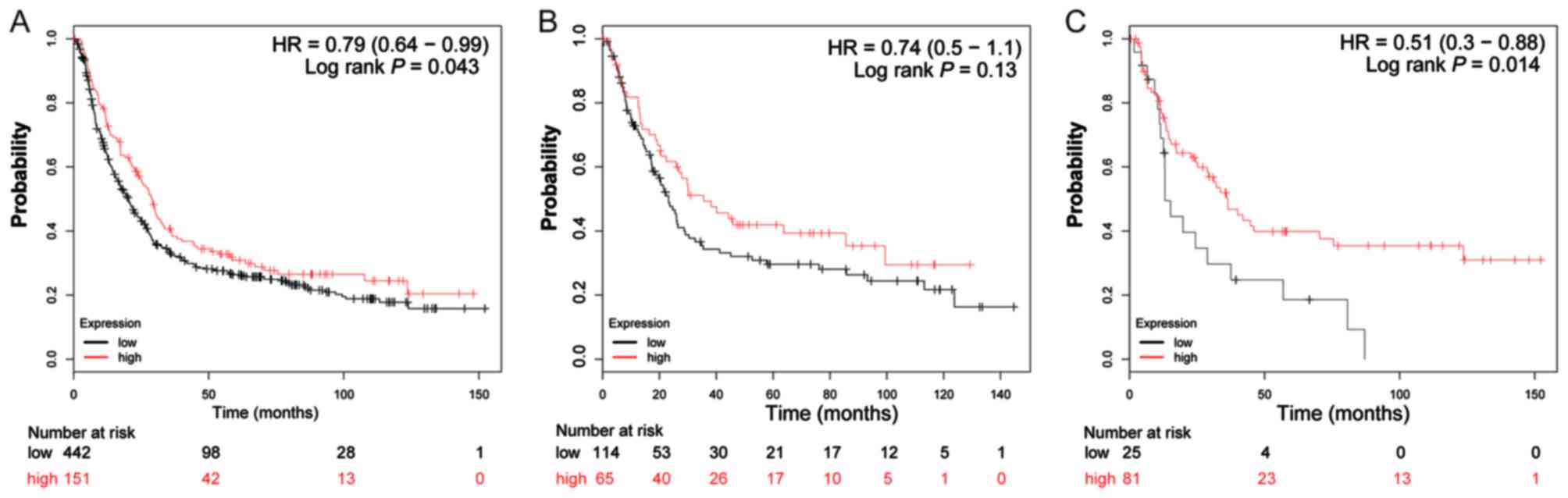
suggested that, overall, patients with GC with a high expression of
ADH1A (log-rank P=0.043; HR=0.79; 95% CI: 0.64–0.99;
Fig. 4A), ADH1B (log-rank
P=1.9×10−5; HR=0.65; 95% CI: 0.53–0.79; Fig. 5A) and ADH5 (log-rank P=0.0011;
HR=0.73; 95% CI: 0.60–0.88; Fig. 8A)
were significantly associated with a favorable prognosis, compared
with those with a low expression of these genes. Similar results
are also observed in ADH4 (Fig.
7A) and ADH7 (Fig. 10A),
however the log-rank P-values did not reach statistical
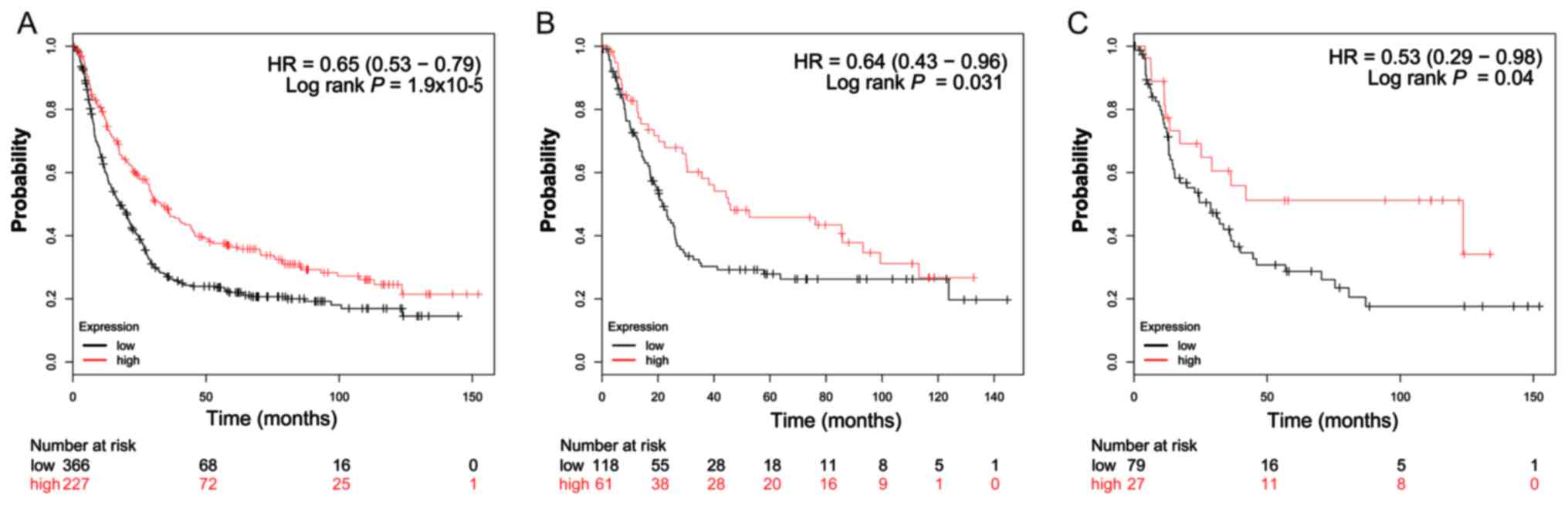
significance. Subgroup survival analysis of patients with
intestinal-type GC demonstrated that high expression of
ADH1B (log-rank P=0.031; HR=0.64; 95% CI: 0.43–0.96;
Fig. 5B) significantly decreased the
risk of mortality, compared with patients with a low expression of
this gene. Furthermore, the increased expression of ADH1A,
ADH1C and ADH4 also demonstrated a similar outcome, but
with a non-significant log-rank P-values (Figs. 4B, 6B
