Introduction
Osteosarcoma (OS) is a common malignant bone cancer
type, with approximately 3,000,000 new cases annually (1). This type of cancer often occurs in
adolescents and children, and individuals over the age of 50 years
are also susceptible to the disease (2). OS can be treated through surgery,
chemotherapy and radiation and it is reported that the newly
developed immunotherapies, such as chimeric antigen
receptor-engineered T cells or immune checkpoint obstruction, can
be employed to treat OS (3).
Combination chemotherapies play an important role in the treatment
of OS (4). Although almost all of the
patients can accept the resection surgery, the OS recurrences
remain high and often metastasize (5), and less than 20% of patients survive
four years after recurrence (6,7).
Therefore, it is imperative to develop a novel therapeutic strategy
to treat osteosarcoma more effectively.
MicroRNAs (miRNAs) are 21–22 nucleotide long
non-coding RNAs and more than 1,000 miRNAs are expressed and
function in human cells (8). miRNAs
have been proven to play important roles in many biological
processes, such as apoptosis, proliferation and differentiation
(9). miRNAs regulate translation or
splicing of their target mRNAs by binding to their 3′-UTRs
(10). miR-26a is one of these
miRNAs. Ma et al reported that the downregulation of miR-26a
increased hepatocellular carcinoma growth and pulmonary metastasis
(11). It was reported that miR-26a
was significantly downregulated in bladder cancer (BC) tissues, and
that the upregulation of miR-26a could suppress BC cell migration
and invasion by modulating procollagen-lysine, 2-oxoglutarate
5-dioxygenase (12). However, no
relative evidence of miR-26a in osteosarcoma cells has been
documented thus far.
The high-mobility group A (HMGA) family is comprised
of three proteins: HMGA1a, HMGA1b and HMGA2 (13). HMGA proteins do not possess
transcriptional ability, but they can change the chromatin
structure to regulate the expression of various genes (14,15). HMGA
family proteins play pivotal roles in various biological processes,
such as proliferation, differentiation, and chromatin structure
(16). The expression levels of HMGA
proteins are very low in normal tissues and cells. By contrast, a
higher expression of HMGA is the hallmark of cancer (17,18). Puca
et al reported that HMGA1 was upregulated in colon tumor
stem cell lines compared with the normal tissues (19). They identified that HMGA1 silencing
could contribute to stem cell quiescence by increasing p53 levels
(19). The overexpression of HMGA1
could increase cell proliferation and the stemness-related genes of
the A2780 ovarian cancer stem line (20). Recent research has shown that HMGA1
acted as a target gene of miR-142 in regulating OS cell growth
(21). However, whether HMGA1 was a
target of miR-26a in regulating OS cell migration and invasion
remains to be determined.
In the present study, we aimed to detect the role of
miR-26a and HMGA1 in OS. We also explored the mechanism of miR-26a
in regulating OS progression. The results showed that miRNA-26a was
frequently downregulated in OS tissues and cells and the
re-expression of miR-26a could suppress cell migration and invasion
in vitro. Moreover, we identified that the expression of
HMGA1 was decreased by miR-26a through targeting its 3′-UTR and
knockdown of HMGA1 inhibited the migration and invasion of OS
cells. Taken together, our study provides a new potential
therapeutic target for OS treatment.
Materials and methods
Tissues and cell lines
Fifty-two pairs of human OS samples were obtained
from patients who underwent surgery at the People's Hospital of
Weifang (Weifang, China) from January, 2010 to December, 2016. All
the patients provided written informed consent to participate in
the study. Tissues obtained were immediately frozen in liquid
nitrogen, stored at −80°C. This study was approved by the Ethics
Committee of People's Hospital of Weifang.
Osteosarcoma cell line U2OS differentiated cell
groups have high migratory capacity, and the normal human
osteoblastic cell line hFOB 1.19 rarely divide, or divide at the
limit of 39.5°C low migratory capacity. The cells were cultured in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Biowest, South America Origin), 100 U/ml penicillin
and 100 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA;
Merck KGaA, Munich, Germany, respectively) in a humidified
incubator with 5% CO2 at 37°C.
Oligonucleotide transfection
The miR-26a mimic, inhibitor or negative control was
transfected into cells for 48 h using Lipofectamine®2000
according to the manufacturer's protocols. The primer sequences
used were: miR-26a mimic forward, UUCAAGUAAUCCAGGAUAGGCU, and
reverse, CCUAUCCUGGAUUACUUGAAUU. miR-26a inhibitor forward,
ATGTTCTTTAATGTCGGGAGC, and reverse, AA AGAATTCTGCCCGTGAC. To
overexpress HMGA1, vectors containing HMGA1 were transfected into
cells for 48 h using RNAiMAX (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
protocols.
