Introduction
Pancreatic cancer is one of the most malignant
tumors and is characterized by a poor prognosis. An increasing
number of patients are diagnosed with de novo pancreatic
cancer each year (1). The majority of
patients with pancreatic cancer when diagnosed with distant
metastasis, require comprehensive treatment, including
chemotherapy. However, patients who have undergone radical
resection have experience a poor prognosis and a high rate of
recurrence (2,3). Additionally, patients still require
conventional adjuvant chemotherapy in order to minimize the risk of
postoperative recurrence and metastasis. Therefore, chemotherapy
currently serves an important role in the comprehensive treatment
of pancreatic cancer and gemcitabine-based chemotherapy, a
first-line chemotherapy option, has been indicated to markedly
prolong survival time in patients with pancreatic cancer (4).
However, multidrug-resistance (MDR) in pancreatic
cancer often occurs during chemotherapy treatment due to the
biological characteristics of the tumors, leading to a decline in
the clinical efficacy of chemotherapy over time (5). Of the mechanisms of pancreatic cancer
drug resistance, the most important is that the transporters on the
tumor cell membrane mediate drug efflux and inactivation (6). Abnormal expression of ATP-binding
cassette (ABC) transporters [ATP binding cassette subfamily B
member 1 (ABCB1), ATP binding cassette subfamily C (ABCC) and ATP
binding cassette subfamily G member 2 (ABCG2)] in patients with
resistant pancreatic cancer has confirmed that these transporters
are associated with MDR in pancreatic cancer (7).
Previous epigenetic studies have revealed that the
level of ABC transporter promoter methylation in pancreatic cancer
is negatively correlated with chemoresistance in patients with
pancreatic cancer (8,9). ABC transporter promoter methylation has
also served as an indicator of drug resistance in certain types of
solid tumor (10). However, promoter
methylation studies concerning MDR in pancreatic cancer are
insufficient. There have been a number of relevant studies
suggesting that the level of ABC transporter promoter methylation
in pancreatic cancer is negatively correlated with drug resistance
in pancreatic cancer (11,12). However, there is a lack of further
quantitative simultaneous analysis and evaluation with regards to
predicting MDR induced by methylation.
During the process of inducing the
gemcitabine-resistant cell lines, PANC-1/Gem and BxPC-3/Gem, the
present study aimed to use methylation-sensitive high-resolution
melting (MS-HRM) to quantitatively and simultaneously detect
promoter methylation of the ABCB1, ABCC and ABCG2 genes, while
monitoring the expression of downstream mRNA and protein
expression. Then, the changes in ABC transporter DNA promoter
methylation of pancreatic cancer were investigated and the
potential for using the promoter methylation level to predict
acquired drug resistance was evaluated in order to provide a
theoretical experimental basis for clinical research concerning the
mechanisms underlying MDR and the treatment of pancreatic
cancer.
Materials and methods
Cell culture and drug preparation
The human pancreatic cancer cell lines, BxPC-3 and
PANC-1, were acquired from the Cell Bank of Type Culture Collection
of Chinese Academy of Sciences (Shanghai, China). The cell lines
were characterized as authentic by short tandem repeat profiling
and were passaged in the laboratory for <6 months following
receipt. All cell lines were grown in Dulbecco's modified Eagle's
medium (HyClone; GE Healthcare, Logan, UT, USA) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 100 mg/ml ampicillin and 100 mg/ml streptomycin.
The cultures were incubated at 37°C in a humidified atmosphere with
95% O2 and 5% CO2. Gemcitabine was purchased
from Eli Lilly and Company (Indianapolis, IN, USA) and dissolved in
sterile saline to form a 50 g/l stock solution.
Establishment of the resistant cell
lines BxPC-3/Gem and PANC-1/Gem
The gemcitabine-resistant cell lines, BxPC-3 and
PANC-1, were incubated in different initial gemcitabine
concentrations of 0, 10 and 20 µM for the PANC-1 cell line (PANC-1,
PANC-1/10, PANC-1/20, respectively) and 0, 6, 20, 40 and 70 µM for
the BxPC-3 cell line (BxPC-3, BxPC-3/6, BxPC-3/20, BxPC-3/40,
BxPC-3/70, respectively), to select cells with natural resistance
to gemcitabine. The cells were incubated in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) without drugs following
cultivation of the BxPC-3 cells and PANC-1 cells in this medium for
72 h. When the cells entered the logarithmic growth phase, they
were passaged twice and cultivated with increasing concentrations
of gemcitabine (1, 2, 5, 10, 20, 50, 100, 200, 500 and 1,000 µM)
over a 10-month period, then resistant cell lines BxPC-3/Gem
(BxPC-3, BxPC-3/6, BxPC-3/20, BxPC-3/40 and BxPC-3/70) and
PANC-1/Gem (PANC-1, PANC-1/10 and PANC-1/20) were acquired. The
cells were then cultivated in RPMI-1640 medium without gemcitabine
for 2 months.
Sensitivity analysis of
gemcitabine-resistant cell lines BxPC-3/Gem and PANC-1/Gem
The logarithmic phase cells were grown in 96-well
plates (4×103/well) for 24 h. Following adherence, cells
were cultured in varying concentrations (1, 2, 5, 10, 20, 50, 100,
200, 500 and 1,000 µM) of gemcitabine for 72 h, with 6 wells per
concentration. After 72 h, the media was removed and 180 µl media
and 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) were added to each well. The media was removed and 200 µl
dimethyl sulfoxide was added to each well 4 h later to dissolve the
formazan crystals. The cells were then agitated on a microplate
shaker for 10 min. Absorbance (A) was read at 530 nm on a
microplate reader. This experiment was repeated 4 times and equal
amounts of DMSO were used as a blank control. The cell inhibition
of each drug was calculated using the following formula:
Inhibition=1-(dosing group A/control group A) ×100%. Data were
plotted on a semi logarithmic curve with drug concentrations on the
X-axis and cell inhibition on the Y-axis. SPSS version 21 software
(IBM Corp., Armonk, NY, USA) was used to calculate the half maximal
inhibitory concentration (IC50) and the resistance index
(RI). The formula used to calculate RI was as follows:
RI=IC50 of resistant cell line/IC50 of
sensitive cell line.
Western blot analysis of ABCB1, ABCC
and ABCG2 protein expression
BxPC-3 and PANC-1 cell lines were untreated.
