Introduction
Pancreatic cancer is one of the most aggressive and
lethal types of cancer (1). The most
common type of pancreatic cancer is adenocarcinoma (accounting for
95%), which originates from the exocrine part of the pancreas and
is classified as pancreatic ductal adenocarcinoma (PDAC). The
prognosis of pancreatic cancer remains poor with a 5-year survival
rate of only 4%, ranking the fourth leading cause of all
cancer-associated mortalities in the USA (2). Chemotherapy remains an effective option,
particularly for advanced pancreatic cancer (3). Unfortunately, pancreatic cancer has been
considered as a relatively chemotherapy-refractory tumor. It may be
difficult for patients with advanced pancreatic cancer to benefit
from chemotherapy. Therefore, there is an immediate requirement to
discover novel therapeutic targets for pancreatic cancer.
DNA double-strand breaks (DSB) trigger genome
rearrangements, and DNA damage response (DDR) is a transduction
cascade that coordinates the signaling and repair of these genomic
lesions (4). The ataxia
telangiectasia and Rad3-related (ATR) protein kinase is a member of
the phosphoinositide 3-kinase-related kinase (PIKK) family
(5). It serves an important role in
DDR and activates checkpoint kinase 1 (CHK1) following replication
fork stalling, leading to cell cycle arrest (6). The overexpression of a dead mutant
version of ATR causes sensitivity to DNA-damaging agents and
defects in cell cycle checkpoints (7). This response is an important mechanism
that facilitates cancer cells in surviving anticancer treatments.
Therefore, ATR may be a potential therapeutic target for the
treatment of pancreatic cancer.
FBXO32 (also known as MAFbx or Atrogin-1) acts as a
ubiquitin E3 ligase and has been demonstrated to target several
proteins for proteasomal degradation (8,9). The
present study demonstrated that FBXO32 acted as an E3 ligase of ATR
in pancreatic cancer cells. These results allowed understanding of
the regulatory mechanism of ATR in human pancreatic cancer and
development of novel therapeutic strategies to treat pancreatic
cancer.
Materials and methods
Plasmids and reagents
Myc-tagged FBXO32 (Myc-FBXO32) was cloned into the
pCMV vector. To construct the Myc-FBXO32 plasmid, the full length
FBXO32 gene was amplified using 293T cells via PCR amplification
and cloned into the pCMV-Myc vector (Takara Bio, Inc., Otsu,
Japan). Myc-FBXO32SR (shRNA-resistant FBXO32 plasmid) was generated
using a KOD-Plus Mutagenesis kit (Toyobo Life Science, Osaka,
Japan). Pancreatic cancer cells were transfected with varying
amounts of Myc-FBXO32 plasmids (3 µg/1×106 cells; 1 µg/1×106 cells
or 3 µg/1×106 cells with transfected plasmids, using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. All
transfections occurred once with each different dose. A total of 24
h post-transfection, cells were collected for subsequent analyses.
Anti-FBXO32 (dilution, 1:500) was purchased from ProteinTech Group,
Inc. (Chicago, IL, USA); anti-ATR (dilution, 1:500), anti-PARP
(dilution, 1:1,000) antibodies were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA); and anti-β-Tubulin (dilution,
1:5,000) was from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Gemcitabine was obtained from Eli Lilly and Company (Indianapolis,
IN, USA) and dissolved in distilled water. Pancreatic cancer cells
were treated with 10 µM gemcitabine and the control group was
treated with equal amounts of distilled water for 24 h. MG132 was
purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany), and
pancreatic cancer cells were treated with 20 µM MG132 for 8 h as
previously described (10).
Cell culture
Pancreatic cancer PANC-1 and MIA PaCa-2 cell lines
were purchased from the American Type Culture Collection (ATCC,
Manassas, VA, USA) and were cultured in 5% CO2 at 37°C in a 95%
humidity incubator. PANC-1 and MIA PaCa-2 cells were cultured in
Dulbecco's modified Eagle's medium (Thermo Fisher Scientific,
Inc.), supplemented with 10% fetal bovine serum (Thermo Fisher
Scientific, Inc.) and 100 U/ml penicillin and 100 µg/ml
streptomycin (Thermo Fisher Scientific, Inc.).
Western blot analysis
Pancreatic cancer PANC-1 and MIA PaCa-2 cells were
lysed by lysis buffer [1% Nonidet P-40, 1× phosphate-buffered
saline (PBS), 0.1% sodium dodecyl sulfate and 1% protease inhibitor
cocktail], followed by protein quantification using the
bicinchoninic acid assay (BCA) method. Samples were diluted in
loading buffer containing DTT and were boiled for 5 min. Equal
amount of protein (100 µg) for each sample was separated by 10%
SDS-PAGE, prior to being transferred onto nitrocellulose membranes.
The membranes were blocked in 5% milk at room temperature for 1 h.
Then, the membranes were immuno-blotted with anti-FBXO32 (cat. no.
PA5-43915; 1:1,000; Thermo Fisher Scientific), anti-ATR (cat. no.
13934; 1:500) and anti-PARP (cat. no. 9532; 1:1,000; both Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-β-Tubulin (cat.
no. sc-5274; 1:5,000) and anti-p53 (cat. no. sc-377567; 1:1,000;
both Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibodies at
4°C overnight. Subsequently, the membrane was wash three times with
1X TBST and incubated with rabbit IgG (cat. no. MR-R100; 1:3,000)
and (mouse IgG; cat. no. MR-M100; 1:3,000) horseradish
peroxidase-conjugated secondary antibodies (both Shanghai
MRbiotech, Co., Ltd., Shanghai, China) for 1 h at room temperature,
and then visualized using SuperSignal West Pico Stable Peroxide
solution (Thermo Fisher Scientific, Inc.). Blots were quantified
with ImageJ software (version 1.8.0; National Institutes of Health,
Bethesda, MD, USA).
