Introduction
Hypopharyngeal cancer is a common malignant tumor,
which has a poor prognosis among head and neck cancer (1,2).
Hypopharyngeal carcinoma originates in the mucosal epithelia in the
hypopharynx (3–5). Hypopharyngeal cancer is invasive, yet
the majority of patients exhibit a lack of evident early symptoms
(6). In addition, the hypopharynx is
not part of routine medical exams, thus the majority of patients
with hypopharyngeal cancer exhibit advanced disease at diagnosis
(7); the 5-year survival rate is
<20% for patients with advanced disease (8). Surgery, chemotherapy and radiotherapy
are used in combination in the clinical treatment of hypopharyngeal
cancer; however, outcomes of these treatments are not satisfactory
(9,10). Therefore, the development of novel
strategies and effective methods to treat hypopharyngeal cancer is
imperative.
Alternative splicing has a powerful role in
regulating gene expression and increasing protein diversity
(11,12). Alternative splicing is performed by
heterogeneous nuclear ribonucleoprotein and splicing factor
proteins (13–15). RNA-binding motif protein 17 (RBM17),
which is a part of the RNA spliceosome complex (16), binds to the single-stranded three AG
dinucleotides at the exon/intron border, and acts in the second
catalytic step of mRNA splicing (17). The N-terminal domain of RBM17 contains
a G-patch that has been implicated in an interaction between
proteins and protein/nucleic acid (18,19), and
the C-terminal domain contains an RNA recognition motif for mRNA
splicing (17). RBM17 is also
involved in DNA repair (20).
Expression of RBM17 is low in normal tissues, including those of
the breast, liver and prostate; however, its overexpression has
been found in a number of solid tumor types including breast,
pancreas and prostate cancer (21).
However, the role of RMP17 in hypopharyngeal carcinoma remains
unclear.
The present study investigated the effects of
RBM17-knockdown on cell proliferation, cell cycle and
apoptosis in the hypopharyngeal carcinoma cell line FaDu using
lentivirus-mediated specific shRNA targeting RBM17.
Materials and methods
Cell culture
FaDu cells were purchased from Cell Bank of Chinese
Academy of Science (Shanghai, China) and cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum, 100 IU/ml penicillin and 100 µg/ml streptomycin (Sangon
Biotech Co. Ltd, Shanghai, China). Cells were maintained in an
incubator with 5% CO2 and 95% humidity at 37°C. FaDu
cells were infected with the lentiviral vectors containing small
interfering RNAs (siRNAs) targeting RBM17 or empty vectors.
Lentiviral construction for shRNA
treatment
RBM17-specific shRNA (5′-ACTTAAGTGTCCTACTAAA-3′;
GenBank NM_032905) and the negative control sequence
(5′-AATTCTCCGAACGTGTCACGT-3′) were cloned into AgeI and EcoRI sites
of the pGV115-green fluorescent protein (GFP) lentiviral vector
(Shanghai Genechem Co., Ltd., Shanghai, China). The plasmids used
were pGV115-GFP-shRBM17 for specific interference of RBM17 and
pGV115-GFP-negative control (NC) for control. Inhibition of RBM17
expression in FaDu cells with RBM17-specific shRNA was performed as
follows. Lentivirus was generated in FaDu cells as described
previously (22). Briefly, FaDu
(2×105 cells/well) cells were seeded into 6-well plates;
when cell growth reached ~80% confluency, appropriate volumes of
lentiviral vectors were transfected into FaDu cells for 48–72 h to
generate lentivirus. Lentivirus was harvested and the viral titer
was measured with a Centricon-plus-20 (EMD Millipore, Billerica,
MA, USA). The cells were used in subsequent experiments when the
rate of infected cells reached 70% at 72 h post-infection.
Human FaDu cells (2×105 cells/well) were
reseeded into 6-well plates and incubated with either a RBM17-shRNA
(1×106 TU) or control-payload lentiviruses
(1×106 TU) for 8–12 h. A total of 72 h post-infection,
infected FaDu cells were observed under a fluorescent imaging
microscope (Olympus Corporation, Tokyo, Japan) at ×100
magnification by counting green cells based on GFP intensity. The
efficiency of this infection was determined by RT-PCR and western
blotting, which were performed according to the subsequent
steps.