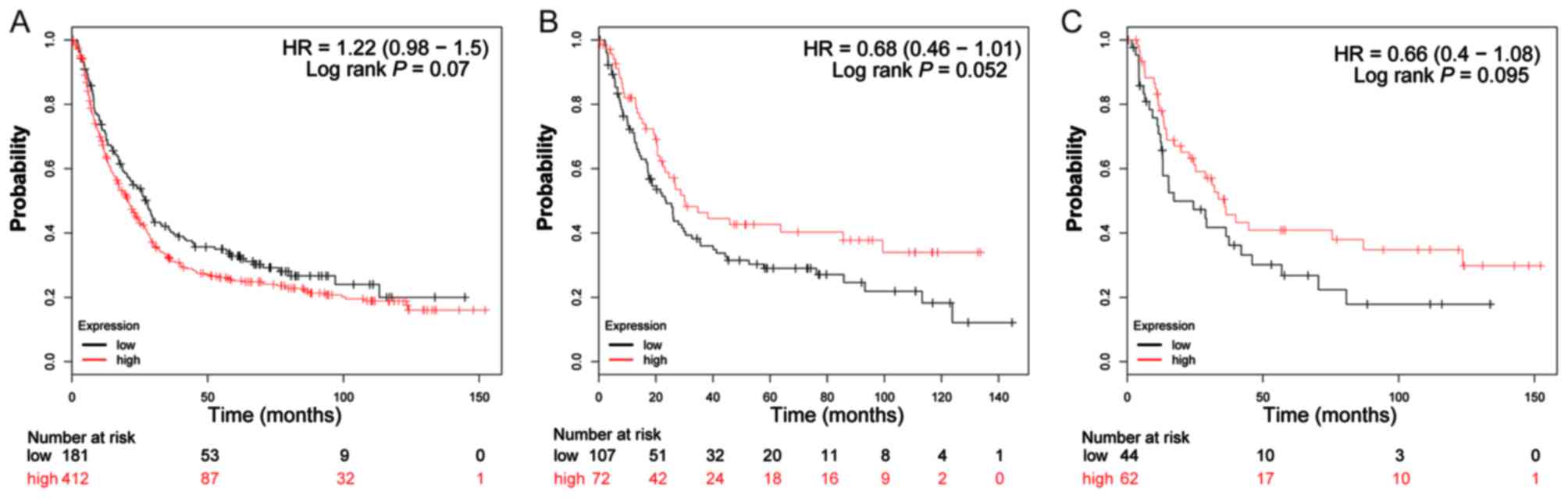
and 7B). Furthermore, the same
analysis performed on subgroups of diffuse-type GC, additionally
suggested that patients with GC with a high expression of
ADH1A (log-rank P=0.014; HR=0.51; 95% CI: 0.30–0.88;
Fig. 4C), ADH1B (log-rank
P=0.04; HR=0.53; 95% CI: 0.29–0.98; Fig.
5C), ADH4 (log-rank P=0.033; HR=0.58; 95% CI: 0.35–0.96;
Fig. 7C) and ADH6 (log-rank
P=0.037; HR=0.59; 95% CI: 0.35–0.97; Fig.
9C) demonstrated significantly decreased risk of mortality,
compared with patients with low expression. In contrast, the high
expression of ADH5 (log-rank P=0.044; HR=1.66; 95% CI:
1.01–2.74; Fig. 8C) was significantly
associated with a poor prognosis for patients with diffuse-type GC
compared with patients with a low expression of this gene.
In addition, the prognostic values of ADH genes in
stratified sampling in distinct patient subgroups of GC were
analyzed. Stratified analyses of patients with GC with different
treatments are presented in Table I.
