Introduction
Cervical cancer is a common malignant tumor
(1) with an incidence of up to
1.2–4.5 per 10,000 delivery women. The incidence of cervical cancer
has shown an increasing trend in Chinese women (2,3).
Cervical intraepithelial neoplasia (CIN) is a type
of precancerous lesion closely related to cervical cancer. With a
high incidence of up to 6.5% (4), CIN
significantly impacts the development of cervical cancer.
Therefore, accurate diagnosis of CIN and better understanding of
the pathogenesis of this disease will definitely improve the
prevention of cervical cancer (5).
However, most studies mainly focused on the treatment of this
disease, and studies on its pathogenesis are relatively rare
(6,7).
In previous years, with the explosion of gene expression data,
bioinformatics-based data digging for gene expression profile
analysis has become a hot research field (8,9).
In the present study, the bioinformatics method was
applied to analyze gene expression data to identify differentially
expressed genes in CIN tissue. Our study provide references for
further studies on the molecular pathogenesis of CIN.
Materials and methods
Acquisition of gene expression
profiling data
Gene expression profiling data with the series
number GSE64217 was obtained from the the Gene Expression Omnibus
(GEO) database. GSE64217 was provided by the Indian Institute of
Technology Kharagpur, School of Medical Science and Technology,
Multimodal Imaging and Computing for Theranostics (West Bengal,
India). The data included 2 cases of CIN, of cervical squamous cell
carcinoma, and 2 of normal tissues. Biopsy samples were collected
during hysterectomy, and half of each sample was analyzed with
optical microscopy (Olympus, Tokyo, Japan) by a pathologist for
histopathological confirmation and the other half was used for
microarray analysis.
Pretreatment of raw data,
identification of differential genes, and preparation of a heat
map
Statistical analysis on chip data was performed
using BRB-ArrayTools 4.3.2 Beta software. Chip data were first
pre-treated using JustRMA algorithm, and filtered and normalized
using median-based method. Chip data were filtered according to the
following criteria: i) No less than two times of difference of
median of genes should be observed in ≥20% of the samples when
comparing the two types of samples; and ii) missed gene expression
data should be ≤50%. The filtered genes were tested with
independent-samples t-test. Classification and comparison of
dataset were performed with the Class comparison tool to identify
differentially expressed genes between CIN and normal tissue
(P<0.00001). Finally, a heat map was drawn using ‘pheatmap’
package in ‘R’ software, and differentially expressed genes were
highlighted.
Gene Ontology (GO) enrichment
analysis
Differentially expressed genes were subjected to GO
enrichment analysis and functional annotation using Database for
Annotation, Visualization and Integrated Discovery (DAVID) and
‘Bingo’ (plug-in of Cytoscape software).
DAVID analysis: DAVID (Database for Annotation,
Visualization and Integration Discovery) software, which integrates
all the major public bioinformatics resources, can be used to
interpret genes related to biological mechanisms by providing
enrichment analysis with standardized genetic terminologies. The
DAVID database aims to provide rapid accessibility of heterogeneous
annotation data from enriched area and enhanced biological
information levels of individual genes specifically to yield a gene
list by enabling high-throughput gene function analysis. DAVID
database can be downloaded for free (https://david.ncifcrf.gov/).
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis
KEGG pathway analysis and functional annotation for
differential genes were performed using KOBAS 3.0, which is the
first hypergeometric distribution-based examination software to
evaluate the significance of enrichment of pathways, and has been
successfully applied in pathway analysis for plants, animals,
bacteria, and other organisms. KOBAS server can be accessed via
https://kobas.cbi.pku.edu.cn.
Protein-protein interaction (PPI)
network analysis
Differential genes were subjected to PPI network
analysis using STRING software. PPR refers to the protein complex
formed by two or more proteins through covalent bond. STRING can be
accessed free of charge via https://string-db.org/.