Western blot analysis
The cells were harvested and lysed with RIPA lysis
buffer (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China). Proteins were separated by 10% SDS-PAGE gel and
transformed onto PVDF membranes (Invitrogen; Thermo Fisher
Scientific, Inc.). The membranes were incubated with the primary
antibody HMGA1, (dilution, 1:1,000; cat. no. ab168260), β-actin,
(dilution, 1:1,000; cat. no. ab8227) (Abcam, Cambridge, UK)
overnight at 4°C after being blocked with 5–10% skimmed milk at
temperature for 2 h. The membrane was then incubated with secondary
antibody (dilution, 1:2,000; cat. no. sc-516102-CM; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) conjugated to horseradish
peroxidase goat anti-mouse at room temperature for 2 h. Signals
were visualized using the enhanced chemiluminescence kit (EMD
Millipore, Billerica, MA, USA). The intensities of individual bands
were analyzed by densitometry using ImageJ software (National
Institutes of Health, Bethesda, MD, USA) and normalized to the
β-actin level.
Reverse transcriptase-quantitative
PCR
Total RNA was extracted from cells or tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instruction. Complementary DNA was
synthesized using PrimeScript RT reagent kit (Takara, Dalian,
China) and RT-qPCR was performed using SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd., Dalian, China) with the Stratagene Mx3000P
real-time PCR system (Agilent Technologies, Inc., Santa Clara, CA,
USA). The primer sequences used were: miR-26a forward,
TTGGATCCGTCAGAAATTCTCTCCC GAGG, and reverse,
GGTCTAGATGTGAACTCTGGTGTT GGTGC. HMGA1 forward, GGCCCAAATCGACCATAAA
GG, and reverse, GGACAAATCATGGCTACCCCT. U6 forward,
GCTTCGGCAGCACATATACTAAAAT, and reverse, CGCTTCACGAATTTGCGTGTCAT.
β-actin forward, CGTGACATTAAGGAGAAGCTG, and reverse, CT
GAAGCATTTGCGGTGGAC. The thermocycler conditions were: 95°C for 5
min, followed by 40 cycles of denaturation at 95°C for 15 sec and
an annealing/elongation step at 60°C for 30 sec. U6 and β-actin
were used as internal controls to standardize miRNA and mRNA,
respectively. The 2−ΔΔCq method was used to detect the
relative expression of miR-26a and HMGA1.
Luciferase reporter assay
HMGA1-3′UTR was amplified and cloned into pmir-GLO
vector (Promega Corporation, Madison, WI, USA). Mutation of
HMGA1-3′UTR was generated using QuikChange Site-Directed
Mutagenesis kit (Stratagene; Agilent Technologies, Inc.). For
luciferase reporter assay, pmir-GLO-HMGA1-3′UTR-WT or mutant
plasmid plus miR-26a mimic or miR-control were transfected into
U2OS cells. After 48 h of transfection, cells were collected and
relative luciferase activity was measured by the dual-luciferase
reporter assay system (Promega Corporation). The firefly luciferase
activity was normalized to Renilla luciferase activities.
Migration and invasion assay in
vitro
Cell migration and invasion assays were performed in
a 24-well Transwell plate (pore size, 8 µm; Corning Incorporated,
Corning, NY, USA). Cells (1×105) were resuspended in 200
µl of serum-free medium and plated onto the upper chamber. The
lower chamber was filled with 15% fetal bovine serum as a
chemoattractant. For the invasion assay, the upper chamber was
pre-coated with Matrigel (dilution, 1:3; BD Biosciences, San Jose,
CA, USA). After incubation for 24 h, the cells on the upper chamber
were removed, and the migratory and invasive cells on the lower
chamber were fixed using formalin and stained with 0.1% crystal
violet 30 min at room temperature. Finally, the cells were
photographed using a Leica DC 300F positive microscope (Leica
Microsystems Imaging Solutions Ltd., Cambridge, UK) at ×20
magnification.