BxPC-3/Gem and PANC-1/Gem cells incubated for 24 h were collected
and lysed with radioimmunoprecipitation lysis buffer [50 mM Tris
(pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1%
SDS, sodium orthovanadate, sodium fluoride,
ethylenediaminetetraacetic acid (EDTA), leupeptin, and 1 nM
phenylmethylsulfonyl fluoride] for 20 min on ice. Cells were then
centrifuged at 20,817 × g and 4°C for 10 min and the supernatant
was collected. The total protein concentration was then detected
with a bicinchoninic acid assay (Beyotime Institute of
Biotechnology, Shanghai, China) and was adjusted to 2.5 µg/µl with
the lysis buffer. Proteins were electrophoresed on 20% SDS-PAGE,
and then a constant current of 30 mA overnight at 4°C was used to
electro-transfer the proteins onto polyvinylidene fluoride
membranes (PVDF; EMD Millipore, Billerica, MA, USA). The PVDF
membranes were blocked with 5% milk at room temperature for 1 h
prior to being incubated with rabbit anti-ABCB1 mAb (cat. no.
13342S), rabbit anti-ABCC mAb (cat. no. 72202S) and rabbit
anti-ABCG2 mAb (cat. no. 42078S) primary antibodies (Cell Signaling
Technology, Inc., Danvers, MA, USA) overnight at 4°C. Following
washing with 1X Tris-buffered saline (TBST; 0.1% Tween-20) 3 times,
horseradish peroxidase-conjugated secondary antibodies (goat
anti-Rabbit IgG (H+L), dilution: 1:1,000; cat. no. A0208, Beyotime
Institute of Biotechnology) were used to incubate the PVDF
membranes for 3 h at room temperature. Following washing with TBST
3 times, enhanced chemiluminescence reagents (GE healthcare,
Chicago, IL, USA; cat. no. RPN2106) were used to detect the bound
antibody complexes and Tanon 5200 multi automatic chemiluminescence
image analysis system (Tanon Science & Technology Co.,
Shanghai, China) was used to image. The same experiment was
implemented on the untreated and gemcitabine-treated PANC-1 cell
lines. An anti-β-Actin mouse monoclonal antibody (dilution,
1:2,500; cat. no. M1000170; Beyotime Institute of Biotechnology)
was used as a loading control.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis of gene expression of
ABCB1, ABCC and ABCG2
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract the total RNA from cultured
cells according to the manufacturer's protocols. The RNA content
was measured by UV spectrophotometry at 260 nm. The sequences of
primers and the size of the sequences are presented in Table I (TaqMan probe; Generay Biotech Co.,
Ltd, Shanghai, China). cDNA was synthesized according to the
protocols of the first cDNA strand synthesis kit (Bioline Reagent
Ltd., London, UK). PCR amplification was performed according to
manufacturer's protocols (MyTaq™; Bioline Reagents
Ltd.). The amplification cycling conditions were as follows: Stage
1: 1 cycle of 95°C for 5 min; stage 2: 40 cycles of 95°C for 15 sec
and 60°C for 45 sec; and stage 3: 1 cycle of 95°C for 15 sec, 60°C
for 1 min, 95°C for 15 sec and 60°C for 15 sec. The PCR products
were analyzed using ABI Prism 7500 SDS Software version 1.4
(Applied Biosystems, Thermo Fisher Scientific, Inc.) and the
expression level of mRNA was calculated by the 2−∆∆Cq
method (13).
 | Table I.Sequences of primers and the size of
the sequences used for RT-qPCR. |
Table I.
Sequences of primers and the size of
the sequences used for RT-qPCR.
| Gene | Sense primer
(5′-3′) | Antisense primer
(5′-3′) | PCR product (bp) |
|---|
| ABCB1 |
TATGCTGGAGCAGTTCCTCA |
CCAGCTCCTCCTCCTTCTTT | 149 |
| ABCC |
GAAGGAAGCAAAGCAAATGG |
CCTGCTGATGTCCCCACTAT | 109 |
| ABCG2 |
CGGAAGGTGTCCTGCTACAT |
CTTGACCATTTCCCTTCTGC | 129 |
| β-actin |
TGCGCAGAAAACAAGATGAG |
GTCACCTTCACCGTTCCAGT | 116 |
Detection of promoter methylation of
ABCB1, ABCC and ABCG2 via MS-HRM
PANC-1 and BxPC-3 cells were grown in 96-well plate
at a density of 5×103 cells per well for 24 h and washed
with phosphate buffered saline 3 times. Total DNA was extracted
from cultured cells and inverted using a DNA extraction kit
(Generay Biotech Co., Ltd.) according to the manufacturer's
protocols. MS-HRM was performed with a Methylated Cytosine Mapping
kit (Shanghai Genmed Co., Ltd., Shanghai, China) in a total volume
of 50 µl containing: 2 µl modified template DNA, 32.5 µl PCR Master
mix (PCR kit; Applied Biosystems, Thermo Fisher Scientific, Inc.),
0.5 µl primer-F, 0.5 µl primer-R, 0.5 µl Taq-1, 0.5 µl
Taq-2 and 13.5 µl PCR grade water. The amplification cycling
conditions were: Stage 1: 1 cycle of 50°C for 2 min; stage 2: 1
cycle of 95°C for 5 min; stage 3: 40 cycles of 95°C for 15 sec and
60°C for 50 sec. The PCR products were analyzed by ABI Prism 7500
SDS Software version 1.4 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Methylation %=100/(1+2 (Cq (CQ) CQ value-Cq (TG)
CQ value)).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS
version 21 (IBM Corp.). A one-way analysis of variance and Student
Newman-Keuls post-hoc test was used to identify statistically
significant differences between groups of data and P<0.05 was
considered to indicate statistically significant differences.
Pearson correlation analysis was used to test whether the
methylation level of the DNA promoter was correlated with the level
of mRNA.
