Introduction
Lung cancer is currently the leading cause of
cancer-associated mortality worldwide (1). The majority of lung cancer cases are
non-small cell lung cancer (NSCLC), which accounts for ~85% of
newly diagnosed lung cancer cases (2). Lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC) are the two most common histologic
subtypes of NSCLC (3).
In recent years, with the progression of molecular
medicine and the emergence of targeted drugs, treatment of NSCLC
started to become individualized molecular targeted ‘precise’
treatment (4). At present, the
individualized molecular targeted therapy for clinical application
is primarily for epidermal growth factor mutation and anaplastic
lymphoma kinase fusion genotypes in lung cancer, both of which have
clear molecular targets, and targeted drug has significantly
improved the clinical efficacy (5,6). However,
the prognosis of LUSC remains poor and identifying effective
prognostic biomarkers for LUSC remains urgent (7–9).
B-cell lymphoma 2 (Bcl-2) is an antiapoptotic
protein, which belongs to the Bcl-2 family. It is located in the
inner mitochondrial membrane and to a lesser extent in cell
membranes (10). The primary function
of Bcl-2 appears to be to inhibit apoptosis (programmed cell death)
and to prolong cell survival by arresting cells in the G0/G1 phase
of the cell cycle (11,12). Previous studies have revealed that
Bcl-2 is highly expressed in several hematologic and solid
malignancies, including acute lymphocytic leukemia (ALL), breast,
prostate, colorectal, lung, stomach, and ovarian cancer (13–15).
It has been confirmed Bcl-2 as an independent
prognostic marker in breast cancer (16), and Bcl-2 is also considered to be a
favorable prognostic marker in NSCLC (17–19), but
Kim et al (20) reported that
Bcl-2 is an adverse prognostic marker. The aforementioned studies
focused on NSCLC or LUAD; however, to the best of our knowledge,
there is no previous study regarding the role of Bcl-2 in LUSC
(21). LUSC and LUAD are very
different in the molecular biological background and clinical
biological characteristics: LUAD has more driver genes, including
EGFR and ALK, which also means that there are more clinical
treatment options, and LUAD is more common in non-smokers, compared
with LUSC (9,22,23).
Over the past decade, high-throughput detection
technology has yielded a mass of tumor data (24), but these datasets are scattered, due
to patient cohort, technology platform and other heterogeneous
variables, thus making it hard to compare (22). In the present study, in order to solve
this problem and to make better use of these public database
(25), 901 LUSC gene expression
profiles were integrated and the association between expression
level and overall survival was analyzed. Furthermore, the
prognostic value of Bcl-2 in LUSC was validated with a tissue
microarray (TMA) using immunohistochemistry (IHC) analysis.
Materials and methods
Data collection and prognostic
association meta-analysis
Data from 16 publicly available microarray studies
on lung cancer with overall survival outcome data from the National
Center for Biotechnology Information Gene Expression Omnibus
(25) and The Cancer Genome Atlas
were curated. Raw CEL files were obtained where possible, and were
normalized, summarized and log-transformed using robust multi-array
average function of the affy R package (https://www.r-project.org/; R version 3.1.2; affy
version 1.44). The probe-based expression was converted into gene
expression profiles, and only cases on patients with squamous cell
carcinoma were retained for subsequent analysis.
For the meta-analysis of the prognostic association
between gene expression and survival outcomes, the statistical
significance was assessed by z-scores via univariate Cox
proportional hazards regression in each of the 16 datasets
(Table I) using the coxph function of
the survival R package (26).
Z-scores represent the number of standard deviation below (or
above) the mean of a normal distribution. In addition, z-scores
conveniently reflect the directionality and significance of
statistical association. In order to obtain the integrated and
robust prognostic landscape, z-scores for a gene were summarized
across all 16 datasets to yield a ‘meta-z-score’ for the prognostic
significance assessment using Lipták's weighted meta-z test with
weights set to the square roots of sample sizes. The prognostic
genes were defined by meta-z-scores filtered for |meta-z| >1.96
(|z|>1.96 is equivalent to a two-tailed P<0.05). Favorable
prognostic genes have meta-z <-1.96 and adversely prognostic
genes have meta-z >1.96.
 | Table I.Details of the 16 LUSC datasets
used. |
Table I.