Statistical analysis
The results are presented as the mean ± standard
deviation (SD). Statistical analysis was conducted using the SPSS
17.0 (SPSS, Inc., Chicago, IL, USA). Each experiment was repeated
at least three times. Student's t-test or post hoc test after
one-way analysis of variance (ANOVA) in SPSS were used to analyze
the differences between the groups. Correlation between mRNA and
miRNA were estimated using the Spearman's correlation method.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miRNA-26a is frequently downregulated
and HMGA1 is upregulated in OS
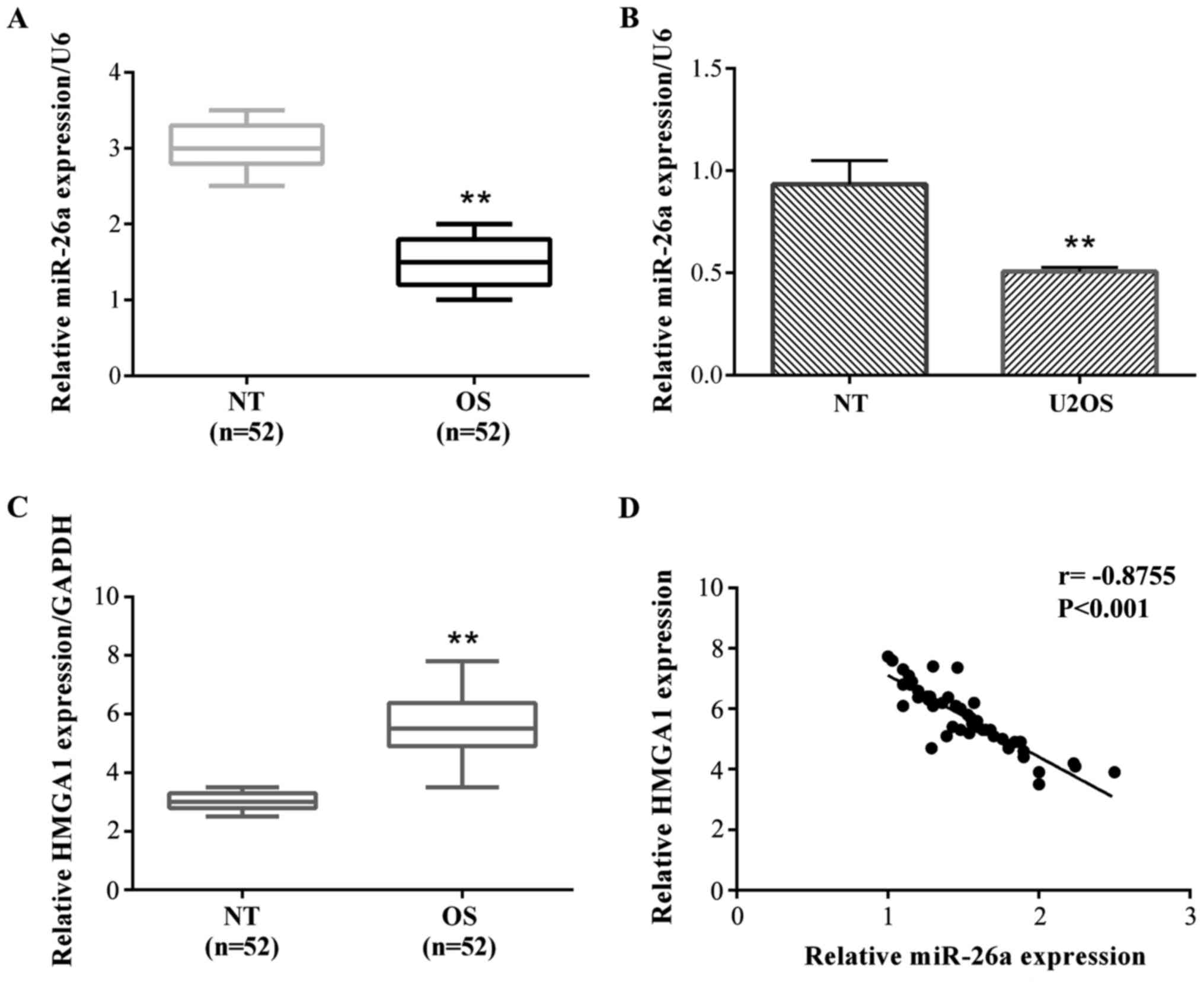
To determine the role of miR-26a in OS, we tested
the expression level of miR-26a in OS and normal tissues in a group
of 52 OS patients by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR). The results showed that a relative
miR-26a expression was frequently decreased in OS tissues compared
with their adjacent non-tumor (NT) tissues (Fig. 1A). Then, we detected the expression of
miR-26a in OS cells. The results showed that miR-26a was also
significantly decreased compared with normal bone cells (hFOB 1.19)
in both OS cell lines (Fig. 1B). We
did not detect the expression of miR-26a in the various tumor
stages.
HMGA1 expression in OS and normal
tissues in a group of 52 OS patients by RT-qPCR
The results stated that the relative HMGA1
expression was frequently increased in OS tissues compared with
their adjacent NT tissues (Fig. 1C).
The relationship between miR-26a and HMGA1 expression was
negatively correlated (Fig. 1D).