Results
Expression of ABCB1, ABCC and ABCG2 in
BxPC-3/Gem and PANC-1/Gem cell lines
PANC-1 and BxPC-3 cell lines treated with a
concentration gradient of gemcitabine were induced to acquire
drug-resistance and inhibition of cell viability was detected using
an MTT assay (Fig. 1). The
IC50 of primary PANC-1 and BxPC-3 cell lines was 65.81
and 6.61 µM, respectively. Additionally, IC50 increased
with an increasing initial gemcitabine concentration of PANC-1
(PANC-1, PANC-1/10 and PANC-1/20) and BxPC-3 (BxPC-3, BxPC-3/6,
BxPC-3/20, BxPC-3/40 and BxPC-3/70), demonstrating that PANC-1 and
BxPC-3 successfully acquired resistance to gemcitabine during the
process of induction. The dynamic changes of ABCB1, ABCC and ABCG2
of PANC-1 and BxPC-3 cell lines with different initial gemcitabine
concentration were detected by western blotting during the process
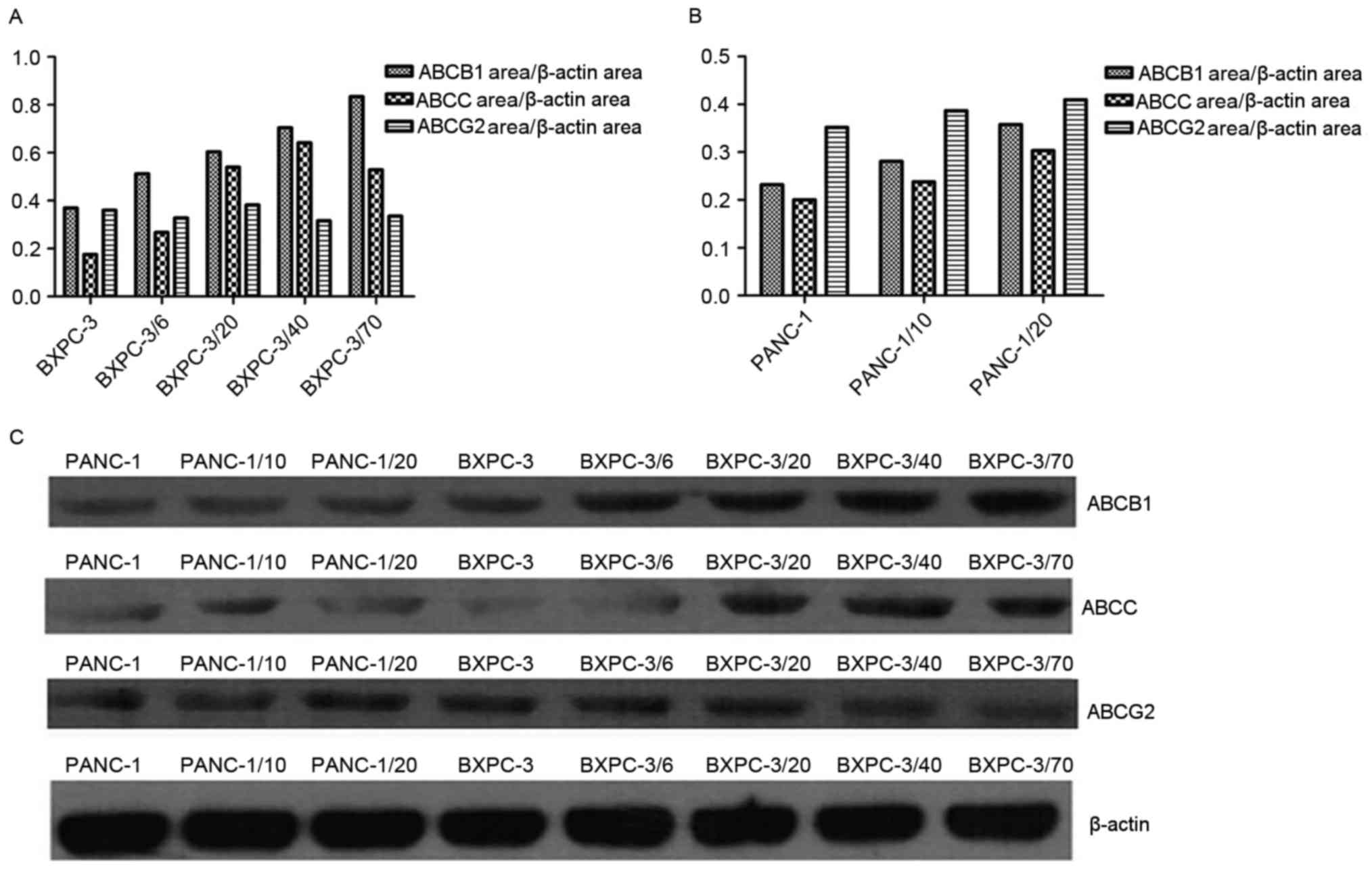
of inducing resistance to gemcitabine (Fig. 2). Compared with the primary culture
cells, expression of ABCC and ABCB1 was significantly increased
with an increase of initial gemcitabine concentration in the
BxPC-3/Gem cell line, while no significant change in expression of
ABCG2 was observed. In PANC-1/Gem, expression of ABCB1, ABCC and
ABCG2 was elevated with an increasing initial gemcitabine
concentration.