Reverse transcription-quantitative
polymerase chain reaction
Total RNA was extracted from PANC-1 cells using
TRIzol® reagent (Life Technologies, Thermo Fisher
Scientific, Inc.), as previously described (11). The cDNA was synthesized using
Superscript II reverse transcriptase (Thermo Fisher Scientific).
qPCR was performed using IQ SYBR Green Supermix (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and an iCycleriQTX detection
system (Bio-Rad Laboratories, Inc.). The thermocycling conditions
were as follows: Denaturing at 95°C for 20 sec; annealing at 58°C
for 30 sec; and extension at 72°C for 30 sec, for 43 cycles. All
signals were normalized against β-actin and the 2-∆∆Cq
method was used to quantify the fold change (12). The sequences of the primers used are
as follows: FBXO32 forward, 5′-GAAGCGCTTCCTGGATGAGA-3′ and
reverse, 5′-GGAATCCAGAATGGCAGTTG-3′; ATR forward,
5′-GCCGCTCCGATCGTGTAC-3′ and reverse,
5′-TTTGTATGCTCTGTGATAACCTTGTTT-3′; β-actin forward,
5′-CCCTGGCTCCTAGCACCAT-3′ and reverse,
5′-AGAGCCACCAATCCACACAGA-3′.
RNA interference
Lentivirus-based control and gene-specific shRNAs
were purchased from Sigma-Aldrich; Merck KGaA. Transfections were
performed using Lipofectamine 2000 (Thermo Fisher Scientific,
Inc.), as described previously (13,14). A
total of 2 µg gene-specific shRNA or shNT (control) were
transfected into 293T cells (5×105 cells). After 48 h transfection,
the cultured medium of 293T cells was collected and applied to
pancreatic cancer cells. Pancreatic cancer cells were cultured in
5% CO2, at 37°C for 48 h, followed by puromycin (0.75 µg/ml;
Sigma-Aldrich; Merck KGaA) selection. Cells were collected 72 h
post-transfection. The knockdown efficiency was confirmed through
western blotting using the aforementioned method. The shRNA
sequences were as follows: shNT,
5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3′; shFBXO32#1,
5′-CCGGCCAAGGAAAGAGCAGTATGGACTCGAGTCCATACTGCTCTTTCCTTGGTTTTTTG-3′;
and shFBXO32#2,
5′-CCGGCTGCCATTCTGGATTCCAGAACTCGAGTTCTGGAATCCAGAATGGCAGTTTTTTG-3′.
Caspase-3 activity measurement
The activity of caspase-3 was measured using a
Caspase-3 Colorimetric Protease Assay Sampler kit (cat. no.
KHZ0022; Thermo Fisher Scientific, Inc.). The PANC-1 cells were
transfected with pcDNA 3.0 or Myc-FBXO32 plasmids or transfected
with shNT (control) or FBXO32-specific shRNA according to the
previously described protocol. A total of 24 h post transfection,
cells were counted and 3–5×106 cells were pelleted per sample.
Cells were then treated with gemcitabine (10 µM) for 24 h. Cells
were then lysed using 50 µl lysis buffer according to the protocol
of the manufacturer (Thermo Fisher Scientific, Inc.), followed by
protein quantification using the BCA method. Each cytosol extracted
was diluted to a concentration of 50–200 µg protein per 50 µl cell
lysis buffer (1–4 mg/ml). 2X reaction buffer (50 µl; containing 10
mM DTT) was added to each sample. The 4 mM DEVD-pNA substrate (5
µl) was added to a final concentration of 200 µM and was incubated
at 37°C for 2 h in the dark. Reactions were measured in a
microplate reader at a wavelength of 405 nm.
Cell proliferation assay
Cell proliferation was monitored by an MTS assay
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocols. The PANC-1 cells were transfected with
pcDNA 3.0 or Myc-FBXO32 plasmids or transfected with shNT (control)
or FBXO32-specific shRNA, as previously described. A total of 24 h
post transfection, Cells were plated onto 96-well plates at a
density of 3,000 cells/well. Cells were treated with different
concentrations (0, 1, 10 and 25 µM) of gemcitabine for 24 h prior
to measurement. Then 20 µl CellTiter 96R AQueous One Solution
Reagent (Promega Corporation) was added to each cell. A total of 50
min after incubation (37°C in a cell incubator), cell proliferation
was measured using a microplate reader at a wavelength of 490
nm.
Cell cycle analysis
The PANC-1 cells were transfected with pcDNA 3.0 or
Myc-FBXO32 plasmids or transfected with shNT (control) or
FBXO32-specific shRNA. According to the previously described
protocol. At 24 h post transfection, cells were treated with
gemcitabine (10 µM) and cultured in 5% CO2 at 37°C in a 95%
humidity incubator for another 24 h. Following treatment with
trypsin, cells were harvested and washed with 1×PBS, prior to being
fixed with 70% ethanol at 4°C overnight. The next day, the cells
were washed with 1X PBS and stained with propidium iodide (10 µg/ml
in 1X PBS) at room temperature for 10 min (Sigma-Aldrich; Merck
KGaA). The cell cycle was analyzed by flow cytometry using a
FACSCalibur system (BD Biosciences, Franklin Lakes, NJ, USA). The
cell cycle fraction data were additionally analyzed using Modfit LT
(Verity Software House, Inc., Topsham, ME, USA).