Overexpression of miR-26a suppresses
cell migration and invasion in vitro
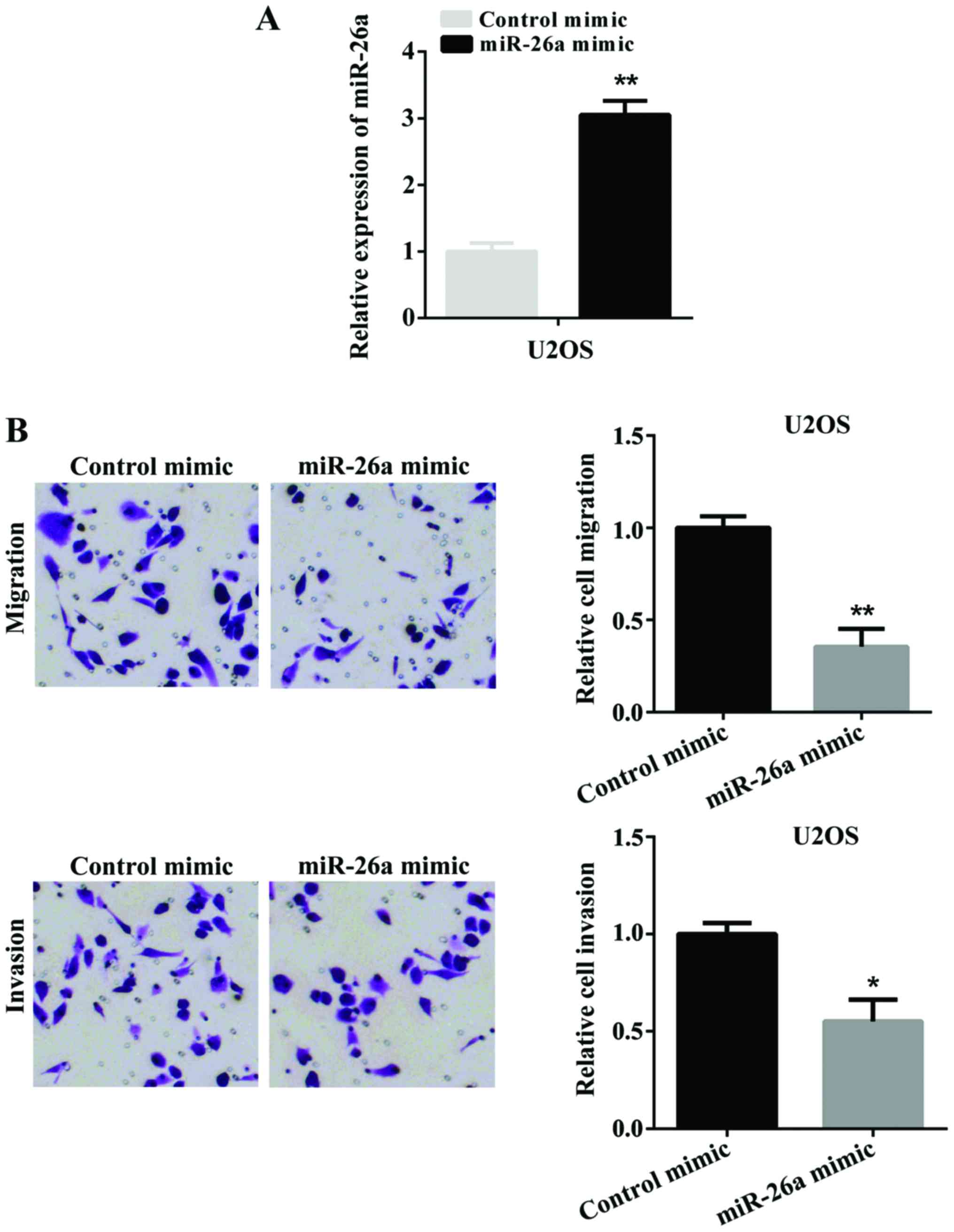
To investigate the potential role of miR-26a in the
migration and invasion of OS, we selected U2OS cells for further
investigation. Firstly, miR-26a mimic was transfected into U2OS
cells to overexpress miR-26a. The successful overexpression of
miR-26a was determined by RT-qPCR (Fig.
2A). Subsequently, we used a Transwell assay to detect the cell
migration and invasion after U2OS cells were transfected with
miR-26a mimic. Fig. 2B shows the
overexpression of miR-26a significantly suppressed cell migration
and invasion. These findings indicated that miR-26a suppressed cell
migration and invasion in vitro.