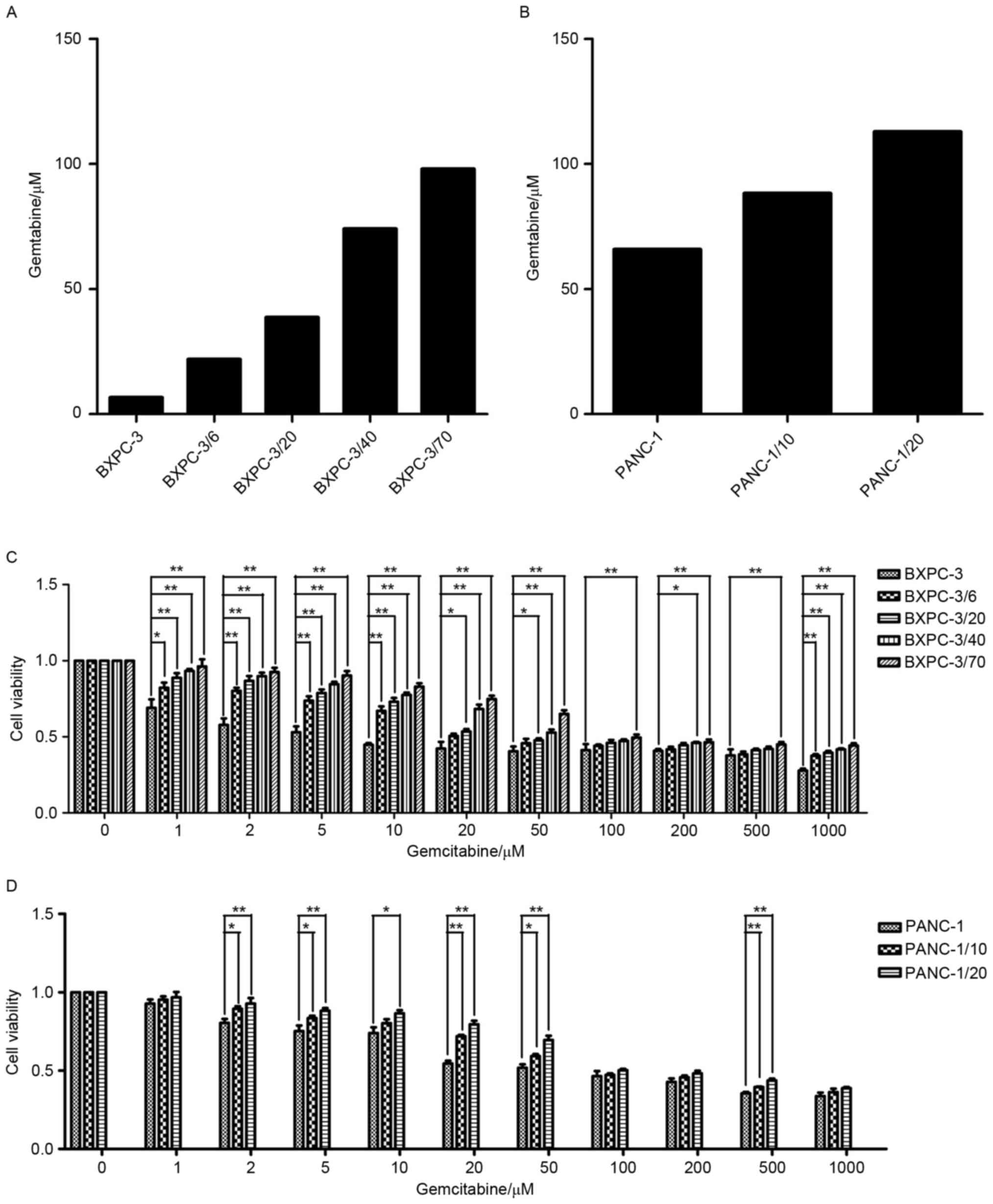 | Figure 1.Establishment of BxPC-3/Gem and
PANC-1/Gem cell lines and analysis of cell proliferation. (A)
BxPC-3 and (B) PANC-1 cells were exposed to with increasing
concentrations of gemcitabine (1, 2, 5, 10, 20, 50, 100, 200, 500
and 1,000 µM) for 72 h and the IC50 were calculated.
Cell viability of (C) BxPC-3 and (D) PANC-1 cells with different
initial gemcitabine concentration were analyzed by MTT assay.
Quantification was performed by assigning a value of 100% to the
untreated group of BxPC-3 and PANC-1 cells. The same volume of
normal saline solution was used as a positive control, while the
same volume of dimethyl sulfoxide was used as a negative control.
*P<0.05, **P<0.01, with comparisons indicated by lines. |
ABCB1, ABCC and ABCG2 mRNA expression
in BxPC-3/Gem and PANC-1/Gem cell lines
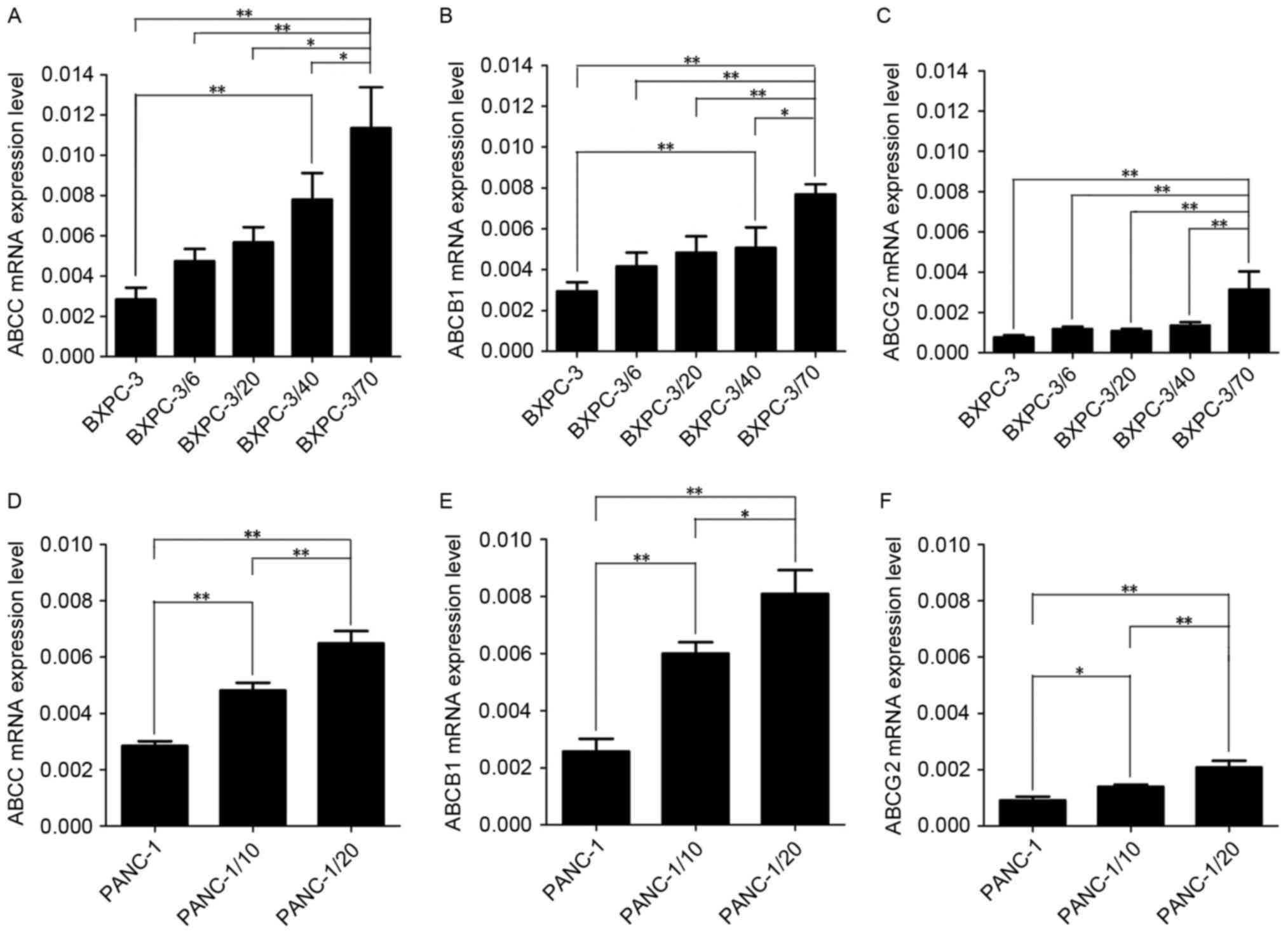
The mRNA expression of the 3 genes, ABCB1, ABCC and
ABCG2, in the BxPC-3/Gem and PANC-1/Gem cell lines was analyzed by
RT-qPCR (Fig. 3). In contrast to
primary culture cells, ABCB1, ABCC and ABCG2 mRNA expression was
increased with a rising initial gemcitabine concentration in the
BxPC-3/Gem and PANC-1/Gem cell lines. Of the 3 drug resistant
genes, expression of ABCB1 and ABCC mRNA was enhanced more markedly
than that of ABCG2.