Details of the 16 LUSC datasets
used.
| Dataset ID | Platform | LUSC number | Country | Reference (PMID) |
|---|
| gse3141 | GPL570 | 53 | USA | 16273092 |
| gse4573 | GPL96 | 130 | USA | 16885343 |
| gse5828 | GPL3877 | 59 | Australia | 17601969 |
| gse11117 | GPL6650 | 14 | Switzerland | 19833826 |
| gse12428 | GPL1708 | 34 | Netherlands | 19334046 |
| gse12472 | GPL1708 | 35 | Netherlands | 20832896 |
| gse14814 | GPL96 | 52 | Canada | 20823422 |
| gse17710 | GPL9053 | 56 | USA | 20643781 |
| gse19188 | GPL570 | 24 | Netherlands | 20421987 |
| gse29013 | GPL570 | 25 | USA | 21742808 |
| gse37745 | GPL570 | 66 | Sweden | 23032747 |
| gse30219 | GPL570 | 61 | France | 23698379 |
| gse41271 | GPL6884 | 80 | USA | 23449933 |
| gse11969 | GPL7015 | 35 | Japan | 16549822 |
| gse50081 | GPL570 | 43 | Canada | 24305008 |
| TCGA | GPL96 | 134 | USA | 22960745 |
Patient tissue samples
A total of 72 patients LUSC who were diagnosed, and
underwent surgery in People's Hospital of Peking University
(Beijing, China) between 2004 and 2010 were enrolled in the present
study. Fresh LUSC tissue from each patient were formalin-fixed,
paraffin-embedded (27), and
constructed into TMAs (Shanghai Outdo Biotech Co., Ltd, Shanghai,
China): Using the tissue chip spotter, the marked tissue is
arranged on the blank wax block according to the design, and then
the slicing machine is used to slice the array wax block
continuously to obtain the tissue chip. Postoperative follow-up has
lasted ≥3 years for all patients. Histopathological evaluation was
performed independently by two pathologists. The clinical stage of
the tumors was evaluated by an experienced pathologist according to
the 7th edition of the American Joint Committee on Cancer (AJCC)
tumor node metastasis (TNM) staging system (28). Complete clinical information,
including age, gender, stage, smoking, follow-up time, and survival
status was collected. The present study was approved by the
Institutional Review Board of the People's Hospital of Peking
University and written informed consent was obtained from each
patient.
IHC analysis
All hematoxylin and eosin slides were centrally
reviewed at the Department of Pathology in People's Hospital of
Peking University according to the histopathological classification
system adopted by the World Health Organization to confirm tumor
type (29). TMA block sections (4-µm
thick) were rewarmed in the oven at 65°C for 3 h, then
deparaffinized in 100% xylene and dehydrated with graded ethanol
washes. Antigen retrieval was performed in a pressure cooker,
followed by the treatment with 3% hydrogen peroxide for 15 min at
room temperature to block endogenous peroxidase activity.
Thereafter, the sections were incubated at 4°C overnight with
anti-Bcl-2 (dilution, 1:20; cat.no. PAB7640; Abnova, Taipei,
China). After being incubated at 37°C for 1 h, these slides were
washed three times in PBS and incubated with horseradish
peroxidase-conjugated anti-rabbit antibody (dilution, 1:1,000; cat.
no. PAB10822; Abnova) for 15 mins at 37°C. The stained specimens
were exposed to the 3,3-diaminobenzidine and counterstained with
hematoxylin for 20 min at room temperature. For the negative
controls, primary antibodies were replaced with PBS.
Bcl-2 staining was microscopically examined (Olympus
Corporation, Tokyo, Japan; inverted fluorescent microscope;
magnification, ×100) and scored by two independent pathologists who
were blind to the clinical data pertaining to the patients. A
semi-quantitatively scoring system (0–3) was used to evaluate the
expression level of Bcl-2. The intensity of the staining was
classified as negative, weak, moderate or strong. Staining
intensity was scored as follows: 0 (negative), 1 (weakly positive),
2 (moderately positive), and 3 (strongly positive). The proportion
of each level of staining cells were estimated A, B, C and D
(between 0–100%). The above two scores were multiplied, the final
score as follows: (0 × A%) + (1 × B%) + (2 × C%) + (3 × D%).