HMGA1 is identified as the target of
miR-26a
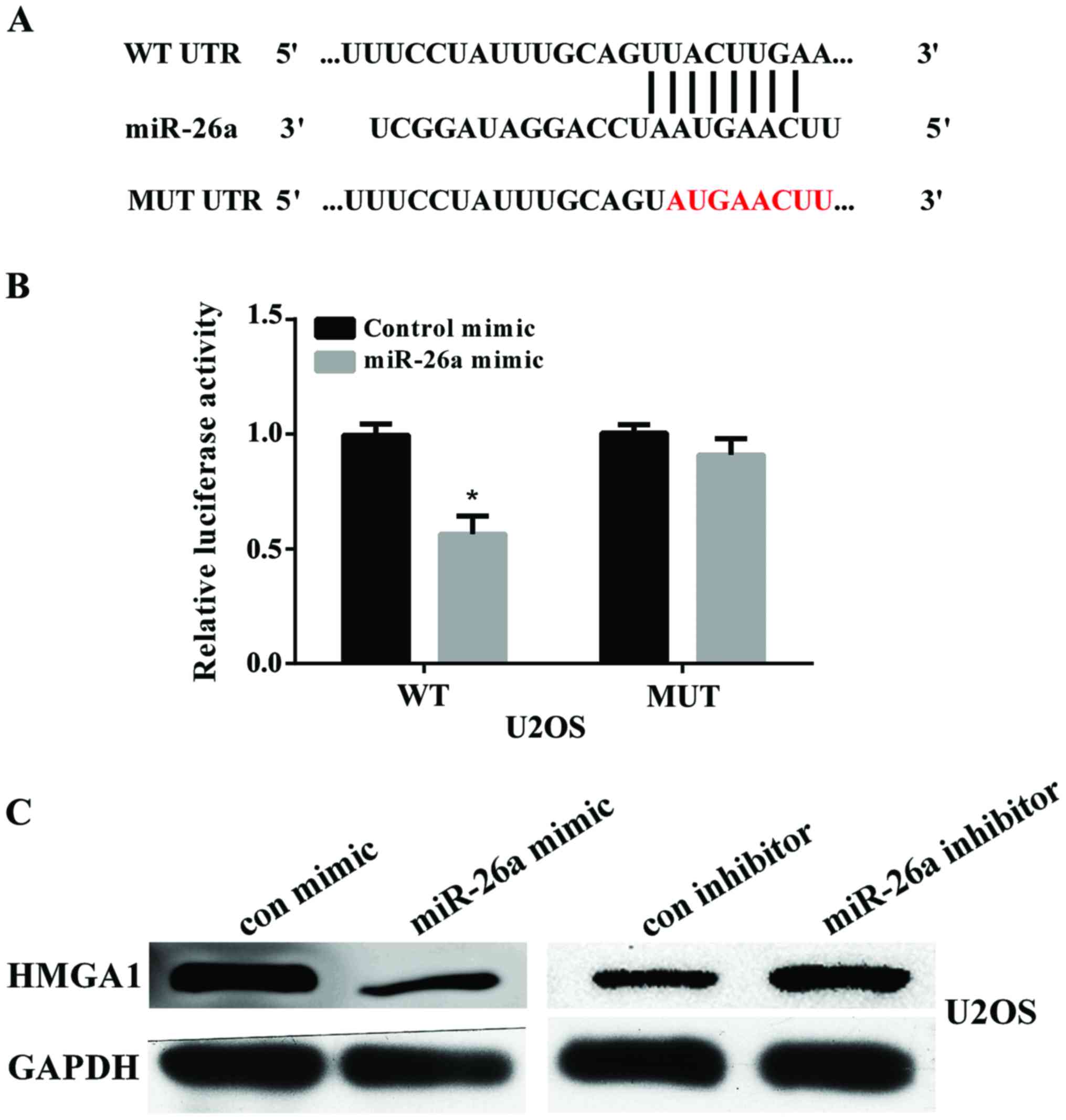
To elucidate the potential mechanism of miR-26a in
suppressing OS migration and invasion, we searched for the
potential target genes of miR-26a using the bioinformatics
algorithm TargetScan (http://www.targetscan.org/vert_71/). HMGA1, containing
a potential binding site at 1317–1324 bp, was selected for
experimental validation (Fig. 3A).
Luciferase reporter assay showed that the luciferase activity was
significantly reduced when U2OS cells were co-transfected with
pmir-GLO-HMGA1-3′UTR (WT) and miR-26a (P<0.01). There was no
effect on cells co-transfected with pmir-GLO-HMGA1-3′UTR (MUT) and
miR-26a (Fig. 3B). We further
examined the protein expression of HMGA1 in U2OS cells after
transfected with miR-26a mimic or miR-26a inhibitor by using
western blot analysis. The results showed that overexpression of
miR-26a reduced HMGA1 expression and knockdown of miR-26a increased
HMGA1 expression (Fig. 3C). These
findings indicated that HMGA1 was a direct target of miR-26a in OS
cells.