Statistical analysis
One-way analysis, followed by Tukey's multiple
comparisons test, was performed for multiple comparisons. Student's
t-test was performed for single comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
FBXO32 regulates ATR expression in
pancreatic cancer cells
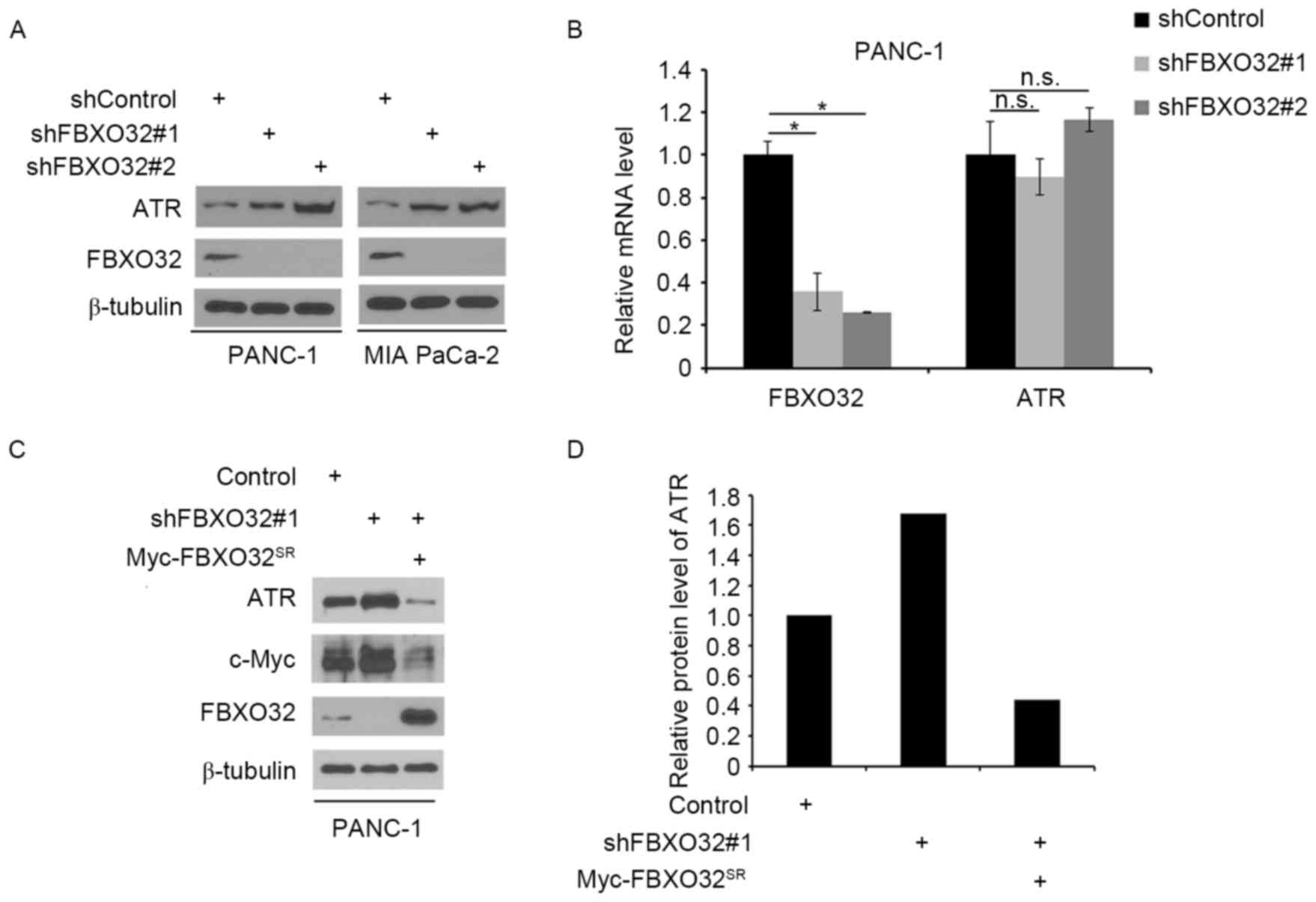
The present study initially examined the association
between FBXO32 and ATR in pancreatic cancer cells. PANC-1 and MIA
PaCa-2 cells were treated with non-specific control or
FBXO32-specific shRNA. Following the effective knockdown of FBXO32,
the protein expression level of ATR was increased in these cells
(Fig. 1A), while knockdown of FBXO32
did not have any effect on ATR mRNA levels in PANC-1 cells
(Fig. 1B). Furthermore, restored
FBXO32 expression through shRNA-resistant FBXO32 plasmid reversed
the effect of FBXO32 knockdown-induced ATR protein level change in
PANC-1 cells. As it has been reported that c-Myc was one of the
FBXO32 targets for degradation (15),
the c-Myc protein levels were examined as a positive control
(Fig. 1C and D). These results
suggested that FBXO32 may regulate the protein level of ATR in
pancreatic cancer cells.