It was observed that the high expression of ADH1A
(P=0.0095), ADH1B (P=1.80×10−6), ADH1C
(P=0.0053), ADH4 (P=0.037), ADH5 (P=0.032) and
ADH7 (P=0.035) significantly decreased the risk of mortality
for patients with GC receiving 5-fluorouracil (5-FU)-based adjuvant
chemotherapy compared with patients with low expression of these
genes. In contrast, high ADH6 expression significantly
increased the risk of mortality for patients with GC receiving
5-FU-based adjuvant chemotherapy compared with patients with low
expression of these genes (P=0.043; Table
I). The strata of patients with GC receiving surgery alone
revealed that the high expression of ADH1B significantly
decreased the risk of mortality for patients with GC compared with
low expression (P=0.0082), whereas no significant associations
between any of the other ADH genes expression and GC prognosis were
identified in the present study following surgery alone.
 | Table I.Associations between the high
expression of ADH genes and different treatments received by
patients with gastric cancer. |
Table I.
Associations between the high
expression of ADH genes and different treatments received by
patients with gastric cancer.
| Gene | Treatment | Cases, n | Hazard ratio (95%
confidence interval) | Log-rank
P-value |
|---|
| ADH1A | Surgery alone | 174 | 0.68
(0.44–1.03) | 0.0660 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 0.59
(0.4–0.88) | 0.0095 |
| ADH1B | Surgery alone | 174 | 0.57
(0.38–0.87) | 0.0082 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 0.41
(0.28–0.6) | <0.0001 |
| ADH1C | Surgery alone | 174 | 0.73
(0.48–1.11) | 0.1400 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 0.57
(0.38–0.85) | 0.0053 |
| ADH4 | Surgery alone | 174 | 0.76
(0.5–1.15) | 0.2000 |
|
| 5-FU-based adjuvant
chemotherapy | 34 | 0.39
(0.16–0.97) | 0.0370 |
| ADH5 | Surgery alone | 174 | 1.27
(0.84–1.93) | 0.2500 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 0.64
(0.43–0.96) | 0.0320 |
| ADH6 | Surgery alone | 174 | 1.23
(0.8–1.91) | 0.3400 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 1.48
(1.01–2.16) | 0.0430 |
| ADH7 | Surgery alone | 174 | 1.34
(0.8–2.24) | 0.2700 |
|
| 5-FU-based adjuvant
chemotherapy | 153 | 0.66
(0.45–0.97) | 0.0350 |
Stratified analysis by HER2 status, presented in
Table II, indicate that high
expression of ADH1B (P=0.0013) and ADH5 (P=0.0041)
were significantly associated with a favorable prognosis in
patients with HER2-negative GC. Similar results may also be
observed with high expression of ADH1A (P=0.032) and
ADH1B (P=0.0033) being significantly associated with a
favorable prognosis in patients with GC with a positive HER2 status
(Table II).
 | Table II.Association between the high
expression of ADH genes and the HER2 status of patients with
gastric cancer. |
Table II.
Association between the high
expression of ADH genes and the HER2 status of patients with
gastric cancer.
| Gene | HER2 status | Cases, (n) | Hazard ratio (95%
confidence interval) | Log-rank
P-value |
|---|
| ADH1A | Negative | 298 | 0.78
(0.59–1.03) | 0.0820 |
|
| Positive | 295 | 0.71
(0.52–0.97) | 0.0320 |
| ADH1B | Negative | 298 | 0.61
(0.45–0.83) | 0.0013 |
|
| Positive | 295 | 0.61
(0.44–0.85) | 0.0033 |
| ADH1C | Negative | 298 | 1.19
(0.9–1.58) | 0.2300 |
|
| Positive | 295 | 1.32
(0.97–1.8) | 0.0790 |
| ADH4 | Negative | 195 | 0.8
(0.56–1.15) | 0.2200 |
|
| Positive | 153 | 0.77
(0.51–1.16) | 0.2100 |
| ADH5 | Negative | 298 | 0.67
(0.51–0.88) | 0.0041 |
|
| Positive | 295 | 0.79
(0.6–1.03) | 0.0830 |
| ADH6 | Negative | 298 | 1.16
(0.87–1.56) | 0.3100 |
|
| Positive | 295 | 0.8
(0.59–1.08) | 0.1400 |
| ADH7 | Negative | 298 | 0.81
(0.58–1.11) | 0.1900 |
|
| Positive | 295 | 0.77
(0.58–1.02) | 0.0710 |
Stratified analysis by pathological grade, presented
in Table III, indicate that the
high expression of ADH1C was significantly associated with
the decreased risk of mortality for patients with GC with
well-differentiated tumors (P=0.046) compared with low expression,
whereas high ADH5 expression was significantly associated
with an increased risk of mortality compared with low expression
(P=0.028). The stratified analysis in GC clinical stages suggests
that high expression of ADH1A (P=0.025), ADH4
(P=0.035), ADH5 (P=0.0059) and ADH6 (P=0.022) in stage 2
patients, and high expression of ADH5 (P=0.033) in stage 3
patients demonstrated a significantly increased risk of mortality
compared with low expression (Table
IV). In contrast, high expression of ADH7 was
significantly associated with a decreased risk of mortality in
patients with stage 1 GC (P=0.028; Table
IV).
 | Table III.Association between the high
expression of ADH genes and the pathological grades of patients
with gastric cancer. |
Table III.