Results
Identification of differential genes
and preparation of the heat map
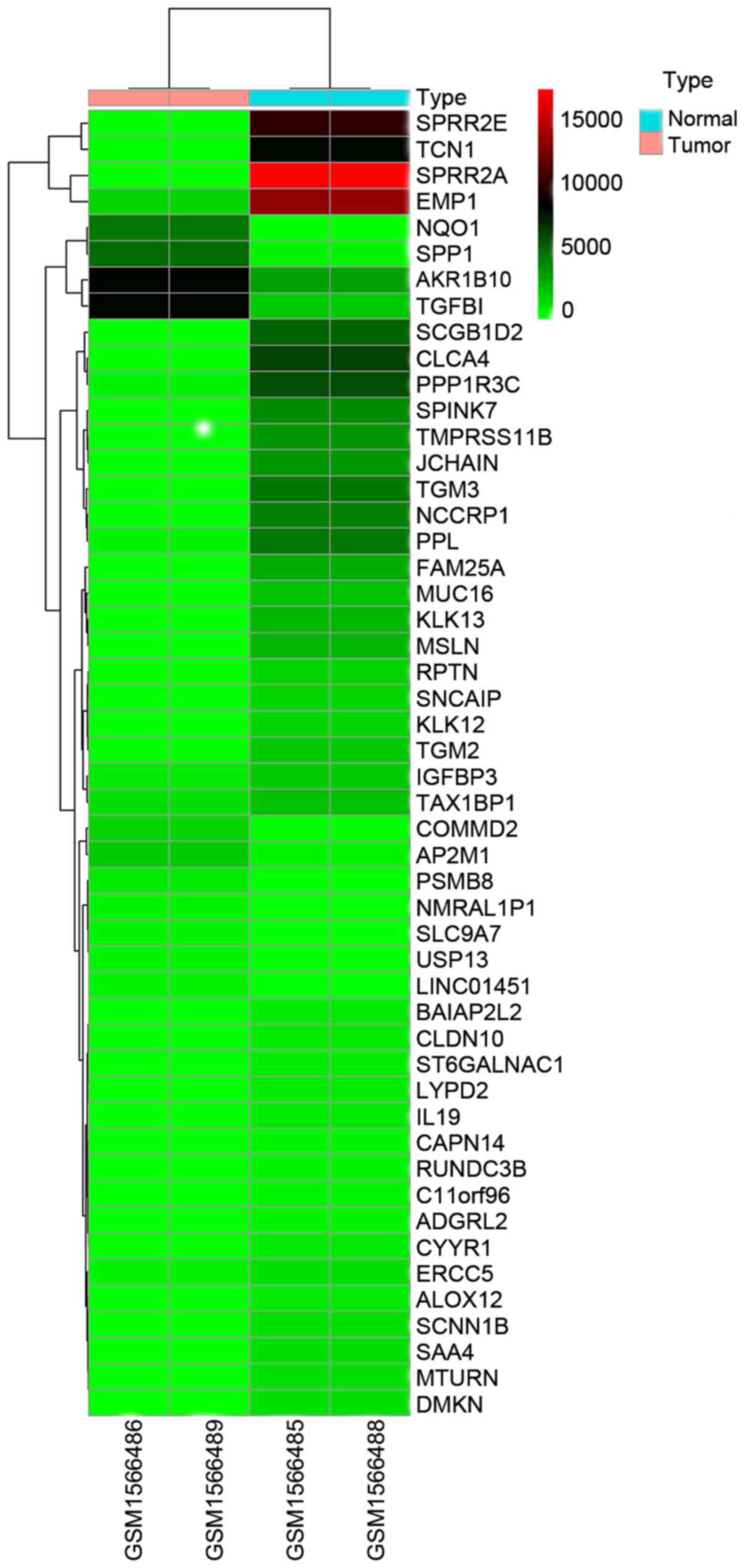
A total of 287 differential genes were obtained
based on GSE64217, and 170 genes were significantly upregulated and
118 genes were significantly downregulated (P<0.00001,
fold-change >6). Representative differential genes are presented
in Table I. Fifty differential genes
with the lowest P-values were analyzed in the heat map (Fig. 1).
 | Table I.Major differential genes. |
Table I.
Major differential genes.