Cell proliferation assay
FaDu cells were infected with RBM17 shRNA lentivirus
(shRBM17) or non-silencing shRNA lentivirus, and 2×103
cells were seeded with 100 µl medium/well into 96-well plates. Cell
growth and viability was evaluated on days 1, 2, 3, 4 and 5. For
cell growth, FaDu cells at the logarithmic phase after being
infected with either the shCtrl or shRBM17 lentivirus and the
plates were counted using the Cellomics ArrayScan™ VT1 automated
reader (Cellomics, Inc.; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) for each day. In each well, ≥800 cells were analyzed. Each
experiment was performed in triplicates. For cell viability, at the
given time, 20 µl MTT (5 mg/ml; Sangon Biotech Co., Ltd.) was added
into each well and plates were incubated for 4 h at 37°C. The
medium in each well was then removed and crystals were dissolved by
the addition of 150 µl dimethyl sulfoxide (Sangon Biotech Co.,
Ltd.) in each well. Following a 10 min incubation at room
temperature, the absorbance was measured at 570 nm.
Reverse transcription-quantitative
polymerase chain reaction
Total RNA was isolated from the FaDu cell lines
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Total RNA was used in a reverse transcription
reaction to synthesize cDNA according to the manufacturer's
protocol using RevertAid® First Strand cDNA Synthesis
Kit (MBI Fermantas; Thermo Fisher Scientific, Inc.). PCR primers
were designed by Beacon Designer 7 software (Premier Biosoft
International, Palo Alto, CA, USA). Primer sequences were as
follows: RBM17 forward, 5′-TCAAATCCGCTGACTGAAATAC-3′ and reverse,
5′-ACCTCCCATTCAAGTCAACAA-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. Quantitative PCR was performed
according to Takara SYBR® Master Mix kit instructions
(Takara Biotechnology Co., Ltd, Dalian, China) as following: 95°C
for 15 sec, followed by 45 cycles of 95°C for 5 sec and 60°C for 20
sec. The 2−ΔΔCq method was used to analyze relative
changes in gene expression (23).
Flow cytometry analysis of cell cycle
distribution
For cell cycle analysis, cells infected with
RBM17-shRNA lentivirus or NC lentivirus were seeded in 6-well
plates and cultured at 37°C for 5 days prior to analysis. Cells
were collected by centrifugation at 1,200 × g for 5 min at 4°C,
washed twice with ice-cold PBS, fixed with cold 70% ethanol for 1 h
at 4°C, and stained with 50 µg/ml propidium iodide (PI)
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in the presence of
100 µg/ml RNase (Sangon Biotech Co., Ltd.) at 37°C for 30 min.
Cells were analyzed by flow cytometry using a FACSCalibur™ flow
cytometer (BD Biosciences, San Diego, CA, USA), according to the
manufacturer's protocol. Data was analyzed using FlowJo Software
(version 10; FlowJo LLC, Ashland, OR, USA).
Analysis of apoptosis by flow
cytometry
To analyze apoptosis, cells were stained with
binding buffer containing annexin V-allophycocyanin (cat. no.
88-8007; eBioscience, San Diego, CA, USA) at 25°C in the dark for
10 min. Cells were analyzed by flow cytometry using a FACSCalibur
flow cytometry (BD Biosciences) according to the manufacturer's
protocol. Data was analyzed using FlowJo Software (version 10;
FlowJo LLC).
Western blot analysis
FaDu cells were lysed and homogenized on ice in
lysis buffer (cat. no. C500001; Sangon Biotech). Homogenates were
centrifuged at 12,000 × g for 20 min at 4°C. The protein
concentration of each sample was measured using Modified BCA
Protein Assay Kit (cat. no. C503051; Sangon Biotech). Equal volumes
(~20 µg total soluble proteins of supernatants) were separated on
12% SDS-PAGE gels and transferred onto polyvinylidene difluoride
membranes. Membranes were incubated with rabbit polyclonal antibody
specific for RBM17 (1:500; cat. no. 101441; Abcam, Cambridge, UK)
and monoclonal antibody specific for GAPDH (1:2,000, cat. no.
sc-32233; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
overnight at 4°C. Membranes were then rinsed 3 times and incubated
with horseradish peroxidase-conjugated goat anti-rabbit IgG
antibody (1:10,000, cat. no. sc-2005; Santa Cruz Biotechnology,
Inc.) at room temperature for 1 h. Membranes were visualized using
an EasyBlot ECL kit (Bio Basic Inc., Markham, ON, Canada).