Statistical analysis
Statistical analyses were performed using the R
statistical language with the ‘survival’ package. Briefly, the
Chi-square test was performed to analyze the association between
Bcl-2 expression and clinicopathological features. In the
univariate survival analyses, the difference in median overall
survival (OS) time between groups of patients was analyzed using
the log-rank test. The independent prognostic factors of OS were
further identified by multivariate Cox proportional hazards
regression models. The hazard ratios (HRs) and 95% confidence
intervals (CIs) of the prognostic factors were calculated.
Kaplan-Meier survival curves were constructed for survival analyses
and differences were tested using the log-rank test. Bcl-2
expression was categorized as high or low using the median score.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical features of patients with
LUSC
A TMA containing 72 LUSC cases was utilized to
perform IHC staining. Overall, 10 female patients and 62 male
patients, with an age range of 41–86 years (mean age, 67.4 years)
were included in the current study. According to the 7th edition of
the AJCC TNM staging system, 53 patients (73.6%) were at early
stages (38 stage I and 15 stage II) and 19 patients (26.4%) were at
advanced stages (18 stage III and 1 stage IV). The diameter of the
tumor of 16 patients (22.5%) was <3 cm, while that of the
remaining 55 patients (77.5%) was ≥3 cm. There were 25 patients
(34.7%) with positive lymph node metastasis and 47 patients (65.3%)
exhibited negative lymph node metastasis. The primary
clinicopathological characteristics of these patients are listed in
Table II. Generally, the overall
follow-up durations ranged between 3.9 and 84.3 months. Forty-five
patients were alive at the end of the follow-up and the OS rate was