Methylation of ABCB1, ABCC and ABCG2
in BxPC-3/Gem and PANC-1/Gem cell lines
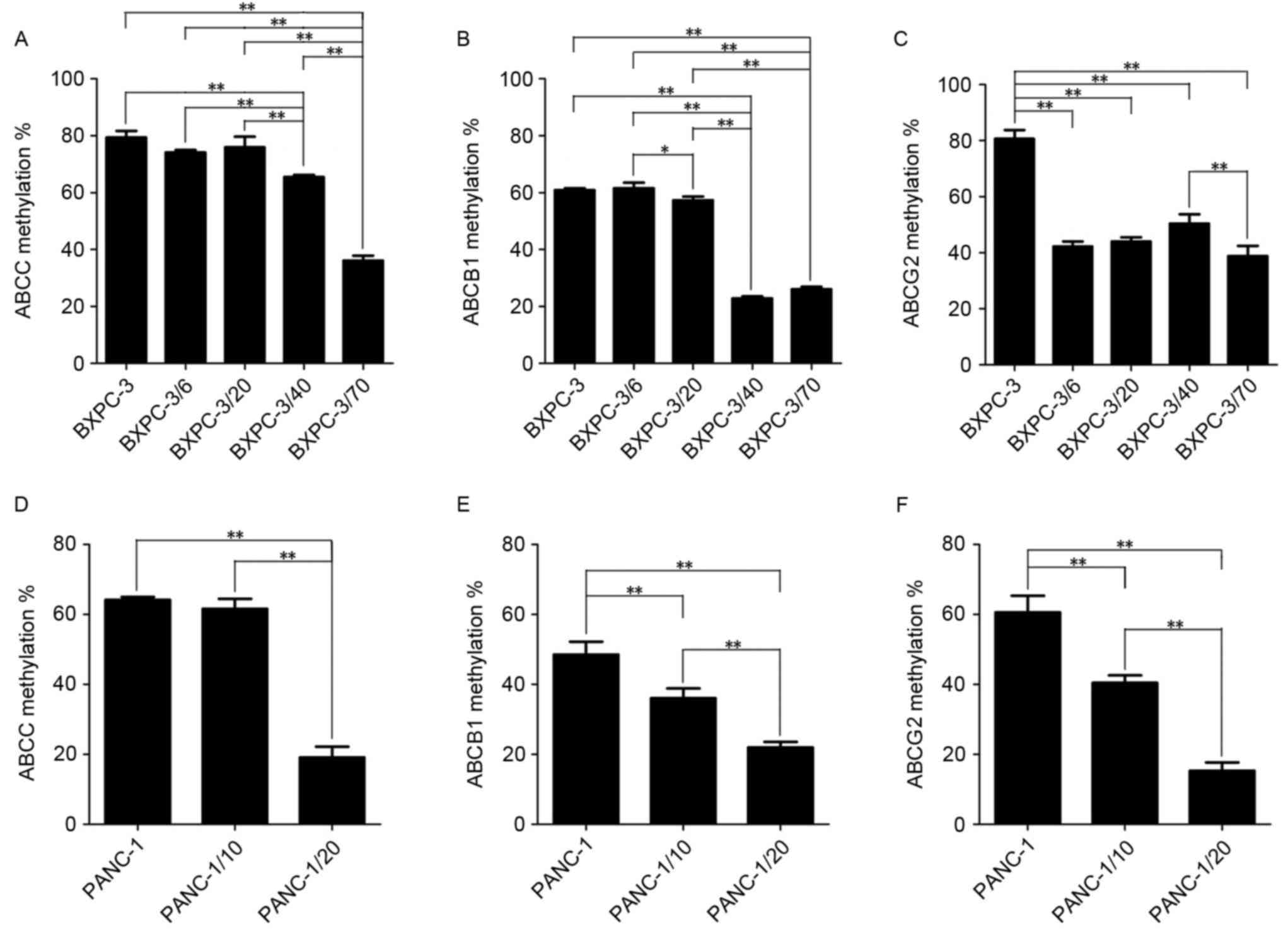
In order to further understand the expression of
ABCB1, ABCC and ABCG1 in BxPC-3/Gem and PANC-1/Gem cell lines, the
promoter methylation of the 3 ABC transporter genes was detected
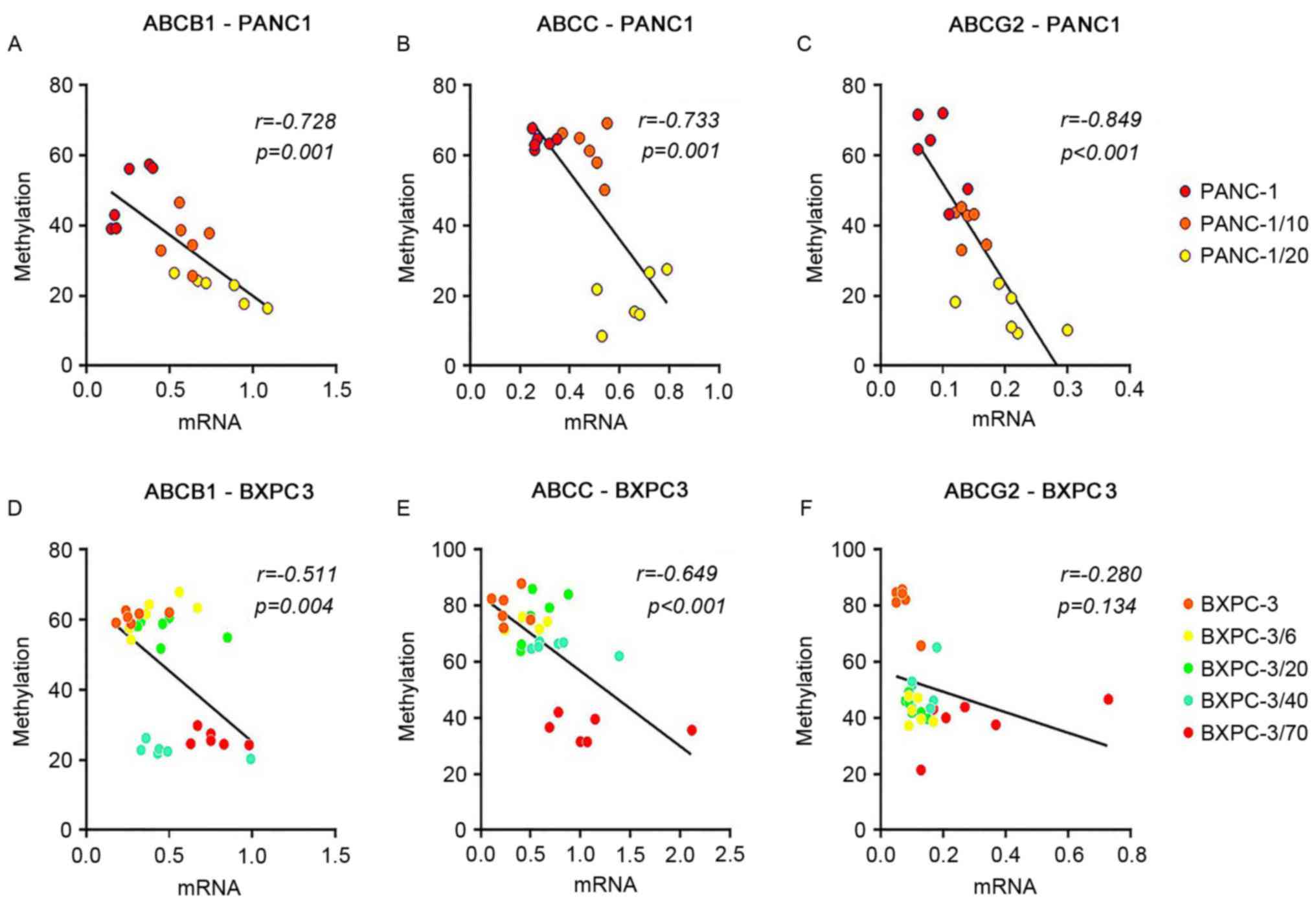
quantitatively and synchronously through MS-HRM (Fig. 4). A correlation analysis between
PANC-1 cell lines (PANC-1, PANC-1/10, PANC-1/20) and promoter
methylation was conducted and between BxPC-3 cell lines (BxPC-3,
BxPC-3/6, BxPC-3/20, BxPC-3/40, BxPC-3/70) and promoter methylation
(Fig. 5). The results demonstrated
that promoter methylation of ABCB1, ABCC and ABCG2 was decreased
with elevation of the initial gemcitabine concentration in
BxPC-3/Gem and PANC-1/Gem cell lines compared with primary culture
cells, suggesting that expression of ABCB1, ABCC and ABCG2 was
increased in the process of inducing gemcitabine resistance in
PANC-1 and BxPC-3 cell lines. It was also revealed that ABCB1
promoter methylation was significantly reduced when the initial
gemcitabine concentration was 40 in the BxPC-3/Gem cell line,
suggesting that drug-resistance of BxPC-3 was markedly increased
when initial gemcitabine concentration ranged from 20 to 40, and
the same as for ABCG2 promoter methylation when initial gemcitabine
concentration ranged from 0 to 6 in the PANC-1/Gem cell line.
Discussion
ABC transporters, which are transmembrane proteins
present on the tumor cell membrane, act as a drug efflux pump to
reduce the intracellular concentration of chemotherapeutic agents
by binding and hydrolyzing ATP, resulting in multidrug resistance
(14). Previous studies have
suggested that the ABCB1 (15,16), ABCC
(17) and ABCG2 (18–20) are
abnormally overexpressed in tumor cells of patients with pancreatic
cancer, which is clinically relevant to the reductive reaction to
chemotherapy of tumors, MDR and a poor clinical prognosis. However,
there is a lack of experimental data regarding the dynamic changes
of the expression of these 3 proteins during the process of