Statistical analyses
Data were expressed as mean ± standard deviation.
Student's t-test was performed to analyze differences between two
groups using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA).
P<0.05 were considered to indicate a statistically significant
difference.
Results
RBM17 mRNA and protein is knocked down
in cells infected with RBM17-shRNA
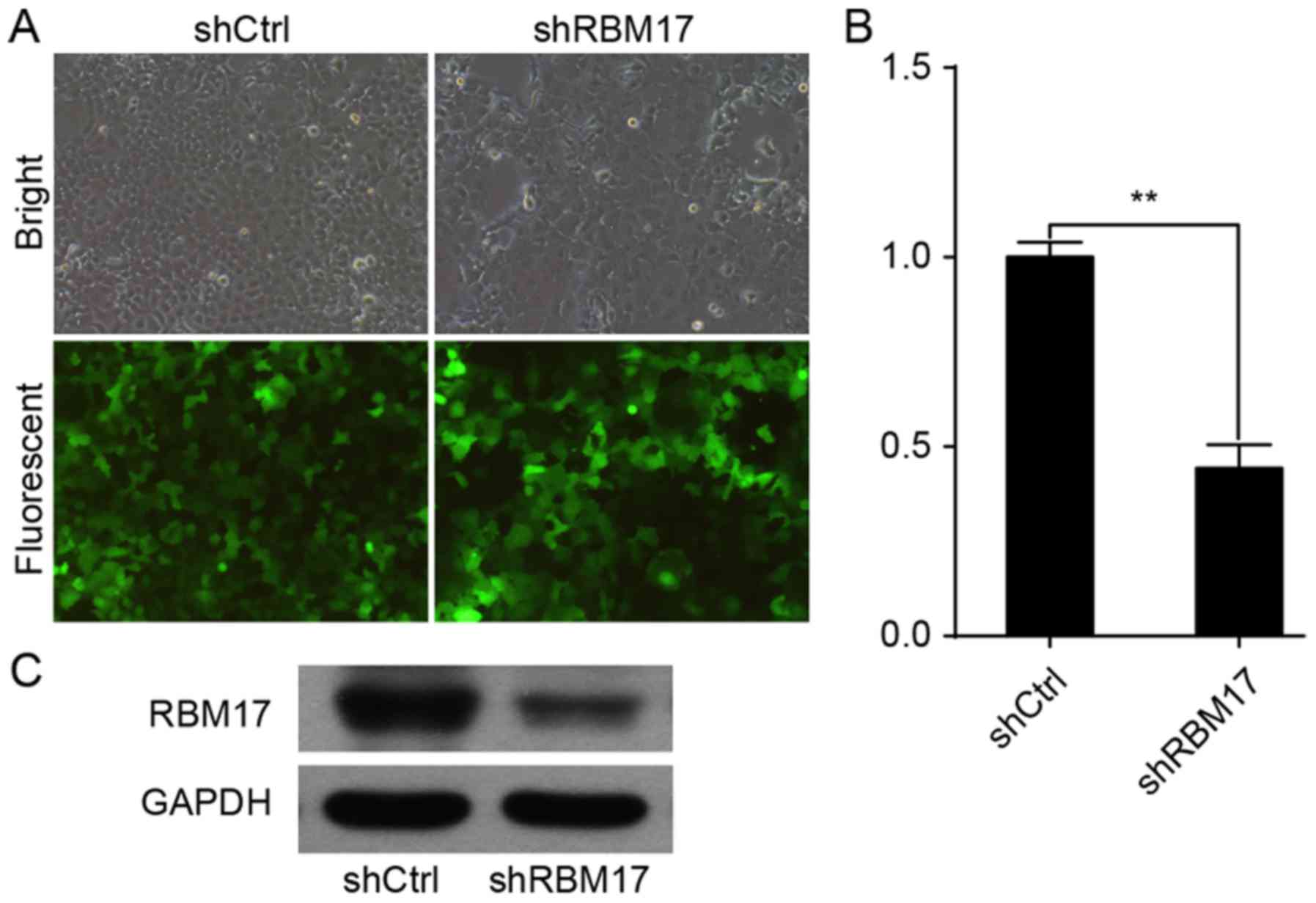
A high infection efficiency was observed by
measuring GFP expression via fluorescence microscopy (Fig. 1A). Expression of RBM17 mRNA in cells
infected with RBM17-siRNA was significantly decreased compared with
control cells (P<0.01; Fig. 1B).