62.5% in the present study.
 | Table II.Summary of patient characteristics
(n=72). |
Table II.
Summary of patient characteristics
(n=72).
| Clinicopathological
features | No. of
patients | Percentage of
patients |
|---|
| Sex |
|
|
|
Male | 62 | 86.1 |
|
Female | 10 | 13.9 |
| Age, years |
|
|
|
<60 | 19 | 26.4 |
|
≥60 | 53 | 73.6 |
| Tumor size, cm |
|
|
|
<3 | 17 | 23.6 |
| ≥3 | 55 | 76.4 |
| Smoking |
|
|
|
<20 | 21 | 29.2 |
|
≥20 | 51 | 70.8 |
| Stage |
|
|
| I | 38 | 52.8 |
| II | 15 | 20.8 |
|
III | 18 | 25.0 |
| IV | 1 | 1.4 |
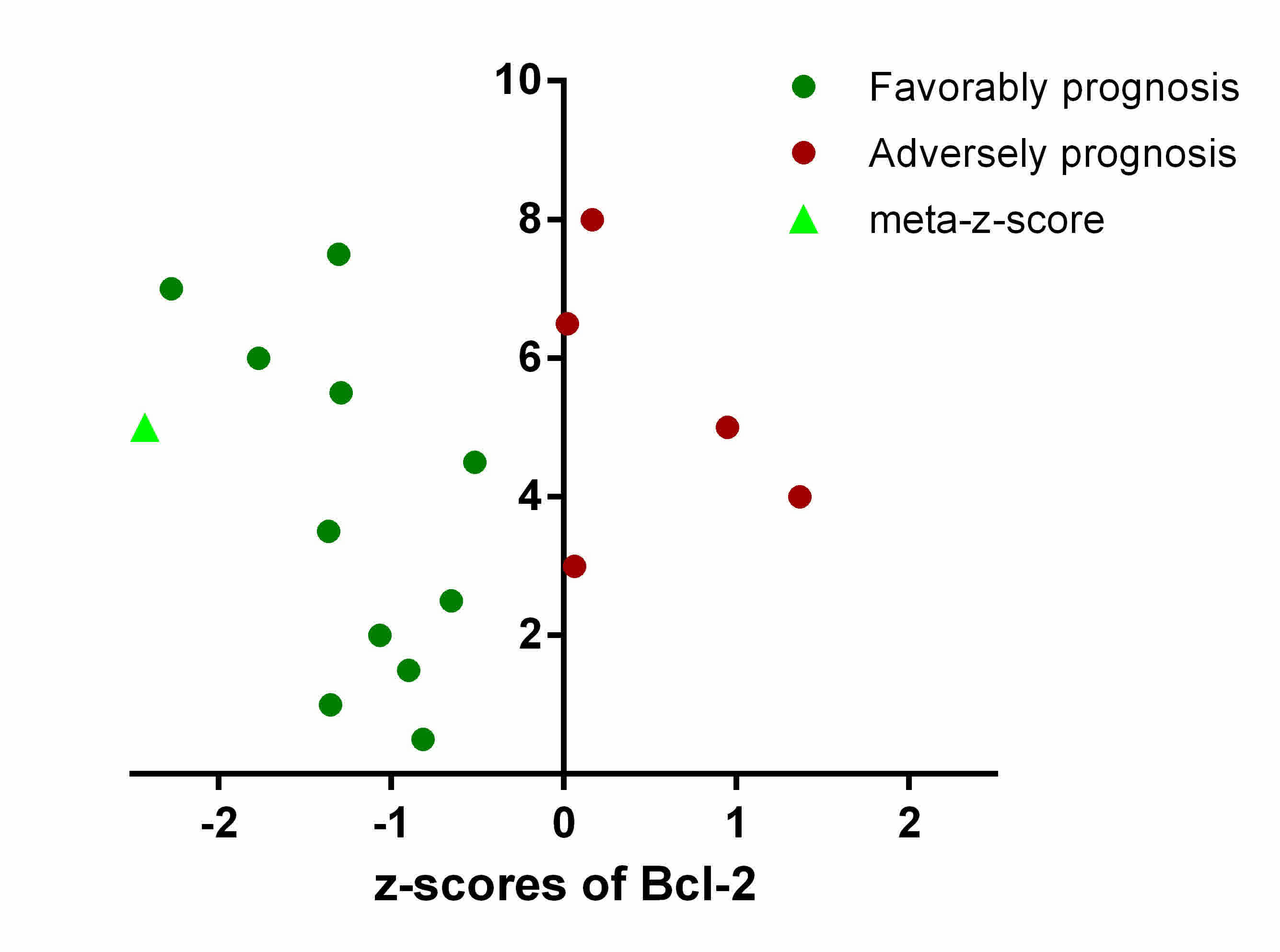
Meta-analysis of prognostic
significance of Bcl-2 in patients with LUSC
To gain a comprehensive and robust insight into the
prognostic significance of Bcl-2 in LUSC, the LUSC tumor gene
expression profiles and survival data of 901 patients from 16
datasets were assembled, and integrated. The prognostic association
between the expression level of Bcl-2 and OS were independently
evaluated in 16 datasets using z-scores.
To minimize the confounding influence of batch
effects and other limitations derived from pooling raw data or
merging expression data across multiple studies, weighted Z-tests
were used to combine independent z-scores of Bcl-2 into a
‘meta-z-score’. In different cohorts, adverse and favorable
prognostic significance was associated with Bcl-2 expression, with
the final meta-z-score being −2.43 for Bcl-2 (Fig. 1). This suggested that high Bcl-2
expression level is associated with longer survival times, and
Bcl-2 may serve as a prognostic biomarker in predicting the OS rate
of patients with LUSC.
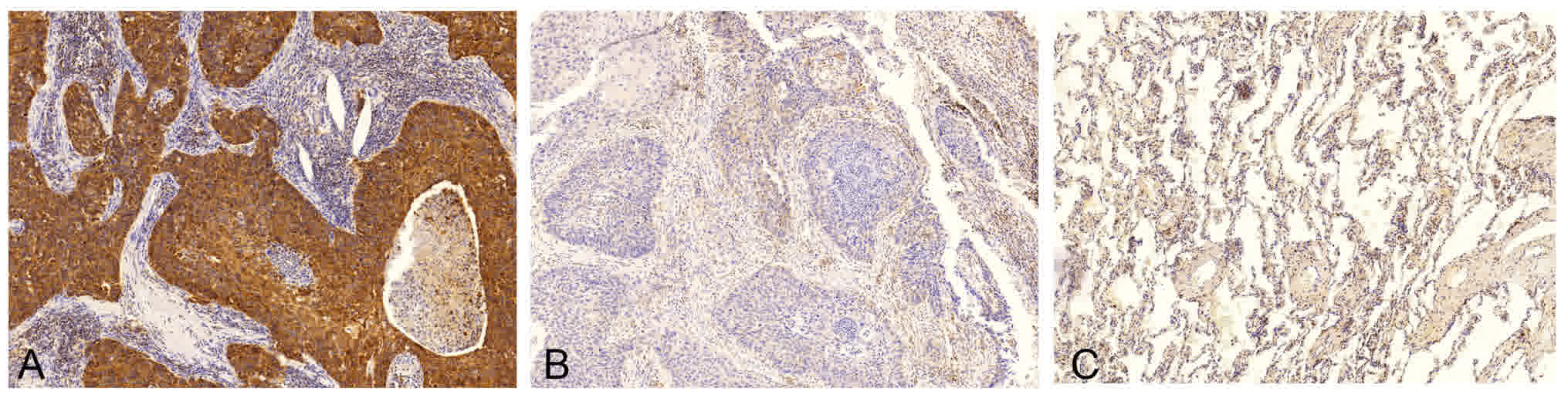
Association between the IHC expression
of Bcl-2 and clinicopathological features
The expression of Bcl-2 protein was analyzed in 72
patients with LUSC using IHC (Fig.