Effect of HMGA1 on OS cell migration
and invasion regulated by miR-26a in vitro
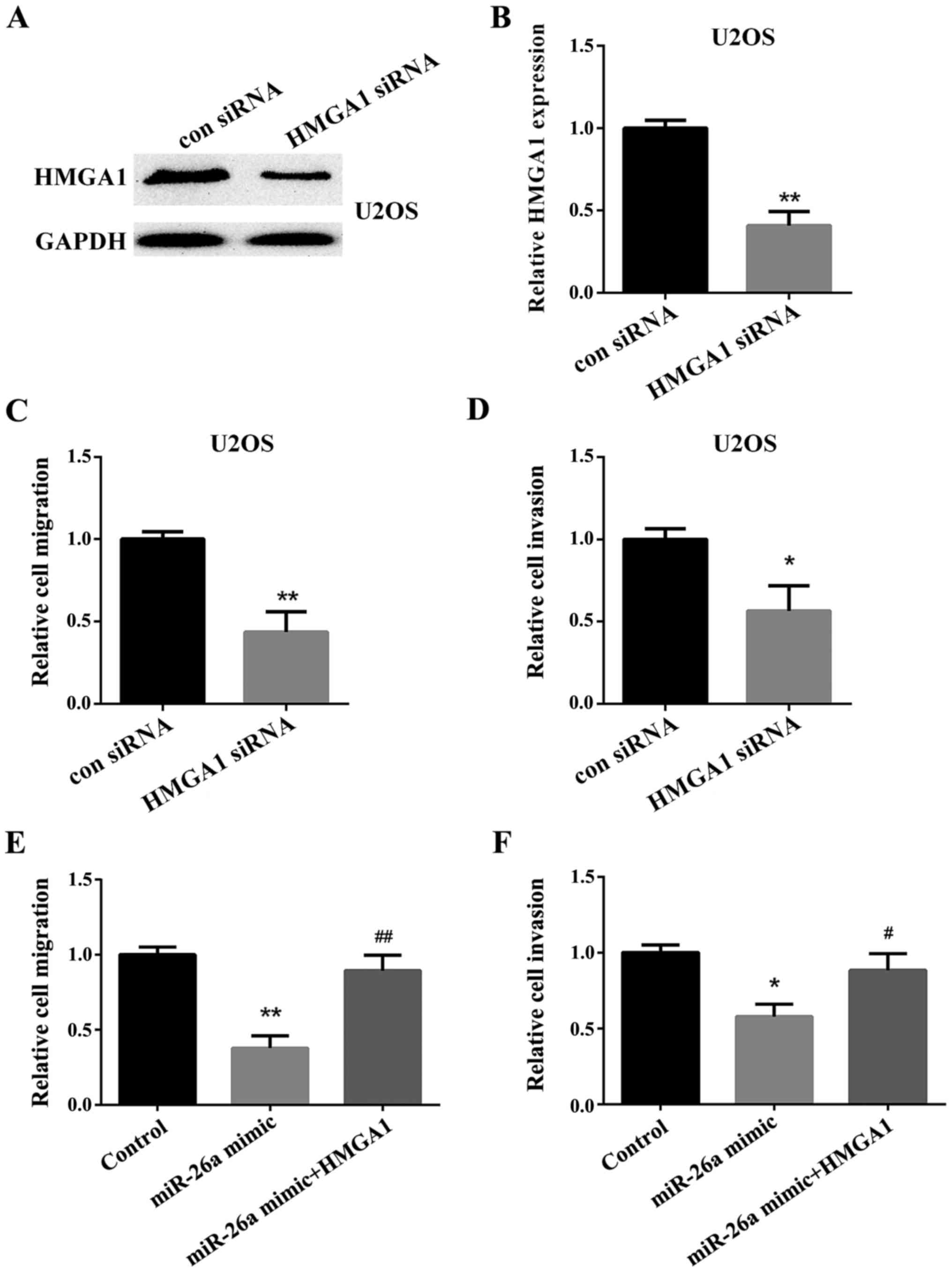
Firstly, the HMGA1 siRNA or negative control was
transfected into the U2OS cells to investigate the effect of HMGA1
on the OS cells. The expression of HMGA1 was detected by western
blot analysis (Fig. 4A) and RT-qPCR
(Fig. 4B). We then used a Transwell
assay to test the relative cell migration and invasion in U2OS
cells. The results showed that the migration and invasion of U2OS
cells transfected with the HMGA1 siRNA was significantly decreased
compared with the cells transfected with the negative control
(Fig. 4C and D). Then, we
investigated the effect of HMGA1 on OS cell migration and invasion
downregulated by miR-26a. We found that HMGA1 could reverse the
inhibitory effect of miR-26a on OS cell migration and invasion
(Fig. 4E and F).
Discussion
OS is the most common pattern of bone tumor
(22). Although efforts have been
made to explore the underlying mechanism of OS tumorigenesis, the
survival rate of OS remains low (23). Therefore, it is imperative to identify
new biomarkers for the development of effective therapeutic methods
for OS.
In the present study, we explored the function of
miR-26a and HMGA1 on human OS. Firstly, we examined the expression
level of miR-26a in OS tissues and cells. The results showed that
the expression level of miR-26a was significantly decreased in OS
tissues and cells compared with the normal ones. Then we
overexpressed miR-26a by transfecting the miR-26a mimic into the
U2OS cells. We found that overexpression of the miR-26a could
suppress the invasion and migration of U2OS cells. Using the
TargetScan online prediction and luciferase reporter assay, we
further identified and confirmed that HMGA1 was the target of
miR-26a, and miR-26a could suppress the expression of HMGA1.
The miR-26 family is comprised of miR-26a and
miR-26b. They play important roles in biological processes, such as
cell cycle and differentiation (24,25). The
miR-26a has been identified downregulated in various human cancers.