Western blot analysis revealed that RBM17 protein levels were
reduced in cells infected with RBM17-siRNA compared with control
cells (P<0.01; Fig. 1C).
Knockdown of RBM17 inhibits growth of
hypopharyngeal carcinoma cells
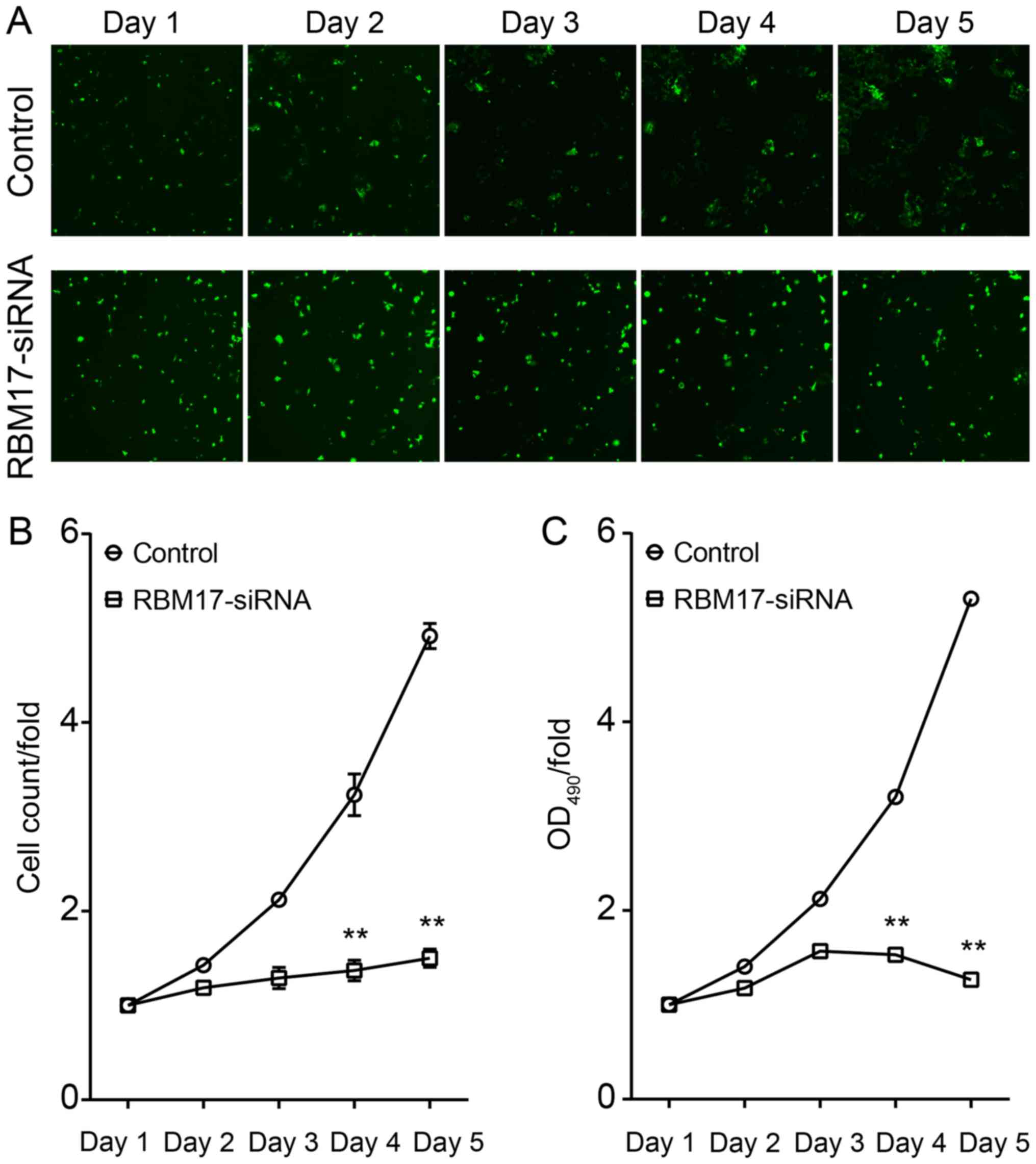
To analyze the effect of regulation of RBM17 levels
on the proliferation of hypopharyngeal carcinoma cells, the
aforementioned FaDu cells infected with lentivirus were analyzed by
Cellomics every day for 5 days. Results from day 4 and 5 of the
assay revealed that proliferation of FaDu cells was significantly
inhibited (P<0.01; Fig. 2),
indicating that knockdown of RBM17 reduces the proliferative
ability of hypopharyngeal carcinoma cells.