Association between the high
expression of ADH genes and the pathological grades of patients
with gastric cancer.
| Gene | Pathological
grades | Cases (n) | Hazard ratio (95%
confidence interval) | Log-rank
P-value |
|---|
| ADH1A | Well
differentiated | 32 | 0.42
(0.14–1.24) | 0.1100 |
|
| Moderately
differentiated | 67 | 0.68
(0.32–1.46) | 0.3200 |
|
| Poorly
differentiated | 165 | 0.7
(0.46–1.05) | 0.0860 |
| ADH1B | Well
differentiated | 32 | 0.67
(0.26–1.73) | 0.4100 |
|
| Moderately
differentiated | 67 | 0.56
(0.28–1.12) | 0.0960 |
|
| Poorly
differentiated | 165 | 0.79
(0.51–1.22) | 0.2800 |
| ADH1C | Well
differentiated | 32 | 0.39
(0.15–1.02) | 0.0460 |
|
| Moderately
differentiated | 67 | 0.66
(0.3–1.45) | 0.3000 |
|
| Poorly
differentiated | 165 | 1.35
(0.9–2.03) | 0.1400 |
| ADH4 | Well
differentiated |
5 | NA | NA |
|
| Moderately
differentiated | 67 | 0.77
(0.4–1.48) | 0.4300 |
|
| Poorly
differentiated | 121 | 0.68
(0.42–1.11) | 0.1200 |
| ADH5 | Well
differentiated | 32 | 2.68
(1.07–6.7) | 0.0280 |
|
| Moderately
differentiated | 67 | 1.46
(0.76–2.83) | 0.2500 |
|
| Poorly
differentiated | 165 | 1.21
(0.81–1.8) | 0.3500 |
| ADH6 | Well
differentiated | 32 | 1.37
(0.46–4.06) | 0.5700 |
|
| Moderately
differentiated | 67 | 0.63
(0.3–1.33) | 0.2200 |
|
| Poorly
differentiated | 165 | 1.41
(0.94–2.1) | 0.0920 |
| ADH7 | Well
differentiated | 32 | 2.45
(0.82–7.31) | 0.0980 |
|
| Moderately
differentiated | 67 | 0.56
(0.29–1.07) | 0.0750 |
|
| Poorly
differentiated | 165 | 0.81
(0.54–1.22) | 0.3100 |
 | Table IV.Association between the high
expression of ADH genes and the clinical stages of patients with
gastric cancer. |
Table IV.
Association between the high
expression of ADH genes and the clinical stages of patients with
gastric cancer.
| Gene | Clinical
stages | Cases (n) | Hazard ratio (95%
confidence interval) | Log-rank
P-value |
|---|
| ADH1A | 1 | 39 | 1.76
(0.56–5.5) | 0.3200 |
|
| 2 | 49 | 4.6
(1.07–19.77) | 0.0250 |
|
| 3 | 217 | 1.15
(0.83–1.6) | 0.3900 |
|
| 4 | 74 | 0.62
(0.36–1.07) | 0.0840 |
| ADH1B | 1 | 39 | 0.56
(0.17–1.84) | 0.3300 |
|
| 2 | 49 | 3.02
(0.89–10.25) | 0.0620 |
|
| 3 | 217 | 0.85
(0.59–1.22) | 0.3800 |
|
| 4 | 74 | 1.23
(0.7–2.16) | 0.4700 |
| ADH1C | 1 | 39 | 2.41
(0.65–8.88) | 0.1700 |
|
| 2 | 49 | 0.62
(0.27–1.44) | 0.2700 |
|
| 3 | 217 | 1.21
(0.85–1.71) | 0.2800 |
|
| 4 | 74 | 0.75
(0.43–1.31) | 0.3100 |
| ADH4 | 1 | 34 | 2.91
(0.84–10.15) | 0.0790 |
|
| 2 | 44 | 2.78
(1.03–7.51) | 0.0350 |
|
| 3 | 109 | 1.45
(0.89–2.38) | 0.1400 |
|
| 4 | 66 | 0.72
(0.37–1.39) | 0.3200 |
| ADH5 | 1 | 39 | 0.45
(0.14–1.46) | 0.1700 |
|
| 2 | 49 | 3.19
(1.33–7.64) | 0.0059 |
|
| 3 | 217 | 1.43
(1.03–1.99) | 0.0330 |
|
| 4 | 74 | 1.56
(0.85–2.87) | 0.1500 |
| ADH6 | 1 | 39 | 2.29
(0.63–8.35) | 0.2000 |
|
| 2 | 49 | 2.7
(1.12–6.51) | 0.0220 |
|
| 3 | 217 | 0.89
(0.61–1.28) | 0.5100 |
|
| 4 | 74 | 0.59
(0.29–1.18) | 0.1300 |
| ADH7 | 1 | 39 | 0.74
(0.94–58.3) | 0.0280 |
|
| 2 | 49 | 0.56
(0.23–1.33) | 0.1800 |
|
| 3 | 217 | 0.85
(0.59–1.23) | 0.3900 |
|
| 4 | 74 | 0.69
(0.37–1.27) | 0.2300 |
Discussion
ADH and ALDH are the most important enzymes for
ethanol metabolism in vivo (31). Ethanol is initially metabolized by ADH
isoenzymes to acetaldehyde, then the acetaldehyde is further
metabolized to acetic acid using ALDH isoenzymes (32). GO enrichment analysis in the present
study revealed that ADH genes are involved in ethanol oxidation,
ADH and retinol dehydrogenase activity, alcohol metabolic and
retinoid metabolic processes; consistent with what is already known
about these genes (33,34). KEGG analysis in the present study
suggested that ADH genes were involved in retinol metabolism, drug
metabolism via cytochrome P450 enzymes, metabolism of xenobiotics
by cytochrome P450 and chemical carcinogenesis pathways. These
results suggest that ADH genes are involved in ethanol, drug and
toxic chemical redox metabolism processes in vivo. ADH
isoenzymes include five class proteins that are encoded by seven
genes. Class I ADH isoenzymes are encoded by ADH1A, ADH1B
and ADH1C, whilst class II ADH isoenzymes are encoded by
ADH4. Class III ADH, class IV ADH and class V ADH isoenzymes
are encoded by ADH5, ADH6 and ADH7, respectively
(35,36).