2), and it was revealed that Bcl-2 protein was primarily
localized to the cell membrane and cytoplasm of lung SCC cells,
which is consistent with a previous study (18). In contrast, adjacent bronchial mucosa
and alveolar epithelial cells were Bcl-2-negative. To minimize the
bias of IHC scoring, a scoring standard was set up and two
independent researchers scored all of IHC staining samples.
Considering the overall positive rate of Bcl-2 expression observed
in this study, patients with LUSC were divided into two groups as
follows: Score ≤1 as the low expression group and score >1 as
the high expression group. The positive rate of Bcl-2 expression in
the current study was 91.7% (66/72), with 54.2% (39/72) of patients
exhibiting weak expression (score ≤1) and 45.8% (33/72) of patients
exhibiting strong expression (score >1).
The association between Bcl-2 protein expression and
the clinicopathological parameters of patients with LUSC was
analyzed using the Chi-square test (Table III). The results revealed that high
Bcl-2 expression was significantly associated with heavy smoking
(P<0.05). No statistically significant difference was identified
between Bcl-2 expression and other clinical parameters, including
age, gender, tumor diameter, TNM stage or lymph node
metastasis.
 | Table III.Association between the
immunohistochemical expression of Bcl-2 and clinicopathological
features. |
Table III.
Association between the
immunohistochemical expression of Bcl-2 and clinicopathological
features.
|
| Bcl-2 |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variables (n=72) | Low | High | χ2 | P-value |
|---|
| Age, years |
|
|
|
|
|
<60 | 10 | 9 |
2.40×10−31 | >0.99 |
|
≥60 | 29 | 24 |
|
|
| Sex |
|
|
|
|
|
Female | 35 | 27 | 0.39 | 0.53 |
|
Male | 4 | 6 |
|
|
| AJCC7 stage |
|
|
|
|
|
I–II | 39 | 32 | 0.01 | 0.93 |
|
III–IV | 0 | 1 |
|
|
| Smoking |
|
|
|
|
|
<20 | 7 | 14 | 4.17 | 0.04 |
|
≥20 | 31 | 18 |
|
|
| Tumor size, cm |
|
|
|
|
|
<3 | 11 | 5 | 4.75 | 0.19 |
|
3–5 | 13 | 15 |
|
|
|
5–7 | 8 | 10 |
|
|
| ≥7 | 7 | 2 |
|
|
| Lymph node |
|
|
|
|
| N0 | 24 | 23 | 1.45 | 0.48 |
| Metastasis |
|
|
|
|
| N1 | 6 | 6 |
|
|
| N2 | 9 | 4 |
|
|
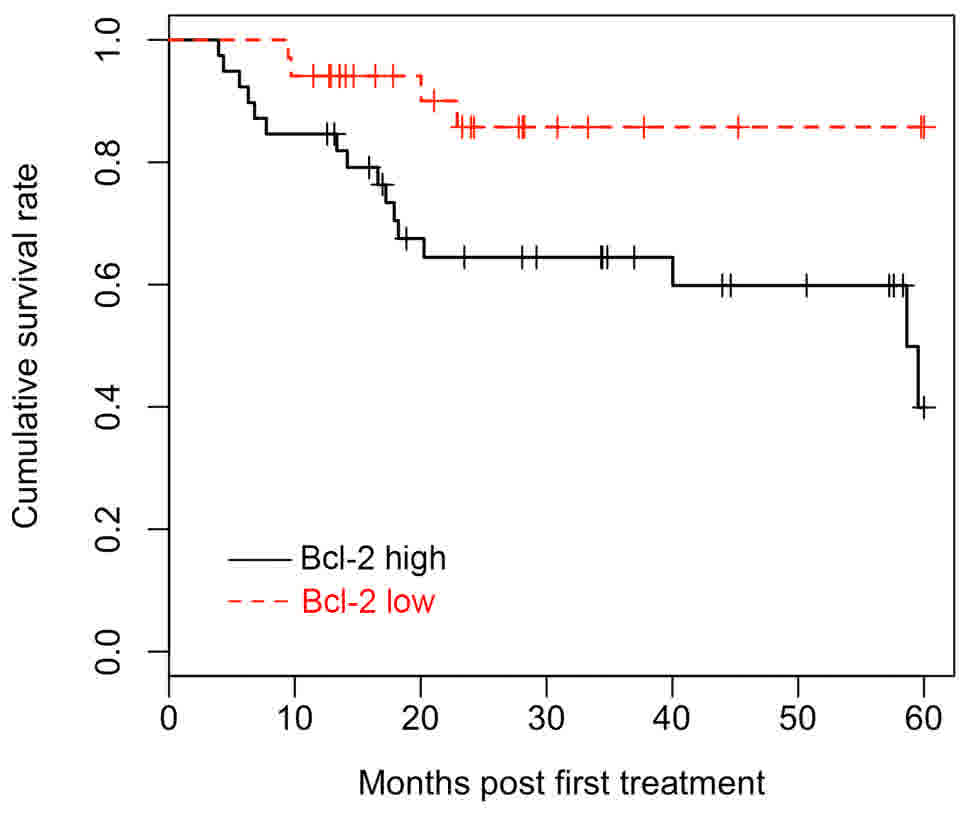
Survival analysis
All 72 patients were included in the survival
analysis and the multivariate Cox proportional hazards model was
applied to determine the effect of Bcl-2 expression on survival.
The Kaplan-Meier survival curves demonstrated that patients with