Downregulation of RBM17 increases the
proportion of human hypopharyngeal carcinoma cells in
G2/M phase
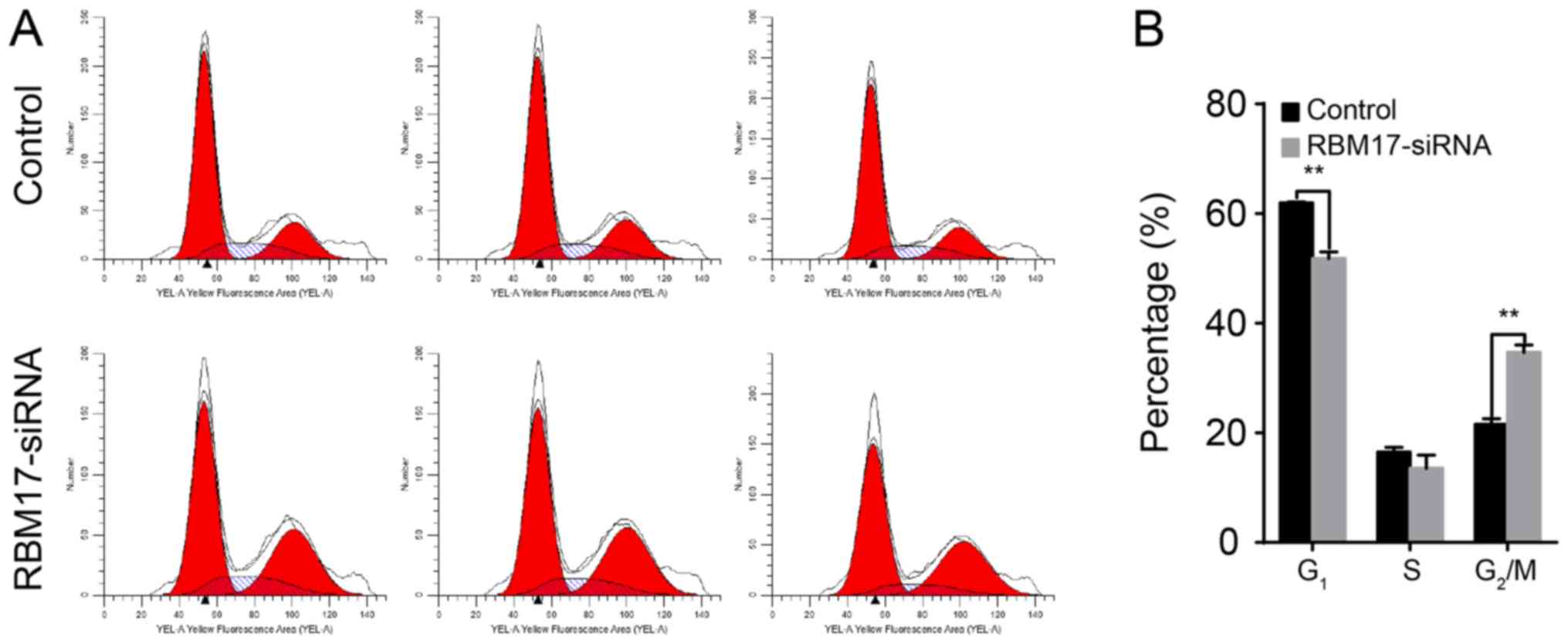
To demonstrate the effects of RBM17-knockdown on the
cell cycle of hypopharyngeal carcinoma cells, a flow cytometry
assay was performed. FaDu cells transfected with RBM17-specific
shRNA had a higher proportion of cells in G2/M phase
than did the control cells (P<0.01; Fig. 3).