pancreatic cancer cells acquiring drug resistance.
In the present study, resistance to gemcitabine was
successfully induced in the human primary pancreatic cancer cell
lines PANC-1 and BxPC-3 via the concentration gradient method.
During the process of inducing drug resistance, it was revealed
that the expression of ABCB1 and ABCC proteins in PANC-1 and
BxPC-3, and the transcription of corresponding upstream mRNA,
indicates a synchronous increase in line with the increased initial
gemcitabine concentration. This result has confirmed the
aforementioned relationship between ABCB1, ABCC and pancreatic
cancer drug resistance (15–17), which also indicates that ABCB1 and
ABCC are involved in the formation of the acquired drug-resistance
in these pancreatic cancer cell line. Compared with ABCB1 and ABCC,
the expression of ABCG2 in PANC-1 and BxPC-3 did not exhibit a
marked increasing trend with an elevated initial gemcitabine
concentration, and the increase in mRNA transcription of ABCG2 was
not as marked as that of ABCB1 and ABCC. In studies of pancreatic
cancer, ABCG2, which mainly exists in side population (SP) cells of
pancreatic cancer and is highly expressed, had been recognized as a
potential marker of tumor stem cells and assists SP cells in
acquiring a more powerful drug efflux capacity than that of non-SP
cells (21,22). The ratio of SP cells in the PANC-1
cell line is low (23) and the cells
may not have proliferated to a certain percentage or demonstrated
any survival advantage in drug-resistant cells due to the short
time span of the experiment, which may have been the cause of the
lack of statistically significant differences in the expression of
ABCG2. Therefore, ABCC and ABCB1 in ABC transporters may serve a
more important function than ABCG2 in the early stage of
establishing drug-resistant pancreatic cancer cell lines.
Methylation of DNA is one of the most essential
biological epigenetic modifications. The methylation of a specific
gene promoter on the DNA cut back has a direct impact on mRNA
transcription and protein expression, resulting in the abnormal
expression of the protein encoded by the gene (24,25).