LUSC with high Bcl-2 expression had a significantly favorable OS
time (Fig. 3). Univariate survival
analyses were employed to identify the difference between patients
with LUSC with different Bcl-2 expression levels. The log-rank test
revealed that Bcl-2 expression levels, clinical stages and tumor
size were significantly associated with OS (P<0.05). In
addition, multivariate analysis indicated that Bcl-2 protein
expression is an independent prognostic factor for patients with
LUSC (HR, 0.295; CI, 0.097–0.904; P<0.05). Detailed data are
listed in Table IV.
 | Table IV.Cox proportional hazard regression
model analysis. |
Table IV.
Cox proportional hazard regression
model analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | 95% CI | Log-rank | P>|z| | 95% CI | HR | P>|z| |
|---|
| Bcl-2
expression | 0.098–0.919 | 19.790 | 0.001 | 0.097–0.904 | 0.295 | 0.033 |
| Age | 0.576–6.713 | 1.210 | 0.281 |
|
|
|
| Gender | 0.555–5.016 | 0.849 | 0.357 |
|
|
|
| Tumor size | 0.561–9.964 | 11.632 | 0.009 | 0.719–2.217 | 1.276 | 0.416 |
| Stage | 0.822–9.827 | 14.123 | 0.010 | 0.801–2.624 | 1.142 | 0.219 |
Discussion
The majority of patients with LUSC are diagnosed at
advanced stages because there are no clinical symptoms or effective
biomarkers (30). Additionally,
numerous patients with lung cancer are diagnosed at advanced stage;
therefore, it is essential to seek highly sensitive and specific
molecular markers of lung cancer for early diagnosis (31).
Bcl-2 as a prognostic marker of lung cancer,
including small cell lung cancer, has been reported in numerous
studies, but the results remain conflicted and controversial
(17,21). Furthermore, due to small sample sizes,
detection methods inconsistencies and other limitations, it is
difficult to compare the results (25). A meta-analysis published by Zhang
et al (21) summarizes 50
articles that investigated the prognostic value of Bcl-2 in NSCLC,
which, to the best of our knowledge, is currently the most
comprehensive study. However, these datasets were divided into
seven subtypes according to clinical or pathological stages and
none of these studies were specific to LUSC.
To get a comprehensive analysis of Bcl-2 expression
and prognosis, a robust survival meta-z approach was used to
integrate multiple LUSC datasets from known databases (30). This method provided a larger sample
size, reducing the potential for errors from single datasets.