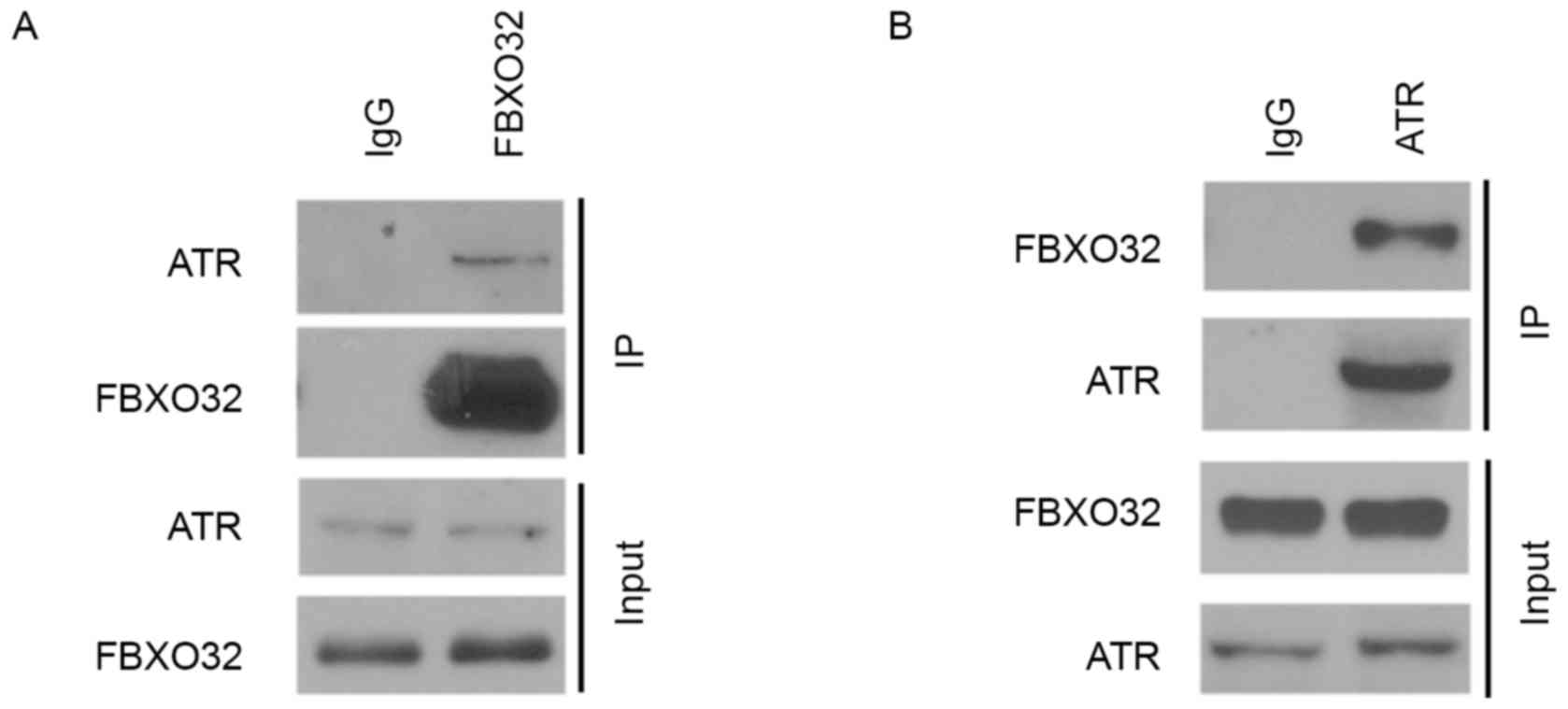
FBXO32 interacts with ATR in
pancreatic cancer cells
It has been reported previously that FBXO32 was an
E3 ubiquitin ligase that regulates the ubiquitination of substrates
(15,16). Since substrate binding is a key event
for E3 ligase-mediated ubiquitination and subsequent proteasome
degradation, the present study examined the interaction between
FBXO32 and ATR using a co-immunoprecipitation (co-IP) assay. The
interaction between endogenous ATR and FBXO32 in PANC-1 cells was
confirmed by reciprocal co-IP assays (Fig. 2A and B). These results suggested that
FBXO32 interacts with ATR in pancreatic cancer cells and these data
promoted the investigation into whether or not FBXO32 functions as
an E3 ubiquitin ligase of ATR in pancreatic cancer cells.