Lu et al (26) identified that
the miR-26a inhibits OS tumor growth in vivo and in
vitro by regulating Jagged1, which is consistent with our
study. The expression of miR-26a was decreased in esophageal
squamous cell carcinoma (ESCC) tumor tissues compared with the
adjacent normal tissues and the miR-26a could suppress the
proliferation and metastasis of ESCC (27). In addition, miR-26a also plays a role
in the metabolism of lipids and glucose, modulating insulin
sensitivity in type 2 diabetes (28).
The miR-26a also plays a crucial role in pancreatic cancer
(29), cholangiocarcinoma (30), breast cancer (31) and non-small lung cancer (32). Yu et al (33) reported that miR-26a was significantly
upregulated in pituitary cancer tissues. There may exist other
potential downstream targets of miR-26a in human OS.
HMGA1 can regulate various genes due to its ability
to alter the chromatin structure. It is reported that the CCNE2 was
a downstream target of HMGA1. The HMGA1 and CCNE2 regulate the cell
migration through the Hippo core kinases in breast cancer (34). The HMGA1 activated cell stemness and
crucial migration-related genes. The HMGA1 gene expression was
related with the poor prognosis in breast cancer (35). HMGA1 overexpression is a hallmark of
human cancer and exhibits a pivotal role in cell transformation
(36). Our research is also
consistent with that report. In the present study, we identified
that the knockdown of HMGA1 significantly suppressed the migration
and invasion of U2OS cells. In medulloblastoma (MB), cdc25A is a
target of HMGA1 and HMGA1 interacts with cdc25A promoter. The
knockdown of HMGA1 suppresses the MB cell invasion, migration and
growth (37). The mutation of HMGA1
gene can influence its functions.
There are some drawbacks to the study. The number of
patient samples is relatively small. Our research lacks clinical
experiments. Thus, further study is required to address these
aspects.
In conclusion, we identified that miRNA-26a was
frequently downregulated in OS tissues and cells. The
overexpression of miR-26a suppresses cell migration and invasion
in vitro. Moreover, the HMGA1 is a target of miR-26a and the
knockdown of HMGA1 inhibits the migration and invasion of U2OS
cells. The miR-26a/HMGA1 axis provides new insight into the
pathogenesis of OS and a biomarker for the treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and CS contributed to the conception of the
study; BM and YW performed the data analyses; XM sorted out the
experimental data; YL performed the data analyses and wrote the
manuscript; PY helped perform the analysis with constructive
discussions. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
People's Hospital of Weifang (Weifang, China). All the patients
provided written informed consent to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jo VY and Fletcher CD: WHO classification
of soft tissue tumours: An update based on the 2013 (4th) edition.
Pathology. 46:95–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrari S and Serra M: An update on
chemotherapy for osteosarcoma. Expert Opin Pharmacother.
16:2727–2736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gorlick R, Janeway K, Lessnick S, Randall
RL, Marina N and Committee COGBT: COG Bone Tumor Committee:
Children's oncology group's 2013 blueprint for research: Bone
tumors. Pediatr Blood Cancer. 60:1009–1015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.PubMed/NCBI
|
|
8
|
Krichevsky AM and Gabriely G: miR-21: A
small multi-faceted RNA. J Cell Mol Med. 13:39–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma DN, Chai ZT, Zhu XD, Zhang N, Zhan DH,
Ye BG, Wang CH, Qin CD, Zhao YM, Zhu WP, et al: MicroRNA-26a
suppresses epithelial-mesenchymal transition in human
hepatocellular carcinoma by repressing enhancer of zeste homolog 2.
J Hematol Oncol. 9:12016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyamoto K, Seki N, Matsushita R, Yonemori
M, Yoshino H, Nakagawa M and Enokida H: Tumour-suppressive
miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by
regulating PLOD2 in bladder cancer. Br J Cancer. 115:354–363. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson KR, Lehn DA and Reeves R:
Alternative processing of mRNAs encoding mammalian chromosomal
high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol.
9:2114–2123. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grosschedl R, Giese K and Pagel J: HMG
domain proteins: Architectural elements in the assembly of
nucleoprotein structures. Trends Genet. 10:94–100. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thanos D and Maniatis T: The high mobility
group protein HMG I (Y) is required for NF-kappa B-dependent virus
induction of the human IFN-beta gene. Cell. 71:777–789. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shah SN and Resar LM: High mobility group
A1 and cancer: Potential biomarker and therapeutic target. Histol
Histopathol. 27:567–579. 2012.PubMed/NCBI
|
|
17
|
Chiappetta G, Avantaggiato V, Visconti R,
Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti
V, Santoro M, et al: High level expression of the HMGI (Y) gene
during embryonic development. Oncogene. 13:2439–2446.