Additionally, the results were validated using IHC on a 72-sample
TMA. The results suggested that Bcl-2 was significantly associated
with the OS of patients with LUSC and may also serve as an
independent prognostic factor as supported by multivariate
analysis. Furthermore, Bcl-2 was associated with tumor size and TNM
stage (Table IV), which are similar
to the results observed in breast cancer (32).
Notably, there may be a dual role for Bcl-2 in
cancer (33). Since Bcl-2 has
anti-apoptotic effects based on in vitro and in vivo
experiments, it is expected that high Bcl-2 expression may lead to
worse prognosis, rather than prolonged survival (11). However, high expression of Bcl-2 was
demonstrated to be a favorable prognosis factor in LUSC in the
present study, suggesting that Bcl-2 may be involved in a feedback
loop for cell regulation, and the exact role of Bcl-2 in the
regulation of apoptosis may depend on the cell environment
(34).
In conclusion, the results of the present study
suggest that high Bcl-2 expression in patients with LUSC indicates
favorable prognosis, indicating Bcl-2 could be a potential
prognostic biomarker for LUSC.
Acknowledgements
Not applicable.
Funding
This study is founded by the National High
Technology Research and Development Program of China (863 Program)
(grant no. ss2014AA020602 and ss2014AA020604).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CF, FY, XY, MQ and JunW designed the study; JiaW and
SH performed the bioinformatics analysis. SG and JinW conducted the
TAM staining experiments. CF, XY and MQ wrote and revised the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the People's Hospital of Peking University and
written informed consent was obtained from each patient.
Consent for publication
Written informed consent was obtained from all
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis DR, Check DP, Caporaso NE, Travis WD
and Devesa SS: US lung cancer trends by histologic type. Cancer.
120:2883–2892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang P, Cheng CL, Chang YH, Liu CH, Hsu
YC, Chen JS, Chang GC, Ho BC, Su KY, Chen HY and Yu SL: Molecular
gene signature and prognosis of non-small cell lung cancer.
Oncotarget. 7:51898–51907. 2016.PubMed/NCBI
|
|
6
|
Tan Q, Wang G, Huang J, Ding Z, Luo Q, Mok
T, Tao Q and Lu S: Epigenomic analysis of lung adenocarcinoma
reveals novel DNA methylation patterns associated with smoking.
Onco Targets Ther. 6:1471–1479. 2013.PubMed/NCBI
|
|
7
|
Botling J, Edlund K, Lohr M, Hellwig B,
Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et
al: Biomarker discovery in non-small cell lung cancer: Integrating
gene expression profiling, meta-analysis, and tissue microarray
validation. Clin Cancer Res. 19:194–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilkerson MD, Yin X, Hoadley KA, Liu Y,
Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski
MA, et al: Lung squamous cell carcinoma mRNA expression subtypes
are reproducible, clinically important, and correspond to normal
cell types. Clin Cancer Res. 16:4864–4875. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Raponi M, Zhang Y, Yu J, Chen G, Lee G,
Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y and Beer DG:
Gene expression signatures for predicting prognosis of squamous
cell and adenocarcinomas of the lung. Cancer Res. 66:7466–7472.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lawson MH, Cummings NM, Rassl DM, Vowler
SL, Wickens M, Howat WJ, Brenton JD, Murphy G and Rintoul RC: Bcl-2
and β1-integrin predict survival in a tissue microarray of small
cell lung cancer. Br J Cancer. 103:1710–1715. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dutta C, Day T, Kopp N, van Bodegom D,
Davids MS, Ryan J, Bird L, Kommajosyula N, Weigert O, Yoda A, et
al: BCL-2 suppresses PARP1 function and nonapoptotic cell death.
Cancer Res. 72:4193–4203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Janumyan Y, Cui Q, Yan L, Sansam CG,
Valentin M and Yang E: G0 function of BCL2 and BCL-xL requires BAX,
BAK, and p27 phosphorylation by Mirk, revealing a novel role of BAX
and BAK in quiescence regulation. J Biol Chem. 283:34108–34120.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daniel JC and Smythe WR: The role of Bcl-2
family members in non-small cell lung cancer. Semin Thorac
Cardiovasc Surg. 16:19–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bouchalova K, Kharaishvili G, Bouchal J,
Vrbkova J, Megova M and Hlobilkova A: Triple negative breast
cancer-BCL2 in prognosis and prediction. Review. Curr Drug Targets.