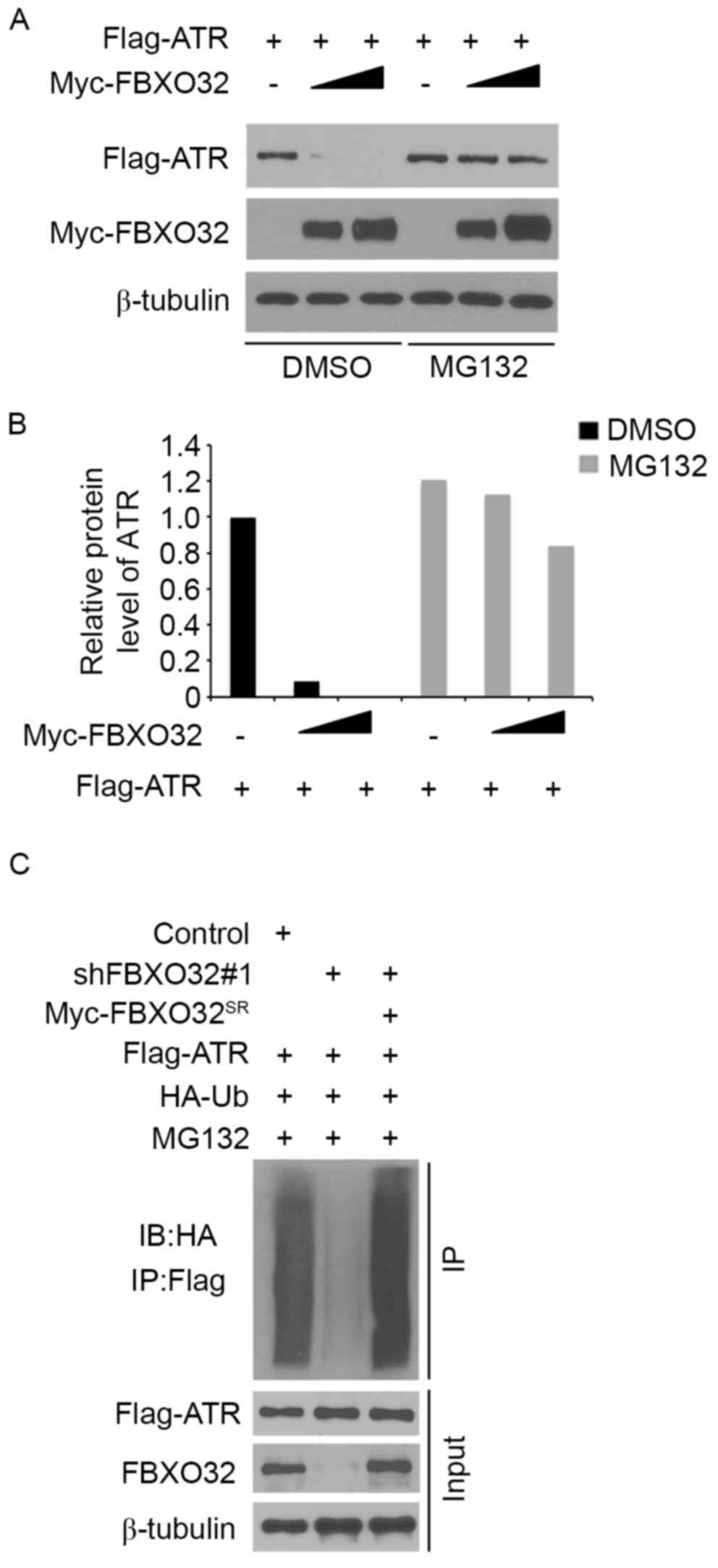
ATR is a degradation substrate of the
E3 ubiquitin ligase FBXO32
The present study systematically investigated
whether FBXO32 degrades ATR protein in pancreatic cancer cells. ATR
was co-transfected with 1 µg/1×106 cells or 3 µg/1×106 cells
transfected with FBXO32 plasmids in pancreatic cancer PANC-1 cells.
Ectopically expressed ATR was downregulated by co-expression of
FBXO32 and this process was blocked by treatment with the
proteasome inhibitor MG132 (Fig. 3A and
B), which indicated that FBXO32 decreased ATR protein
expression levels via the proteasome pathway. In order to determine
whether FBXO32 regulates ATR polyubiquitination, endogenous FBXO32
was knocked down in PANC-1 cells, leading to decreased
polyubiquitination of ATR (Fig. 3C),
while restored FBXO32 expression through the shRNA-resistant FBXO32
plasmid increased the polyubiquitination of ATR. Taken together,
these results suggested that ATR is a degradation substrate of E3
ubiquitin ligase FBXO32 in pancreatic cancer cells.
FBXO32 regulates the DDR induced by
gemcitabine in pancreatic cancer
Gemcitabine is one of the first-line therapeutic
agents for pancreatic cancer (12).
Gemcitabine inhibits ribonucleotide reductase and is an active
chemotherapeutic agent that disrupts DNA replication (17). Gemcitabine activated checkpoint
signaling pathways and acts as a strong activator of ATR (17,18).
Treatment with an ATR inhibitor increased the anticancer activity
of gemcitabine in pancreatic cancer cells (18). In order to investigate the role of
FBXO32 in the regulation of DDR induced by gemcitabine in
pancreatic cancer, pancreatic cancer cells were treated with
gemcitabine alone or in combination with FBXO32 overexpression or
knockdown. It was revealed that overexpression of FBXO32 led to
increased expression of cleaved PARP (a pro-apoptotic protein) and
P53 protein, as well as an increase in the caspase-3 activity
induced by gemcitabine (Fig. 4A and
B) in pancreatic cancer PANC-1 cells. Next, a cell viability
assay was performed in FBXO32-overexpressing PANC-1 cells treated
with different concentrations of gemcitabine. The results
demonstrated that lower concentrations of gemcitabine were required
to suppress the cell proliferation in FBXO32-overexpressing PANC-1
cells, compared with the normal group (Fig. 4C). The present study indicated that
FBXO32 serves an important role in sensitizing pancreatic cancer
cells to gemcitabine treatment. By contrast, the knockdown of
FBXO32 decreased cleaved PARP and p53 expression, decreased
caspase-3 activity and required a higher concentration of
gemcitabine to suppress the viability of PANC-1 cells when treated
with gemcitabine (Fig. 4D-F).
Furthermore, Fig. 4G demonstrates
that treatment of PANC-1 cells with gemcitabine resulted in S-phase
cell cycle arrest and increased ratio of cells in the S phase.
Overexpression of FBXO32 enhanced the effect of gemcitabine while
knockdown of FBXO32 attenuated this effect in PANC-1 cells. Taken
together, these results indicated that FBXO32 regulates the DDR
induced by gemcitabine in pancreatic cancer.