Knockdown of RBM17 in human
hypopharyngeal carcinoma cells increases apoptosis
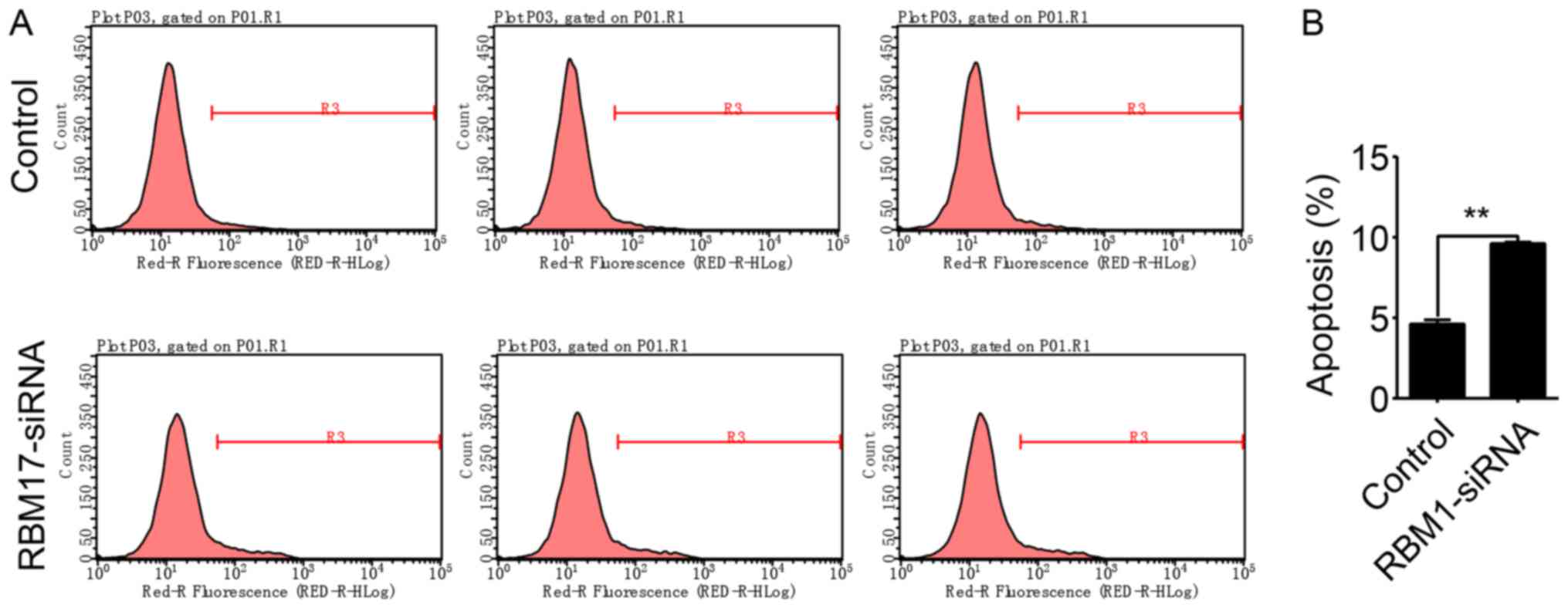
To observe the interplay between RBM17 and
apoptosis, apoptosis was measured by flow cytometry in FaDu cells
in which RBM17 was knocked down (Fig.
4A). The proportion of apoptotic FaDu cells was significantly
increased in shRBM17 cells compared with control cells (P<0.01;
Fig. 4B), indicating that RBM17
expression is a determinant of apoptosis in human hypopharyngeal
carcinoma cells.