A previous study has reported that ADH isoenzymes
possess diagnostic value in multiple malignant neoplasms and that
ADH isoenzymes may be used as cancer biomarkers (37). In addition, previous studies
demonstrated a significant increase in the activity of serum class
I ADH isoenzymes in patients with renal cell cancer (38), colorectal cancer (CRC) (12), endometrial cancer (39), brain cancer (40,41) and
cervical cancer patients (42)
compared with healthy patients, and demonstrate a diagnostic value
in these cancer types. Similarly, class III isoenzymes also
demonstrate a diagnostic value in pancreatic cancer (43), whereas, class IV ADH isoenzymes show a
diagnostic value in GC (14) and
esophageal cancer (13). A
significant increase in total ADH isoenzymes was additionally
observed in the serum of patients with these cancer types. In
addition, class I ADH isoenzymes are upregulated in liver cancer
(44), esophageal cancer (45) and bladder cancer (46), however, the diagnostic value of class
I ADH in these cancer types requires further exploration.
Similarly, as in the serum results of patients with malignant
neoplasms, ADH isoenzyme activity has been identified in tumor
tissues (37). Studies have
additionally reported that class I ADH isoenzymes are significantly
increased in the tumor tissue of cervical cancer (47), ovarian cancer (48), endometrial cancer (49), brain cancer (50), liver cancer (44,51), CRC
(52,53) and renal cell carcinoma (54) compared with healthy tissue, whereas
ALDH isoenzymes in these cancer types did not demonstrate a
statistically significant difference between cancer and healthy
tissue. However, a downregulation of class I ADH was observed in
breast cancer tissues (55), whereas
the expression of other ADH isoenzymes remained unchanged. ADH and
ALDH are the most important enzymes for ethanol metabolism, and
acetaldehyde is a product of ADH metabolized alcohol and ALDH
further metabolizes it to acetic acid (31). As is well known, acetaldehyde is the
most toxic ethanol metabolite and a cancer-causing agent (7). The metabolism of acetaldehyde depends on
the balance between ADH and ALDH; therefore, these results suggest
the activity of ADH and an unchanged level of ALDH may result in
the accumulation of acetaldehyde, that may result in tumorigenesis.
In addition, other classes of ADH isoenzyme upregulation have
additionally been observed in other cancer types, including class
III ADH for pancreatic cancer tissue (56) and class III ADH for esophageal cancer
(57). In addition, a marked upward
trend of ADH IV was exhibited in patients with GC according to the
advancement of tumor progression, and the other isoenzymes
additionally revealed an upward trend in accordance with tumor
progression; however, the changes did not demonstrate a statistical
significance (14). Consequently,
there was an increased trend of total ADH activity according to GC
progression (14). These results
suggest that disturbances of ADH isoenzyme activity serve a notable
function in alcohol-associated neoplasms, and may be potential
diagnostic biomarkers. In the present study, ADH isoenzyme
expression between GC tumor and adjacent non-tumor tissues was
further analyzed, and revealed that the mRNA expression level of
ADH1A, ADH1B, ADH1C and ADH7 were significantly
downregulated in tumor tissues compared with non-tumor tissues,
whereas ADH5 was significantly upregulated in tumor tissues
compared with non-tumor tissues. The comparison of tumor and
adjacent non-tumor tissues suggests that the dysregulation of
ADH1A, ADH1B, ADH1C, ADH5 and ADH7 were associated
with tumorigenesis in GC.