1996.PubMed/NCBI
|
|
18
|
Pierantoni GM, Agosti V, Fedele M, Bond H,
Caliendo I, Chiappetta G, Lo Coco F, Pane F, Turco MC, Morrone G,
et al: High-mobility group A1 proteins are overexpressed in human
leukaemias. Biochem J. 372:145–150. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Puca F, Colamaio M, Federico A, Gemei M,
Tosti N, Bastos AU, Del Vecchio L, Pece S, Battista S and Fusco A:
HMGA1 silencing restores normal stem cell characteristics in colon
cancer stem cells by increasing p53 levels. Oncotarget.
5:3234–3245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim DK, Seo EJ, Choi EJ, Lee SI, Kwon YW,
Jang IH, Kim SC, Kim KH, Suh DS, Seong-Jang K, et al: Crucial role
of HMGA1 in the self-renewal and drug resistance of ovarian cancer
stem cells. Exp Mol Med. 48:e2552016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu G, Wang J, Jia Y, Shen F, Han W and
Kang Y: MiR-142-3p functions as a potential tumor suppressor in
human osteosarcoma by targeting HMGA1. Cell Physiol Biochem.
33:1329–1339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wan J and Zhang X, Liu T and Zhang X:
Strategies and developments of immunotherapies in osteosarcoma.
Oncol Lett. 11:511–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Velletri T, Xie N, Wang Y, Huang Y, Yang
Q, Chen X, Chen Q, Shou P, Gan Y, Cao G, et al: P53 functional
abnormality in mesenchymal stem cells promotes osteosarcoma
development. Cell Death Dis. 7:e20152016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kota J, Chivukula RR, O'Donnell KA,
Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P,
Torbenson M, Clark KR, et al: Therapeutic microRNA delivery
suppresses tumorigenesis in a murine liver cancer model. Cell.
137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu J, Song G, Tang Q, Yin J, Zou C, Zhao
Z, Xie X, Xu H, Huang G, Wang J, et al: MiR-26a inhibits stem
cell-like phenotype and tumor growth of osteosarcoma by targeting
Jagged1. Oncogene. 36:231–241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D,
Wang L, Wang YJ, Zong Y, Wu YD, et al: MiR-26a and miR-144 inhibit
proliferation and metastasis of esophageal squamous cell cancer by
inhibiting cyclooxygenase-2. Oncotarget. 7:15173–15186. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu X, Dong B, Tian Y, Lefebvre P, Meng Z,
Wang X, Pattou F, Han W, Wang X, Lou F, et al: MicroRNA-26a
regulates insulin sensitivity and metabolism of glucose and lipids.
J Clin Invest. 125:2497–2509. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X,
Tsark WM, Riggs AD, Ku HT and Huang W: MicroRNA-26a targets ten
eleven translocation enzymes and is regulated during pancreatic
cell differentiation. Proc Natl Acad Sci USA. 110:17892–17897.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang P and Lv L: miR-26a induced the
suppression of tumor growth of cholangiocarcinoma via KRT19
approach. Oncotarget. 7:81367–81376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma X, Dong W, Su Z, Zhao L, Miao Y, Li N,
Zhou H and Jia L: Functional roles of sialylation in breast cancer
progression through miR-26a/26b targeting ST8SIA4. Cell Death Dis.
7:e25612016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu S, Wang T, Yang Z, Li Y, Li W, Wang T,
Wang S, Jia L, Zhang S and Li S: miR-26a desensitizes non-small
cell lung cancer cells to tyrosine kinase inhibitors by targeting
PTPN13. Oncotarget. 7:45687–45701. 2016.PubMed/NCBI
|
|
33
|
Yu C, Li J, Sun F, Cui J, Fang H and Sui
G: Expression and clinical significance of miR-26a and pleomorphic
adenoma gene 1 (PLAG1) in invasive pituitary adenoma. Med Sci
Monit. 22:5101–5108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pegoraro S, Ros G, Ciani Y, Sgarra R,
Piazza S and Manfioletti G: A novel HMGA1-CCNE2-YAP axis regulates
breast cancer aggressiveness. Oncotarget. 6:19087–19101. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pegoraro S, Ros G, Piazza S, Sommaggio R,
Ciani Y, Rosato A, Sgarra R, Del Sal G and Manfioletti G: HMGA1
promotes metastatic processes in basal-like breast cancer
regulating EMT and stemness. Oncotarget. 4:1293–1308. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Esposito F, De Martino M, Petti MG,
Forzati F, Tornincasa M, Federico A, Arra C, Pierantoni GM and
Fusco A: HMGA1 pseudogenes as candidate proto-oncogenic competitive
endogenous RNAs. Oncotarget. 5:8341–8354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lau KM, Chan QK, Pang JC, Ma FM, Li KK,
Yeung WW, Cheng AS, Feng H, Chung NY, Li HM, et al: Overexpression
of HMGA1 deregulates tumor growth via cdc25A and alters
migration/invasion through a cdc25A-independent pathway in
medulloblastoma. Acta Neuropathol. 123:553–571. 2012. View Article : Google Scholar : PubMed/NCBI
|