Previous studies on other malignant tumors have proposed that the
methylation of ABC transporter gene promoters is negatively
correlated with chemoresistance in tumor cells (26–28). In a
study on pancreatic cancer, Chen et al (29) observed that the promoter methylation
level of the ABC transporter family (ABCB1, ABCC and ABCG2) in a
gemcitabine-resistant cell line (SW1990/GZ) was significantly lower
than that in the primary cell line (SW1990), while resistance to
gemcitabine of the cell line increased 33.3 times. The present
study revealed that, during the process of establishing gemcitabine
resistance in PANC-1 and BxPC-3 cell lines, the promoter
methylation level of the ABC transporter family drug resistance
genes, ABCB1, ABCC and ABCG2, decreased gradually with an increased
initial gemcitabine concentration, and an increase in the
expression of the corresponding downstream mRNA and protein. This
was consistent with the effect of epigenetic methylation
modification, suggesting that the promoter hypomethylation
modification of the chemo-resistance genes, ABCB1, ABCC and ABCG2,
was also involved in the acquisition of the drug resistance in the
pancreatic cancer cells. This provides evidence of the feasibility
of predicting MDR of pancreatic cancer through independent
indicators-the promoter methylation level of ABCB1, ABCC and ABCG2.
However, the specific mechanisms underpinning this require further
experimental verification and discussion.
The present study discovered that the change of the
resistant gene methylation not only reveals drug resistance of
pancreatic cancer, but may also be used as an evaluation indicator
to assess the drug resistance of tumors that are treated with
chemotherapy. Previously, pancreatic tumor tissues were obtained
mainly from surgical resection and thus, the majority of tumor
samples were from patients with resectable tumors (30). However, there is no clinical
significance in detecting the methylation of resistant genes for
such patients. In recent years, with the development of medical
technology, endoscopic ultrasound and fine needle aspiration is
widely applied in clinical practice and the tumor tissues of
patients with unresectable tumors who always require long-term
chemotherapy are accessible using this technique (31). Additionally, it is also possible to
obtain tumor cells from the blood and to use these to detect
resistance gene methylation. Therefore, the detection of resistance
gene methylation may be used to monitor tumor drug resistance in
patients undergoing chemotherapy, so as to aid clinicians in
modulating chemotherapy regimens according to the changes of tumor
drug resistance.
Certain studies have proposed the novel concept of
cancer evolution, wherein the tumor follows the same selection
principle of nature, as with other living creatures (32). With the mutation of tumor cell genes,
tumor cells that are resistant to treatment survive and expand, and
are able to evolve and adapt. At present, owing to the advanced
sequencing technology and accumulation of a large volume of
clinical data, scholars have drawn the map of cancer evolution and
have revealed the origin of the drug resistance (33). According to the map of cancer
evolution, the trunk mutations of tumor cell genes may be
discovered during the process of cancer evolution as these types of
mutation are present in all tumor cells. Notably, there are also
branch mutations and these types of mutation are only present in
certain tumor cells. We hypothesized that the epigenetic changes of
the ABC transporter family is one of the trunk mutations in the
present study. Through the detection of ABC transporter family
methylation, changes in the chemoresistance of pancreatic cancer
cells were observed, which in turn may be a closely monitored
process of tumor evolution and adaptation and, as mentioned
previously, may provide further basis for clinicians to adjust
chemotherapy regimens in sufficient time. Future studies should
also focus on further exploring branch mutations in pancreatic
cancer to refine the evolutionary map of pancreatic cancer and
provide more treatment options for clinicians.
It was also observed that not all resistance genes
were altered to the same degree as tumor chemoresistance, and
certain resistance genes served a leading function while others
served a minor function or no function at all. These findings may
also aid clinicians in determining more accurate therapies to
target tumors. Although there is a shortcoming of the present
study, namely the low detecting precision of MS-HRM, this method is
economic, efficient, convenient and also has the advantage of
distinguishing tumors from normal tissue samples more accurately
and thus, is capable of being widely used in clinical practice
(34). However, the lack of animal
experiments is a drawback of the present study and animal
experiments were not completed for a number of reasons. Firstly,
the survival time of genetically engineered mouse models of
pancreatic cancer is relatively short, and achieving a change in
tumor drug resistance often requires a longer time period and
therefore it was difficult to observe the variety of drug
resistance in the tumors. Additionally, it is well known that
pancreatic cancer is a stromal-rich malignant tumor and therefore,
if a subcutaneous transplantation tumor model of pancreatic cancer
in nude mice were to be selected, it would not be able to
completely simulate human pancreatic tumor tissue and the results
would be unreliable. Future studies will aim to find a suitable
mouse model and to undertake animal experiments.
To conclude, it is presumed that the pathway to
reduce the reaction of pancreatic cancer cells to drugs includes
the increased expression of downstream mRNA and protein through the
promoter hypomethylation modification of the ABCB1, ABCC and ABCG2
genes in the ABC transporter family and the enhanced drug efflux
capacity of the transporters. Additionally, with the evolution of
drug resistance in pancreatic cancer cells, the promoter
methylation of ABCB1, ABCC and ABCG2 gradually decreases.
Therefore, the promoter methylation level is capable of
quantitatively and dynamically reflecting the progression of
chemo-resistance in pancreatic cancer cells.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by The
National Natural Science Foundation of China (grant no.
81201896).
Availability of data and materials
All data generated and analyzed during the present
study are included in the published article and available for
use.
Authors' contributions
LY and JG performed all the data analysis and
drafted the manuscript. YM and XZ performed basic experiments and
collected the data. XW helped to edit the manuscript and provided
support for experiments. DF and CJ performed independent reviews of
all studies and data, and JL designed this study and executed the
analysis.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gillen S, Schuster T, Meyer Zum
Büschenfelde C, Friess H and Kleeff J: Preoperative/neoadjuvant
therapy in pancreatic cancer: A systematic review and meta-analysis
of response and resection percentages. PLoS Med. 7:e10002672010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Neoptolemos JP, Stocken DD, Friess H,
Bassi C, Dunn JA, Hickey H, Beger H, Fernandez-Cruz L, Dervenis C,
Lacaine F, et al: A randomized trial of chemoradiotherapy and
chemotherapy after resection of pancreatic cancer. N Engl J Med.
350:1200–1210. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tanaka M, Okazaki T, Suzuki H, Abbruzzese
JL and Li D: Association of multi-drug resistance gene
polymorphisms with pancreatic cancer outcome. Cancer. 117:744–751.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polgar O and Bates SE: ABC transporters in
the balance: Is there a role in multidrug resistance? Biochem Soc
Trans. 33:241–245. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Douet V, Heller MB and Le Saux O: DNA
methylation and Sp1 binding determine the tissue-specific
transcriptional activity of the mouse Abcc6 promoter. Biochem
Biophys Res Commun. 354:66–71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bram EE, Stark M, Raz S and Assaraf YG:
Chemotherapeutic drug-induced ABCG2 promoter demethylation as a
novel mechanism of acquired multidrug resistance. Neoplasia.