The diagnostic value of ADH isoenzymes has been
investigated in multiple cancer types, particularly in studies by
Jelski et al and Orywal and Szmitkowski (14,37). These
previous studies reveal ADH isoenzyme activity in malignant
neoplasms, particularly for alcohol-associated neoplasms. However,
the prognostic values of these isoenzymes have rarely been reported
in previous studies. A study by Wei et al (11) demonstrated that ADH4 mRNA and
protein expression in hepatocellular carcinoma (HCC) was
significantly downregulated in tumor tissues compared with adjacent
non-tumor tissues. High ADH4 expression was significantly
associated with a favorable prognosis and may be a potential
prognostic marker for patients with HCC (11). The downregulation of ADH genes may
additionally be revealed in hepatitis B (HBV)-associated HCC tumor
tissue, and the high expression of ADH1A, ADH1C, ADH5 and
ADH6 exhibit protective effects in patients with
HBV-associated HCC (58). The genetic
variation of ADH genes has additionally been reported to be
involved in the association between ADH polymorphisms and cancer
survival. A study by Li et al (59) demonstrated that the genetic variation
of ADH1B-rs1229984 was associated with laryngeal cancer
overall survival (OS), and that the genetic algorithm genotype of
rs1229984 decreased the risk of mortality in patients with
laryngeal cancer and may be a prognostic indicator. The survival
analysis performed in the present study demonstrated that the high
expression of ADH1A, ADH1B and ADH5 were associated
with a significantly decreased risk of mortality in all patients
with GC. Furthermore, a similar effect was observed for
ADH1B in patients with intestinal-type GC and ADH1A,
ADH1B, ADH4 and ADH6 for patients with diffuse-type GC.
The results of the present study were consistent with those of a
previous study (11). However,
paradoxically, the present study additionally observed that the
high expression of ADH5 in patients with diffuse-type GC
exhibit a poor prognosis, and ADH5 high expression in GC
tumor tissues appears to be more consistent with the performance of
an oncogene.
The majority of highly expressed ADH genes revealed
a protective effect in patients with GC receiving 5-FU-based
adjuvant chemotherapy, these results were consistent with the
pathway enrichment analysis that indicated that ADH genes were
involved in drug metabolism via the cytochrome P450 pathway. This
is potentially a consequence of ADH participation in drug
metabolism and serves a function in the anticarcinogenic response,
however this assumption requires further investigation. It was
additionally observed that ADH6 is associated with a
significantly increased risk for patients with GC receiving
5-FU-based adjuvant chemotherapy in univariate stratification
analysis. Due to a lack of data from previous studies and
multivariate analysis in the present study, further studies are
required in order to verify the association between ADH6 and
GC prognosis. In addition, ADH1B also demonstrated
protective effects in patients with GC who received surgery
alone.
Overexpression of HER2 is associated with poor GC
prognosis and associated with Lauren classification, tumor size,
lymph node and World Health Organization classification (60). However, to the best of our knowledge,
there are no reports on the association between HER2 and ADH
isoenzymes. The results of the present study revealed that high
expression of ADH1A and ADH1B were associated with a
significantly decreased risk of mortality in patients with
HER2-positive GC, in addition to high ADH1B and ADH5
for patients with HER2-negative GC.
There were limitations in the present study that
need to be recognized. First, the clinical information from the GEO
database was not comprehensive; therefore, this study evaluates the
association between ADH genes expression and OS based on univariate
survival analysis. Second, the sample size of this study was not
large enough to validate the impact of a number of strata with a
small sample size on OS in the stratified analysis. Third, the
results of the present study reveal a significantly positive
association between the mRNA expression of all ADH genes with each
other, and consequently, individual ADH genes may have complex
interactions and finally have a joint effect on the outcome of
patients with GC. Unfortunately, the KM-plotter tool is unable to
analyze the joint effect of ADH gene expression in GC
prognosis.
Despite these limitations, to the best of our
knowledge, the present study is the first study to investigate the
associations between ADH genes expression and OS in patients with
GC, in addition to the prognostic values among the different strata
of GC. These results provide insight into the function of ADH genes
in clinical outcomes of cancer and may demonstrate a clinical
utility for prognosis prediction and decision-making in GC
management.
In summary, the results of the present study
demonstrated that the high expression of ADH1A and
ADH1B mRNA were associated with favorable prognosis in
patients with intestinal-type and diffuse-type GC, whereas the
other ADH genes either resulted in a different OS between these two
types of GC or did not demonstrate a statistically significant
difference in the OS between the groups. Therefore, the prognostic
value of other ADH genes requires further investigation.
The results of the present study suggest that
ADH1A and ADH1B may be potential prognostic
biomarkers of GC. Owing to the small sample size and inability to
perform multivariate analysis, further well-designed and larger
sample size studies are necessary in order to validate these
results.
Acknowledgements
The present study was supported in part by the
National Nature Science Foundation of China (grant no. 81360448),
the Innovation Project of Guangxi Graduate Education (2017), the
Self-raised Scientific Research Fund of the Health and Family
Planning Commission of the Guangxi Zhuang Autonomous Region (grant
no. Z2015198) and the Nanning Scientific Research and Technology
Development Project (Key Research and Development Plan; grant no.
20173018-3). The authors thank the contributors of the Kaplan-Meier
plotter for sharing their data on open access.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen MJ, Chiou YY, Wu DC and Wu SL:
Lifestyle habits and gastric cancer in a hospital-based
case-control study in taiwan. Am J Gastroenterol. 95:3242–3249.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bujanda L: The effects of alcohol
consumption upon the gastrointestinal tract. Am J Gastroenterol.