Discussion
RBM17 is part of the spliceosome complex, and is
involved in the alternate splicing of mRNA with AGs at the
exon/intron border (16). The
expression of RBM17 is limited in the majority of normal tissues,
and is high in epithelial cells and a number of types of cancer
tissue (21). However, the biological
function of RBM17 is poorly understood.
RBM17 has been confirmed to be overexpressed in
multiple cancer types (21,24). A recent study revealed that
overexpression of RBM17 suppresses cell proliferation and adhesion
to fibronectin (25). Furthermore,
the overexpression of RBM17 is known to induce cell migration and
invasion (26). To investigate RBM17
function in the hypopharyngeal carcinoma FaDu cell line, RBM17
expression was knocked down in FaDu cells. The proliferation of
FaDu cells infected with RBM17-shRNA was reduced, and
downregulation of RBM17 increased apoptosis in FaDu cells and the
proportion of cells in G2/M phase. These findings
indicate that RBM17 accelerates the growth rate of FaDu cells.
The results of the present study imply that RBM17
may be associated with cell cycle checkpoints responsible for
maintaining genomic integrity and regulating cellular proliferation
in FaDu cells (27). The
G2 checkpoint responds to DNA damage, with cancer cells
with DNA damage passing through the S phase checkpoint but
remaining at the G2 phase checkpoint (28). A previous study found that RBM17 was
involved in DNA repair (20).
However, an increase in the proportion of cells infected with
RBM17-shRNA in G2/M phase in the present study
indicates the association of RBM17 with the decreased DNA
repair.
In summary, the present study underlines the
potential roles of RBM17 in the human hypopharyngeal carcinoma FaDu
cell line. The results of the current study indicate that knockdown
of RBM17 by shRNA reduces the proliferation of FaDu cells,
and the knockdown of RBM17 increased the proportion of cells
undergoing apoptosis and arrested the cell cycle at the
G2/M phase. These findings provide the basis for further
investigation of the precise mechanism by which RBM17 influences
cell biology in hypopharyngeal carcinoma.
Acknowledgements
The present study was supported by the Key Project
of Anhui Provincial Department of Education (grant no.
KJ2015A284).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gooi Z, Fakhry C, Goldenberg D, Richmon J
and Kiess AP; Education Committee of the American Head and Neck
Society (AHNS), : AHNS Series: Do you know your guidelines?
Principles of radiation therapy for head and neck cancer: A review
of the National Comprehensive Cancer Network guidelines. Head Neck.
38:987–992. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lewis CM, Hessel AC, Roberts DB, Guo YZ,
Holsinger FC, Ginsberg LE, El-Naggar AK and Weber RS: Prereferral
head and neck cancer treatment: Compliance with national
comprehensive cancer network treatment guidelines. Arch Otolaryngol
Head Neck Surg. 136:1205–1211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hong YM, Gan WG and Xu ZH: Significance of
the expression of integrin β1, VEGF and MVD in hypopharyngeal
squamous cell carcinoma. Genet Mol Res. 13:6455–6465. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zvrko E, Mikic A and Vuckovic L:
Clinicopathologic significance of CD105-assessed microvessel
density in glottic laryngeal squamous cell carcinoma. Auris Nasus
Larynx. 37:77–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chien CY, Su CY, Hwang CF, Chuang HC,
Hsiao YC, Wu SL and Huang CC: Clinicopathologic significance of
CD105 expression in squamous cell carcinoma of the hypopharynx.
Head Neck. 28:441–446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ge N, Lin HX, Xiao XS, Guo L, Xu HM, Wang
X, Jin T, Cai XY, Liang Y, Hu WH and Kang T: Prognostic
significance of Oct4 and Sox2 expression in hypopharyngeal squamous
cell carcinoma. J Transl Med. 8:942010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kotwall C, Sako K, Razack MS, Rao U,
Bakamjian V and Shedd DP: Metastatic patterns in squamous cell
cancer of the head and neck. Am J Surg. 154:439–442. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milisavljevic D, Stankovic M, Zivic M,
Popovic M and Radovanović Z: Factors affecting results of treatment
of Hypopharyngeal Carcinoma. Hippokratia. 13:154–160.