| Gene | logFC | P-value | Adjusted P-value |
|---|
| SPRR2A | 17458.83345 | 6.83E-11 | 1.35E-06 |
| SPRR2E | 10240.24442 | 1.30E-10 | 1.35E-06 |
| TGM3 | 4833.325709 | 1.26E-09 | 8.70E-06 |
| SCGB1D2 | 5415.64156 | 2.82E-09 | 1.26E-05 |
| KLK13 | 2711.589154 | 3.03E-09 | 1.26E-05 |
| NCCRP1 | 4315.455575 | 1.11E-08 | 3.84E-05 |
| AKR1B10 | −5251.888875 | 1.38E-08 | 4.09E-05 |
| KLK12 | 1742.502479 | 1.75E-08 | 4.54E-05 |
| MSLN | 2480.833944 | 2.15E-08 | 4.97E-05 |
| RPTN | 1595.155099 | 4.20E-08 | 8.71E-05 |
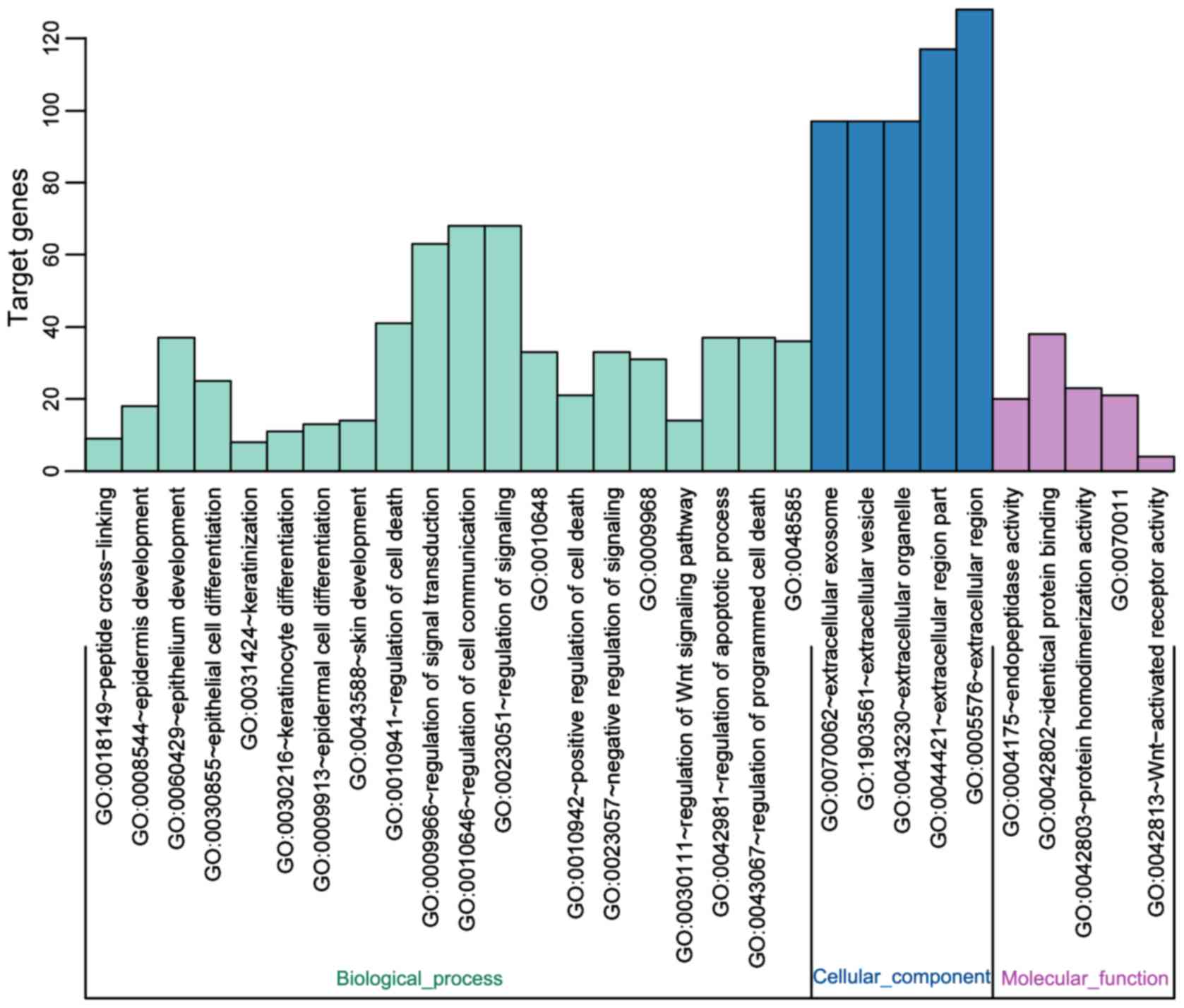
GO enrichment analysis
The list of differential genes was uploaded to DAVID
bioinformatics resource network (https://david.ncifcrf.gov/). The identifier was set as
OFFICIAL_GENE_SYMBOL and list type as Gene List. Other parameters
were all default. The results showed that differential genes were
mainly concentrated in the following fields: ‘Epithelial cell
differentiation’, ‘epithelium development’, and ‘epidermis
development’, which can affect the development and differentiation
of epithelial cells (Fig. 2).
KEGG pathway analysis
KOBAS 3 software was used for KEGG pathway analysis
and functional annotation of differential genes, and five key KEGG
pathways were identified, including ‘glutathione metabolism’,
‘arachidonic acid metabolism’, and ‘pentose phosphate pathway’,
among which ‘glutathione metabolism’ and ‘grachidonic acid
metabolism’ pathways were considered to be the two most important
ones (Table II).
 | Table II.KEGG enrichment outcome of
differential genes. |
Table II.