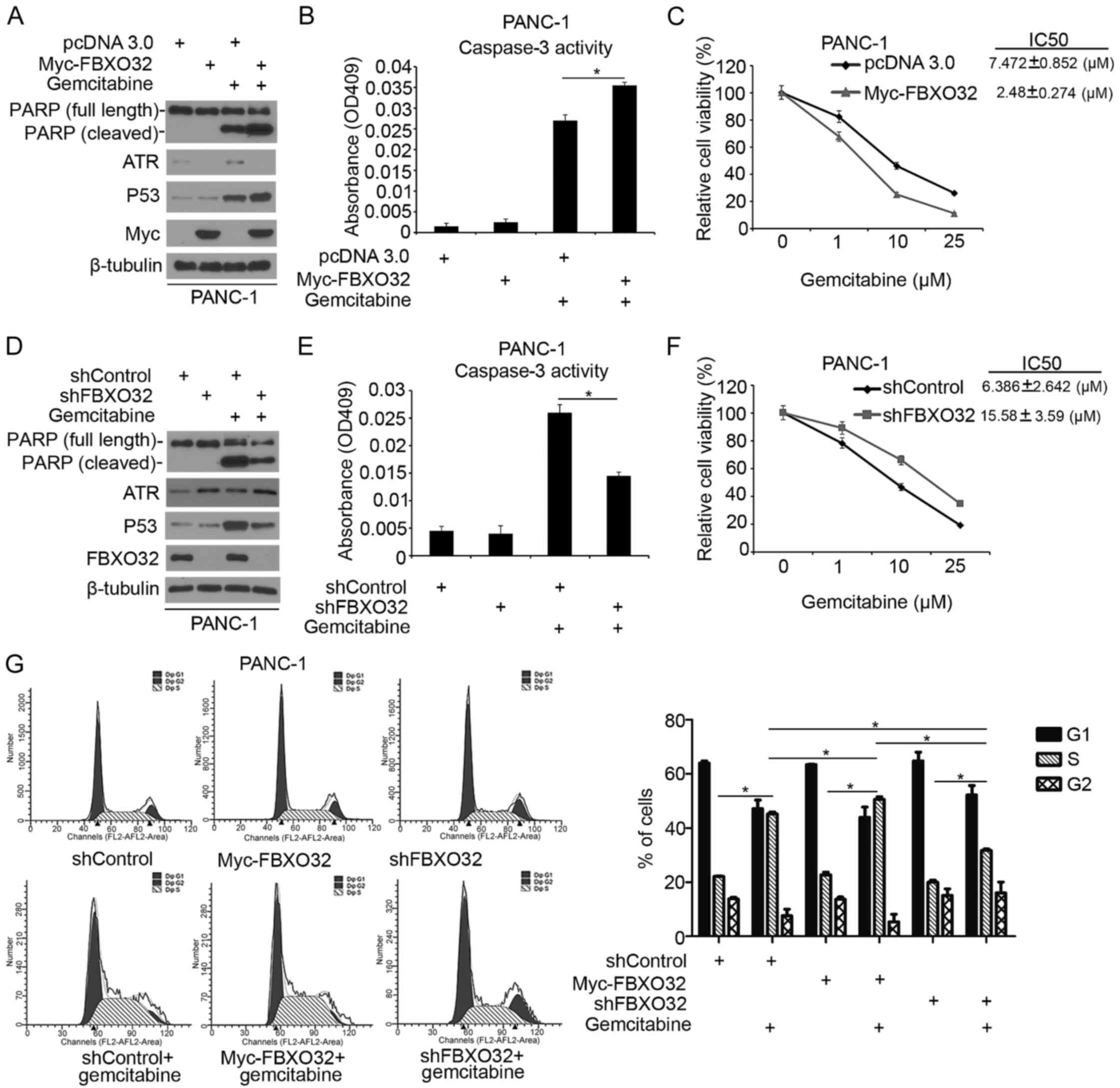 | Figure 4.FBXO32 regulates the DNA damage
response induced by gemcitabine in pancreatic cancer. PANC-1 cells
were transfected with indicated constructs. Cells were treated with
gemcitabine (10 µM) for 24 h prior to harvest. Cells were harvested
for (A) western blot analysis and (B) measurement of caspase-3
activity. Data are presented as the mean ± standard deviation of
two replicate experiments. *P<0.05 for pcDNA 3.0+gemcitabine
treatment vs. Myc-FBXO32+ Gemcitabine treatment. (C) PANC-1 cells
were transfected with indicated constructs. Cells were treated with
different concentrations (0, 1, 10 and 25 µM) of gemcitabine for 24
h prior to measurement. Cell viability was measured using an MTS
assay. Data are presented as the mean ± standard deviation of three
replicate experiments. PANC-1 cells were transfected with control
or one of two independent FBXO32-specific shRNAs. At 24 h after
transfection, cells were treated with gemcitabine (10 µM) for 24 h
prior to harvest. Cells were harvested for (D) western blot
analysis and (E) measurement of caspase-3 activity. Data are
presented as the mean ± standard deviation of two replicate
experiments. *P<0.05 for shControl+gemcitabine treatment vs.
shFBXO32+gemcitabine treatment. (F) PANC-1 cells were transfected
with control or one of two independent FBXO32-specific shRNAs. At
24 h after transfection, cells were treated with different
concentrations (0, 1, 10 and 25 µM) of gemcitabine for 24 h prior
to measurement. Cell viability was measured using an MTS assay.