15:1166–1175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Renouf DJ, Wood-Baker R, Ionescu DN, Leung
S, Masoudi H, Gilks CB and Laskin J: BCL-2 expression is prognostic
for improved survival in non-small cell lung cancer. J Thorac
Oncol. 4:486–491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Martinez-Arribas F, Alvarez T, Del Val G,
Martín-Garabato E, Núñez-Villar MJ, Lucas R, Sánchez J, Tejerina A
and Schneider J: Bcl-2 expression in breast cancer: A comparative
study at the mRNA and protein level. Anticancer Res. 27:219–222.
2007.PubMed/NCBI
|
|
17
|
Martin B, Paesmans M, Berghmans T, Branle
F, Ghisdal L, Mascaux C, Meert AP, Steels E, Vallot F, Verdebout
JM, et al: Role of Bcl-2 as a prognostic factor for survival in
lung cancer: A systematic review of the literature with
meta-analysis. Br J Cancer. 89:55–64. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shibata Y, Hidaka S, Tagawa Y and Nagayasu
T: Bcl-2 protein expression correlates with better prognosis in
patients with advanced non-small cell lung cancer. Anticancer Res.
24:1925–1928. 2004.PubMed/NCBI
|
|
19
|
Porebska I, Kosacka M, Wyrodek E and
Jankowska R: Expression of p53, bcl-2 and nm23 proteins in squamous
cell lung cancer. Pneumonol Alergol Pol. 77:131–137. 2009.(In
Polish). PubMed/NCBI
|
|
20
|
Kim YC, Park KO, Kern JA, Park CS, Lim SC,
Jang AS and Yang JB: The interactive effect of Ras, HER2, P53 and
Bcl-2 expression in predicting the survival of non-small cell lung
cancer patients. Lung Cancer. 22:181–190. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Wang S, Wang L, Wang R, Chen S,
Pan B, Sun Y and Chen H: Prognostic value of Bcl-2 expression in
patients with non-small-cell lung cancer: A meta-analysis and
systemic review. Onco Targets Ther. 8:3361–3369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baty F, Facompré M, Kaiser S, Schumacher
M, Pless M, Bubendorf L, Savic S, Marrer E, Budach W, Buess M, et
al: Gene profiling of clinical routine biopsies and prediction of
survival in non-small cell lung cancer. Am J Respir Crit Care Med.
181:181–188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Zhao W and Zhou X: Integration of
genomic data analysis for demonstrating potential targets in the
subgroup populations of squamous cell lung cancer patients.
Oncotarget. Jun 15–2016.DOI: 10.18632/oncotarget.10072.
|
|
26
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pøhl M, Olsen KE, Holst R, Ditzel HJ and
Hansen O: Tissue microarrays in non-small-cell lung cancer:
Reliability of immunohistochemically-determined biomarkers. Clin
Lung Cancer. 15:222–230.e3. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Micke P, Mattsson JS, Djureinovic D, Nodin
B, Jirström K, Tran L, Jönsson P, Planck M, Botling J and
Brunnström H: The impact of the 4th edition of the WHO
classification of lung tumours on histological classification of
resected pulmonary NSCCs. J Thorac Oncol. 11:862–872. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Q, Hou J, Hu Z, Gu B and Shi Y:
Multiple mutations of lung squamous cell carcinoma shared common
mechanisms. Oncotarget. 7:79629–79636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Der SD, Sykes J, Pintilie M, Zhu CQ,
Strumpf D, Liu N, Jurisica I, Shepherd FA and Tsao MS: Validation
of a histology-independent prognostic gene signature for
early-stage, non-small-cell lung cancer including stage IA
patients. J Thorac Oncol. 9:59–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hwang KT, Woo JW, Shin HC, Kim HS, Ahn SK,
Moon HG, Han W, Park IA and Noh DY: Prognostic influence of BCL-2
expression in breast cancer. Int J Cancer. 131:E1109–E1119. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng Y, Wang L, Qing Y, Li C, Ren T, Li Q,
Li M, Zhang S, Shan J, Wang G, et al: Polymorphisms of BCL2 and BAX
genes associate with outcomes in advanced non-small cell lung
cancer patients treated with platinum-based chemotherapy. Sci Rep.
5:177662015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh K and Briggs JM: Functional
implications of the spectrum of BCL2 mutations in lymphoma. Mutat
Res Rev Mutat Res. 769:1–18. 2016. View Article : Google Scholar : PubMed/NCBI
|