95:3374–3382. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Torre LA, Sauer AM, Chen MS Jr,
Kagawa-Singer M, Jemal A and Siegel RL: Cancer statistics for asian
americans, native hawaiians, and pacific islanders, 2016:
Converging incidence in males and females. CA Cancer J Clin.
66:182–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boffetta P and Hashibe M: Alcohol and
cancer. Lancet Oncol. 7:149–156. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seitz HK and Stickel F: Molecular
mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer.
7:599–612. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hashibe M, McKay JD, Curado MP, Oliveira
JC, Koifman S, Koifman R, Zaridze D, Shangina O, Wünsch-Filho V,
Eluf-Neto J, et al: Multiple ADH genes are associated with upper
aerodigestive cancers. Nat Genet. 40:707–709. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li
Y, Hu Z, He Z, Jia W, Abnet CC, et al: Genome-wide association
analyses of esophageal squamous cell carcinoma in chinese identify
multiple susceptibility loci and gene-environment interactions. Nat
Genet. 44:1090–1097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei RR, Zhang MY, Rao HL, Pu HY, Zhang HZ
and Wang HY: Identification of ADH4 as a novel and potential
prognostic marker in hepatocellular carcinoma. Med Oncol.
29:2737–2743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jelski W, Mroczko B and Szmitkowski M: The
diagnostic value of alcohol dehydrogenase (ADH) isoenzymes and
aldehyde dehydrogenase (ALDH) measurement in the sera of colorectal
cancer patients. Dig Dis Sci. 55:2953–2957. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jelski W, Laniewska-Dunaj M, Niklinski J,
Kozlowski M, Laudanski J and Szmitkowski M: The alcohol
dehydrogenase isoenzyme (ADH IV) as a candidate tumour marker of
esophageal cancer. Acta Biochim Pol. 60:489–493. 2013.PubMed/NCBI
|
|
14
|
Jelski W, Orywal K, Laniewska M and
Szmitkowski M: The diagnostic value of alcohol dehydrogenase (ADH)
isoenzymes and aldehyde dehydrogenase (ALDH) measurement in the
sera of gastric cancer patients. Clin Exp Med. 10:215–219. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jelski W and Szmitkowski M: Alcohol
dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer
diseases. Clin Chim Acta. 395:1–5. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gyorffy B, Lanczky A and Szallasi Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li K, Guo X, Wang Z, Li X, Bu Y, Bai X,
Zheng L and Huang Y: The prognostic roles of ALDH1 isoenzymes in
gastric cancer. Onco Targets Ther. 9:3405–3414. 2016.PubMed/NCBI
|
|
22
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: A real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9:S42008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sobin LH, Gospodarowicz MK and Wittekind
CH; International Union against Cancer, : TNM classification of
malignant tumours. 7th. West Sussex; Chichester, UK: 2009
|
|
28
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and so-called intestinal-type
carcinoma. an attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang G, Hu N, Yang HH, Wang L, Su H, Wang
C, Clifford R, Dawsey EM, Li JM, Ding T, et al: Comparison of
global gene expression of gastric cardia and noncardia cancers from
a high-risk population in china. PLoS One. 8:e638262013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
R Development Core Team: R, . A language
and environment for statistical computing. R Foundation for
Statistical Computing; Vienna: 2008
|
|
31
|
Druesne-Pecollo N, Tehard B, Mallet Y,
Gerber M, Norat T, Hercberg S and Latino-Martel P: Alcohol and
genetic polymorphisms: Effect on risk of alcohol-related cancer.
Lancet Oncol. 10:173–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klyosov AA: Kinetics and specificity of
human liver aldehyde dehydrogenases toward aliphatic, aromatic, and
fused polycyclic aldehydes. Biochemistry. 35:4457–4467. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Crabb DW, Bosron WF and Li TK: Ethanol
metabolism. Pharmacol Ther. 34:59–73. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seitz HK and Oneta CM: Gastrointestinal
alcohol dehydrogenase. Nutr Rev. 56:52–60. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Holmes RS: Alcohol dehydrogenases: A
family of isozymes with differential functions. Alcohol Alcohol
Suppl. 2:127–130. 1994.PubMed/NCBI
|
|
36
|
Holmes R: Alcohol dehydrogenases: Gene
multiplicity and differential functions of five classes of
isozymes. Drug Alcohol Rev. 12:99–110. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Orywal K and Szmitkowski M: Alcohol
dehydrogenase and aldehyde dehydrogenase in malignant neoplasms.
Clin Exp Med. 17:131–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Orywal K, Jelski W, Werel T and
Szmitkowski M: The diagnostic significance of serum alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase activity in
renal cell cancer patients. Exp Mol Pathol. 100:416–420. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Orywal K, Jelski W, Zdrodowski M and
Szmitkowski M: The diagnostic value of alcohol dehydrogenase
isoenzymes and aldehyde dehydrogenase measurement in the sera of
patients with endometrial cancer. Anticancer Res. 33:3725–3730.