Data are presented as the mean ± standard deviation of three
replicate experiments. (G) PANC-1 cells were transfected with
indicated constructs. Cells were treated with distilled water or
gemcitabine (20 µM) for 24 h. The cell cycle was analyzed after
staining with PI. The cell cycle distribution was analyzed using
ModFit LT software. Data from a representative experiment (from a
total of two) are presented. Student's t-test was performed for
single comparisons and one-way analysis of variance, followed by
Tukey's multiple comparisons test was performed for multiple
comparisons. *P<0.05 for comparing S phase in groups as follows:
shControl vs. shControl+gemcitabine treatment, Myc-FBXO32 vs.
Myc-FBXO32+gemcitabine treatment, shFBXO32 vs. shFBXO32+gemcitabine
treatment, shControl+gemcitabine treatment vs.
Myc-FBXO32+gemcitabine treatment, shControl+gemcitabine treatment
vs. shFBXO32+gemcitabine treatment, Myc-FBXO32+gemcitabine
treatment+shFBXO32+gemcitabine treatment. |
Discussion
Oncogene activation has been revealed to generate
replicative stress in early stages of tumor progression (19). The principle of using DNA damage to
kill tumor cells has been applied for decades (20). Chemotherapeutic agents and ionizing
radiation induce DNA damage, thereby activating cell cycle
checkpoint pathways (21). Cell cycle
checkpoint pathways are involved in regulating cell cycle
progression and repairing of damaged DNA (22). This response is an important mechanism
that aids cancer cells in surviving anticancer treatment (22). ATR serves an important role in DDR and
activates CHK1 following replication fork stalling, leading to cell
cycle arrest. As a result, ATR-CHK1 pathway components are
considered to be promising therapeutic targets for enhancing the
effectiveness of replication inhibitors (23). Although significant progress has been
made toward understanding the function and deregulation of ATR in
cancer cells, there has been little investigation into the
modification of ATR after transcription. The present study provided
evidence that FBXO32 interacts with ATR and regulates ATR
expression via ubiquitination and degradation in pancreatic cancer
cells.
FBXO32 contains the F-box domain and functions as
one component of a skp1, cullin, F-box protein ubiquitin ligase
complex (24,25). FBXO32 acts as a muscle-specific E3
ligase by targeting multiple substrates for ubiquitination and
degradation (8,9). Furthermore, FBXO32 has been demonstrated
to be involved in tumorigenesis. FBXO32 expression is decreased in
ovarian cancer (26) and esophageal
squamous cell carcinoma (27).
Overexpression of FBXO32 in ovarian cancer cells inhibits colony
formation in culture and xenograft tumor growth in mice (26). Promoter methylation of FBXO32 is
responsible for the downregulation of FBXO32 in cancer cells
(27). EZH2 has been reported to
suppress FBXO32 expression (28). The
present study demonstrated that FBXO32 acted as an E3 ligase of ATR
in pancreatic cancer cells. Furthermore, FBXO32 interacts with p21
to induce p21 protein degradation and regulates DDR (16). Gemcitabine is one of the first-line
therapeutic agents for pancreatic cancer. It activates checkpoint
signaling pathways and acts as a strong activator of ATR (17). The results of the present study
suggested that FBXO32 regulates the DDR induced by gemcitabine in
pancreatic cancer, partially through inducing ATR degradation.
These observations have revealed important aspects of the function
of FBXO32 in tumorigenesis.
Taken together, the results of the present study
demonstrated that FBXO32 acts as an E3 ubiquitin ligase of ATR and
that it regulates the DDR induced by gemcitabine in pancreatic
cancer, possibly through inducing ATR degradation. Therefore, we
hypothesized that FBXO32 may be a potential therapeutic target for
pancreatic cancer.
Acknowledgements
The present study was supported by the Scientific
Research Training Program for Young Talents of Union Hospital,
Tongji Medical College, Huazhong University of Science and
Technology.
Funding
The present study was supported by the Chinese
National Natural Science Foundation (grant no. 81702374).
Availability of data and materials
The datasets generated/analyzed in the present study
are available upon reasonable request from the corresponding
author.
Authors' contributions
HW and XJ conceived the study. XJ, CY and PF
performed the experiments. SZ and HY performed bioinformatics and
statistics analyses. XJ and HW wrote the manuscript.
Ethics statement and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
The Lancet Oncology: Pancreatic cancer in
the spotlight. Lancet Oncol. 15:2412014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blum R and Kloog Y: Metabolism addiction
in pancreatic cancer. Cell Death Dis. 5:e10652014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin X, Pan Y, Wang L, Ma T, Zhang L, Tang
AH, Billadeau DD, Wu H and Huang H: Fructose-1,6-bisphosphatase
inhibits ERK activation and bypasses gemcitabine resistance in
pancreatic cancer by blocking IQGAP1-MAPK interaction. Cancer Res.
77:4328–4341. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Toledo LI, Murga M and Fernandez-Capetillo
O: Targeting ATR and Chk1 kinases for cancer treatment: A new model
for new (and old) drugs. Mol Oncol. 5:368–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cimprich KA, Shin TB, Keith CT and
Schreiber SL: cDNA cloning and gene mapping of a candidate human
cell cycle checkpoint protein. Proc Natl Acad Sci USA.