11:1359–1370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao P, Yang X, Xue YW, Zhang XF, Wang Y,
Liu WJ and Wu XJ: Promoter methylation of glutathione S-transferase
pi1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma
and its correlation with DNA methyltransferase 1 expression.
Cancer. 115:3222–3232. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pang L, Word B, Xu J, Wang H, Hammons G,
Huang SM and Lyn-Cook B: ATP-binding cassette genes genotype and
expression: A potential association with pancreatic cancer
development and chemoresistance? Gastroenterol Res Pract 2014.
4149312014.
|
|
12
|
Chen M, Xue X, Wang F, An Y, Tang D, Xu Y,
Wang H, Yuan Z, Gao W, Wei J, et al: Expression and promoter
methylation analysis of ATP-binding cassette genes in pancreatic
cancer. Oncol Rep. 27:265–269. 2012.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cascorbi I: Role of pharmacogenetics of
ATP-binding cassette transporters in the pharmacokinetics of drugs.
Pharmacol Ther. 112:457–473. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suwa H, Ohshio G, Arao S, Imamura T,
Yamaki K, Manabe T, Imamura M, Hiai H and Fukumoto M:
Immunohistochemical localization of P-glycoprotein and expression
of the multidrug resistance-1 gene in human pancreatic cancer:
Relevance to indicator of better prognosis. Jpn J Cancer Res.
87:641–649. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu Z, Kleeff J, Shrikhande S, Zimmermann
T, Korc M, Friess H and Büchler MW: Expression of the
multidrug-resistance 1 (MDR1) gene and prognosis in human
pancreatic cancer. Pancreas. 21:240–247. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
König J, Hartel M, Nies AT, Martignoni ME,
Guo J, Büchler MW, Friess H and Keppler D: Expression and
localization of human multidrug resistance protein (ABCC) family
members in pancreatic carcinoma. Int J Cancer. 115:359–367. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee SH, Kim H, Hwang JH, Lee HS, Cho JY,
Yoon YS and Han HS: Breast cancer resistance protein expression is
associated with early recurrence and decreased survival in
resectable pancreatic cancer patients. Pathol Int. 62:167–175.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Xue X, Wei J, An Y, Yao J, Cai H,
Wu J, Dai C, Qian Z, Xu Z and Miao Y: hsa-miR-520 h downregulates
ABCG2 in pancreatic cancer cells to inhibit migration, invasion,
and side populations. Br J Cancer. 103:567–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hamada S, Satoh K, Hirota M, Kanno A,
Umino J, Ito H, Masamune A, Kikuta K, Kume K and Shimosegawa T: The
homeobox gene MSX2 determines chemosensitivity of pancreatic cancer
cells via the regulation of transporter gene ABCG2. J Cell Physiol.
227:729–738. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olempska M, Eisenach PA, Ammerpohl O,
Ungefroren H, Fandrich F and Kalthoff H: Detection of tumor stem
cell markers in pancreatic carcinoma cell lines. Hepatobiliary
Pancreat Dis Int. 6:92–97. 2007.PubMed/NCBI
|
|
22
|
Miranda-Lorenzo I, Dorado J, Lonardo E,
Alcala S, Serrano AG, Clausell-Tormos J, Cioffi M, Megias D,
Zagorac S, Balic A, et al: Intracellular autofluorescence: A
biomarker for epithelial cancer stem cells. Nat Methods.
11:1161–1169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou J, Wang CY, Liu T, Wu B, Zhou F,
Xiong JX, Wu HS, Tao J, Zhao G, Yang M and Gou SM: Persistence of
side population cells with high drug efflux capacity in pancreatic
cancer. World J Gastroenterol. 14:925–930. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bird AP and Wolffe AP: Methylation-induced
repression-belts, braces, and chromatin. Cell. 99:451–454. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Toyota M, Suzuki H, Yamashita T, Hirata K,
Imai K, Tokino T and Shinomura Y: Cancer epigenomics: Implications
of DNA methylation in personalized cancer therapy. Cancer Sci.
100:787–791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding S, Gong BD, Yu J, Gu J, Zhang HY,
Shang ZB, Fei Q, Wang P and Zhu JD: Methylation profile of the
promoter CpG islands of 14 ‘drug-resistance’ genes in
hepatocellular carcinoma. World J Gastroenterol. 10:3433–3440.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chekhun VF, Kulik GI, Yurchenko OV,
Tryndyak VP, Todor IN, Luniv LS, Tregubova NA, Pryzimirska TV,
Montgomery B, Rusetskaya NV and Pogribny IP: Role of DNA
hypomethylation in the development of the resistance to doxorubicin
in human MCF-7 breast adenocarcinoma cells. Cancer Lett. 231:87–93.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tada Y, Wada M, Kuroiwa K, Kinugawa N,
Harada T, Nagayama J, Nakagawa M, Naito S and Kuwano M: MDR1 gene
overexpression and altered degree of methylation at the promoter
region in bladder cancer during chemotherapeutic treatment. Clin
Cancer Res. 6:4618–4627. 2000.PubMed/NCBI
|
|
29
|
Chen M, Xue X, Wang F, An Y, Tang D, Xu Y,
Wang H, Yuan Z, Gao W, Wei J, et al: Expression and promoter
methylation analysis of ATP-binding cassette genes in pancreatic
cancer. Oncol Rep. 27:265–269. 2012.PubMed/NCBI
|
|
30
|
Wray CJ, Ahmad SA, Matthews JB and Lowy
AM: Surgery for pancreatic cancer: Recent controversies and current
practice. Gastroenterology. 128:1626–1641. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krishna NB, LaBundy JL, Saripalli S,
Safdar R and Agarwal B: Diagnostic value of EUS-FNA in patients
suspected of having pancreatic cancer with a focal lesion on CT
scan/MRI but without obstructive jaundice. Pancreas. 38:625–630.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Goymer P: Natural selection: The evolution
of cancer. Nature. 454:1046–1086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang Y, Qiu Y, Minn AJ and Zhang NR:
Assessing intratumor heterogeneity and tracking longitudinal and
spatial clonal evolutionary history by next-generation sequencing.
Proc Natl Acad Sci USA. 113:pp. E5528–E5537. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
34
|
BLUEPRINT consortium, . Quantitative
comparison of DNA methylation assays for biomarker development and
clinical applications. Nat Biotechnol. 34:726–737. 2016. View Article : Google Scholar : PubMed/NCBI
|