2009.PubMed/NCBI
|
|
9
|
Chu PY, Wang LW and Chang SY: Surgical
treatment of squamous cell carcinoma of the hypopharynx: Analysis
of treatment results, failure patterns, and prognostic factors. J
Laryngol Otol. 118:443–449. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chu PY and Chang SY: Reconstruction of the
hypopharynx after surgical treatment of squamous cell carcinoma. J
Chin Med Assoc. 72:351–355. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Black DL: Protein diversity from
alternative splicing: A challenge for bioinformatics and
post-genome biology. Cell. 103:367–370. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng S: IRAS: High-throughput
identification of novel alternative splicing regulators. Methods
Enzymol. 572:269–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hastings ML and Krainer AR: Functions of
SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway.
RNA. 7:471–482. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Manley JL and Tacke R: SR proteins and
splicing control. Genes Dev. 10:1569–1579. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mayeda A, Munroe SH, Cáceres JF and
Krainer AR: Function of conserved domains of hnRNP A1 and other
hnRNP A/B proteins. EMBO J. 13:5483–5495. 1994.PubMed/NCBI
|
|
16
|
Neubauer G, King A, Rappsilber J, Calvio
C, Watson M, Ajuh P, Sleeman J, Lamond A and Mann M: Mass
spectrometry and EST-database searching allows characterization of
the multi-protein spliceosome complex. Nat Genet. 20:46–50. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lallena MJ, Chalmers KJ, Llamazares S,
Lamond AI and Valcárcel J: Splicing regulation at the second
catalytic step by Sex-lethal involves 3′ splice site recognition by
SPF45. Cell. 109:285–296. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Silverman EJ, Maeda A, Wei J, Smith P,
Beggs JD and Lin RJ: Interaction between a G-patch protein and a
spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol
Cell Biol. 24:10101–10110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Svec M, Bauerová H, Pichová I, Konvalinka
J and Strísovský K: Proteinases of betaretroviruses bind
single-stranded nucleic acids through a novel interaction module,
the G-patch. FEBS lett. 576:271–276. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chaouki AS and Salz HK: Drosophila SPF45:
A bifunctional protein with roles in both splicing and DNA repair.
PLoS Genet. 2:e1782006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sampath J, Long PR, Shepard RL, Xia X,
Devanarayan V, Sandusky GE, Perry WL III, Dantzig AH, Williamson M,
Rolfe M and Moore RE: Human SPF45, a splicing factor, has limited
expression in normal tissues, is overexpressed in many tumors, and
can confer a multidrug-resistant phenotype to cells. Am J Pathol.
163:1781–1790. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakoda T, Kasahara N, Hamamori Y and Kedes
L: A high-titer lentiviral production system mediates efficient
transduction of differentiated cells including beating cardiac
myocytes. J Mol Cell Cardiol. 31:2037–2047. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Perry WL III, Shepard RL, Sampath J, Yaden
B, Chin WW, Iversen PW, Jin S, Lesoon A, O'Brien KA, Peek VL, et
al: Human splicing factor SPF45 (RBM17) confers broad multidrug
resistance to anticancer drugs when overexpressed-a phenotype
partially reversed by selective estrogen receptor modulators.
Cancer Res. 65:6593–6600. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Al-Ayoubi AM, Zheng H, Liu Y, Bai T and
Eblen ST: Mitogen-activated protein kinase phosphorylation of
splicing factor 45 (SPF45) regulates SPF45 alternative splicing
site utilization, proliferation, and cell adhesion. Mol Cell Biol.
32:2880–2893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Conaway L, Rutherford Bethard J,
Al-Ayoubi AM, Thompson Bradley A, Zheng H, Weed SA and Eblen ST:
Phosphorylation of the alternative mRNA splicing factor 45 (SPF45)
by Clk1 regulates its splice site utilization, cell migration and
invasion. Nucleic Acids Res. 41:4949–4962. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kuntz K and O'Connell MJ: The G(2) DNA
damage checkpoint: Could this ancient regulator be the Achilles
heel of cancer? Cancer Biol Ther. 8:1433–1439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bucher N and Britten CD: G2 checkpoint
abrogation and checkpoint kinase-1 targeting in the treatment of
cancer. Br J Cancer. 98:523–528. 2008. View Article : Google Scholar : PubMed/NCBI
|