93:2850–2855. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Q, Guntuku S, Cui XS, Matsuoka S,
Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A,
et al: Chk1 is an essential kinase that is regulated by Atr and
required for the G(2)/M DNA damage checkpoint. Genes Dev.
14:1448–1459. 2000.PubMed/NCBI
|
|
7
|
Cliby WA, Roberts CJ, Cimprich KA,
Stringer CM, Lamb JR, Schreiber SL and Friend SH: Overexpression of
a kinase-inactive ATR protein causes sensitivity to DNA-damaging
agents and defects in cell cycle checkpoints. EMBO J. 17:159–169.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lagirand-Cantaloube J, Offner N, Csibi A,
Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT and
Leibovitch SA: The initiation factor eIF3-f is a major target for
atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J.
27:1266–1276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie P, Guo S, Fan Y, Zhang H, Gu D and Li
H: Atrogin-1/MAFbx enhances simulated ischemia/reperfusion-induced
apoptosis in cardiomyocytes through degradation of MAPK
phosphatase-1 and sustained JNK activation. J Biol Chem.
284:5488–5496. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin X, Pan Y, Wang L, Zhang L,
Ravichandran R, Potts PR, Jiang J, Wu H and Huang H: MAGE-TRIM28
complex promotes the Warburg effect and hepatocellular carcinoma
progression by targeting FBP1 for degradation. Oncogenesis.
6:e3122017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian S, Li P, Sheng S and Jin X:
Upregulation of pyruvate kinase M2 expression by fatty acid
synthase contributes to gemcitabine resistance in pancreatic
cancer. Oncol Lett. 15:2211–2217. 2018.PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin X, Yang C, Fan P, Xiao J, Zhang W,
Zhan S, Liu T, Wang D and Wu H: CDK5/FBW7-dependent ubiquitination
and degradation of EZH2 inhibits pancreatic cancer cell migration
and invasion. J Biol Chem. 292:6269–6280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin X, Tian S and Li P: Histone
acetyltransferase 1 promotes cell proliferation and induces
cisplatin resistance in hepatocellular carcinoma. Oncol Res.
25:939–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mei Z, Zhang D, Hu B, Wang J, Shen X and
Xiao W: FBXO32 targets c-Myc for proteasomal degradation and
inhibits c-Myc activity. J Biol Chem. 290:16202–16214. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee
YJ, Jiang X, Tan J, Aau M, Lim CZ and Yu Q: Polycomb protein EZH2
regulates cancer cell fate decision in response to DNA damage. Cell
Death Differ. 18:1771–1779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karnitz LM, Flatten KS, Wagner JM,
Loegering D, Hackbarth JS, Arlander SJ, Vroman BT, Thomas MB, Baek
YU, Hopkins KM, et al: Gemcitabine-induced activation of checkpoint
signaling pathways that affect tumor cell survival. Mol Pharmacol.
68:1636–1644. 2005.PubMed/NCBI
|
|
18
|
Woods D and Turchi JJ: Chemotherapy
induced DNA damage response: Convergence of drugs and pathways.
Cancer Biol Ther. 14:379–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gorgoulis VG, Vassiliou LV, Karakaidos P,
Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr,
Kastrinakis NG, Levy B, et al: Activation of the DNA damage
checkpoint and genomic instability in human precancerous lesions.
Nature. 434:907–913. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matt S and Hofmann TG: The DNA
damage-induced cell death response: A roadmap to kill cancer cells.
Cell Mol Life Sci. 73:2829–2850. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wagner JM and Kaufmann SH: Prospects for
the use of ATR inhibitors to treat cancer. Pharmaceuticals (Basel).
3:1311–1334. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sclafani RA and Holzen TM: Cell cycle
regulation of DNA replication. Annu Rev Genet. 41:237–280. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tao Y, Leteur C, Yang C, Zhang P, Castedo
M, Pierré A, Golsteyn RM, Bourhis J, Kroemer G and Deutsch E:
Radiosensitization by Chir-124, a selective CHK1 inhibitor: Effects
of p53 and cell cycle checkpoints. Cell Cycle. 8:1196–1205. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bodine SC, Latres E, Baumhueter S, Lai VK,
Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K,
et al: Identification of ubiquitin ligases required for skeletal
muscle atrophy. Science. 294:1704–1708. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gomes MD, Lecker SH, Jagoe RT, Navon A and
Goldberg AL: Atrogin-1, a muscle-specific F-box protein highly
expressed during muscle atrophy. Proc Natl Acad Sci USA.
98:14440–14445. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chou JL, Su HY, Chen LY, Liao YP,
Hartman-Frey C, Lai YH, Yang HW, Deatherage DE, Kuo CT, Huang YW,
et al: Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4
target gene and tumor suppressor, is associated with poor prognosis
in human ovarian cancer. Lab Invest. 90:414–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo W, Zhang M, Shen S, Guo Y, Kuang G,
Yang Z and Dong Z: Aberrant methylation and decreased expression of
the TGF-β/Smad target gene FBXO32 in esophageal squamous cell
carcinoma. Cancer. 120:2412–2423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ciarapica R, De Salvo M, Carcarino E,
Bracaglia G, Adesso L, Leoncini PP, Dall'Agnese A, Walters ZS,
Verginelli F, De Sio L, et al: The Polycomb group (PcG) protein
EZH2 supports the survival of PAX3-FOXO1 alveolar rhabdomyosarcoma
by repressing FBXO32 (Atrogin1/MAFbx). Oncogene. 33:4173–4184.
2014. View Article : Google Scholar : PubMed/NCBI
|