2013.PubMed/NCBI
|
|
40
|
Jelski W, Laniewska-Dunaj M, Orywal K,
Kochanowicz J, Rutkowski R and Szmitkowski M: The diagnostic value
of alcohol dehydrogenase (ADH) isoenzymes and aldehyde
dehydrogenase (ALDH) measurement in the sera of patients with brain
tumor. Arch Med Sci. 13:346–352. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jelski W, Laniewska-Dunaj M, Orywal K,
Kochanowicz J, Rutkowski R and Szmitkowski M: The activity of
alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase
(ALDH) in the sera of patients with brain cancer. Neurochem Res.
39:2313–2318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Orywal K, Jelski W, Zdrodowski M and
Szmitkowski M: The diagnostic value of alcohol dehydrogenase
isoenzymes and aldehyde dehydrogenase measurement sera of cervical
cancer patients. Anticancer Res. 36:2265–2269. 2016.PubMed/NCBI
|
|
43
|
Jelski W, Kutylowska E, Laniewska-Dunaj M
and Szmitkowski M: Alcohol dehydrogenase (ADH) and aldehyde
dehydrogenase (ALDH) as candidates for tumor markers in patients
with pancreatic cancer. J Gastrointestin Liver Dis. 20:255–259.
2011.PubMed/NCBI
|
|
44
|
Jelski W, Zalewski B and Szmitkowski M:
Alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase
(ALDH) activity in the sera of patients with liver cancer. J Clin
Lab Anal. 22:204–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jelski W, Kozlowski M, Laudanski J,
Niklinski J and Szmitkowski M: Alcohol dehydrogenase isoenzymes and
aldehyde dehydrogenase activity in the sera of patients with
esophageal cancer. Clin Exp Med. 9:131–137. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Orywal K, Jelski W, Werel T and
Szmitkowski M: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in the sera of
bladder cancer patients. Acta Biochim Pol. 64:81–84. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Orywal K, Jelski W, Zdrodowski M and
Szmitkowski M: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in cervical
cancer. Clin Biochem. 44:1231–1234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Orywal K, Jelski W, Zdrodowski M and
Szmitkowski M: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in ovarian
cancer and ovarian cysts. Adv Med Sci. 58:216–220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Orywal K, Jelski W, Zdrodowski M and
Szmitkowski M: The activity of class I, II, III, and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in endometrial
cancer. J Clin Lab Anal. 24:334–339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Laniewska-Dunaj M, Jelski W, Orywal K,
Kochanowicz J, Rutkowski R and Szmitkowski M: The activity of class
I, II, III and IV of alcohol dehydrogenase (ADH) isoenzymes and
aldehyde dehydrogenase (ALDH) in brain cancer. Neurochem Res.
38:1517–1521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jelski W, Zalewski B and Szmitkowski M:
The activity of class I, II, III, and IV alcohol dehydrogenase
(ADH) isoenzymes and aldehyde dehydrogenase (ALDH) in liver cancer.
Dig Dis Sci. 53:2550–2555. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jelski W, Zalewski B, Chrostek L and
Szmitkowski M: Alcohol dehydrogenase (ADH) isoenzymes and aldehyde
dehydrogenase (ALDH) activity in the sera of patients with
colorectal cancer. Clin Exp Med. 7:154–157. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jelski W, Zalewski B, Chrostek L and
Szmitkowski M: The activity of class I, II, III, and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in colorectal
cancer. Dig Dis Sci. 49:977–981. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Orywal K, Jelski W, Werel T and
Szmitkowski M: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in renal cell
carcinoma. Exp Mol Pathol. 98:403–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jelski W, Chrostek L, Szmitkowski M and
Markiewicz W: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in breast
cancer. Clin Exp Med. 6:89–93. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jelski W, Chrostek L and Szmitkowski M:
The activity of class I, II, III, and IV of alcohol dehydrogenase
isoenzymes and aldehyde dehydrogenase in pancreatic cancer.
Pancreas. 35:142–146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jelski W, Kozlowski M, Laudanski J,
Niklinski J and Szmitkowski M: The activity of class I, II, III,
and IV alcohol dehydrogenase (ADH) isoenzymes and aldehyde
dehydrogenase (ALDH) in esophageal cancer. Dig Dis Sci. 54:725–730.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shang L, Zhu G, Su H, Chen B, Ye X, Chen
X, Xiao K, Li L, Peng M and Peng T: Identification of alcohol
dehydrogenase as a potential prognostic marker in HBV-related
hepatocellular carcinoma. Int J Clin Exp Med. 10:4457–4472.
2017.
|
|
59
|
Li D, Zhang R, Jin T, He N, Ren L, Zhang
Z, Zhang Q, Xu R, Tao H, Zeng G and Gao J: ADH1B and CDH1
polymorphisms predict prognosis in male patients with
non-metastatic laryngeal cancer. Oncotarget. 7:73216–73228.
2016.PubMed/NCBI
|
|
60
|
Jiang W, Jin Z, Zhou F, Cui J and Wang L
and Wang L: High co-expression of Sp1 and HER-2 is correlated with
poor prognosis of gastric cancer patients. Surg Oncol. 24:220–225.
2015. View Article : Google Scholar : PubMed/NCBI
|