KEGG enrichment outcome of
differential genes.
| Term | Count | P-value | FDR |
|---|
| hsa00480: Glutathione
metabolism | 5 | 0.006573368 | 7.715865143 |
| hsa00590: Arachidonic
acid metabolism | 5 | 0.012975437 | 14.70170689 |
| hsa00030: Pentose
phosphate pathway | 3 | 0.067690311 | 57.40275058 |
| hsa05230: Central
carbon metabolism in cancer | 4 | 0.068191912 | 57.68095253 |
| hsa04610: Complement
and coagulation cascades | 4 | 0.081430803 | 64.44748515 |
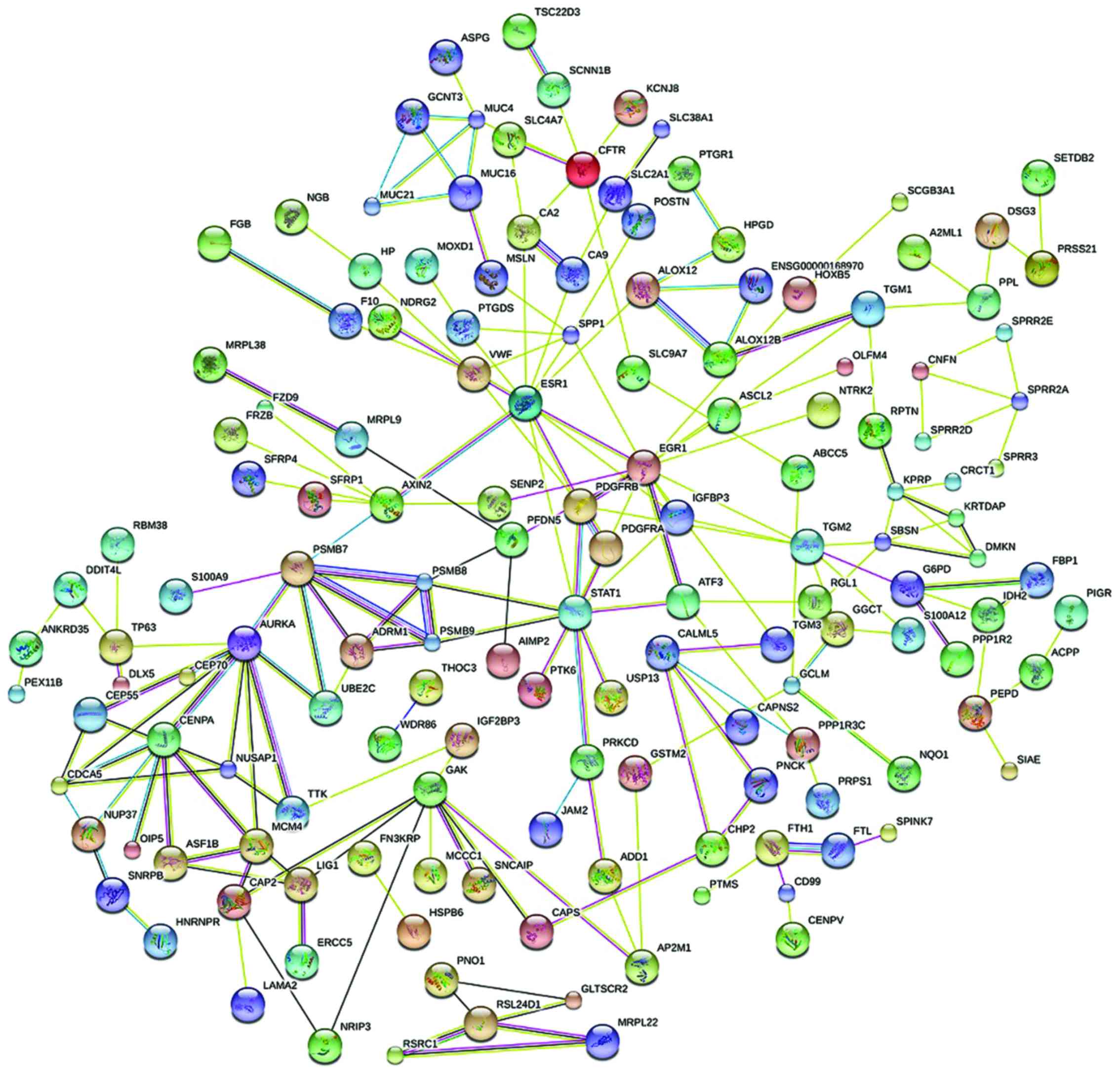
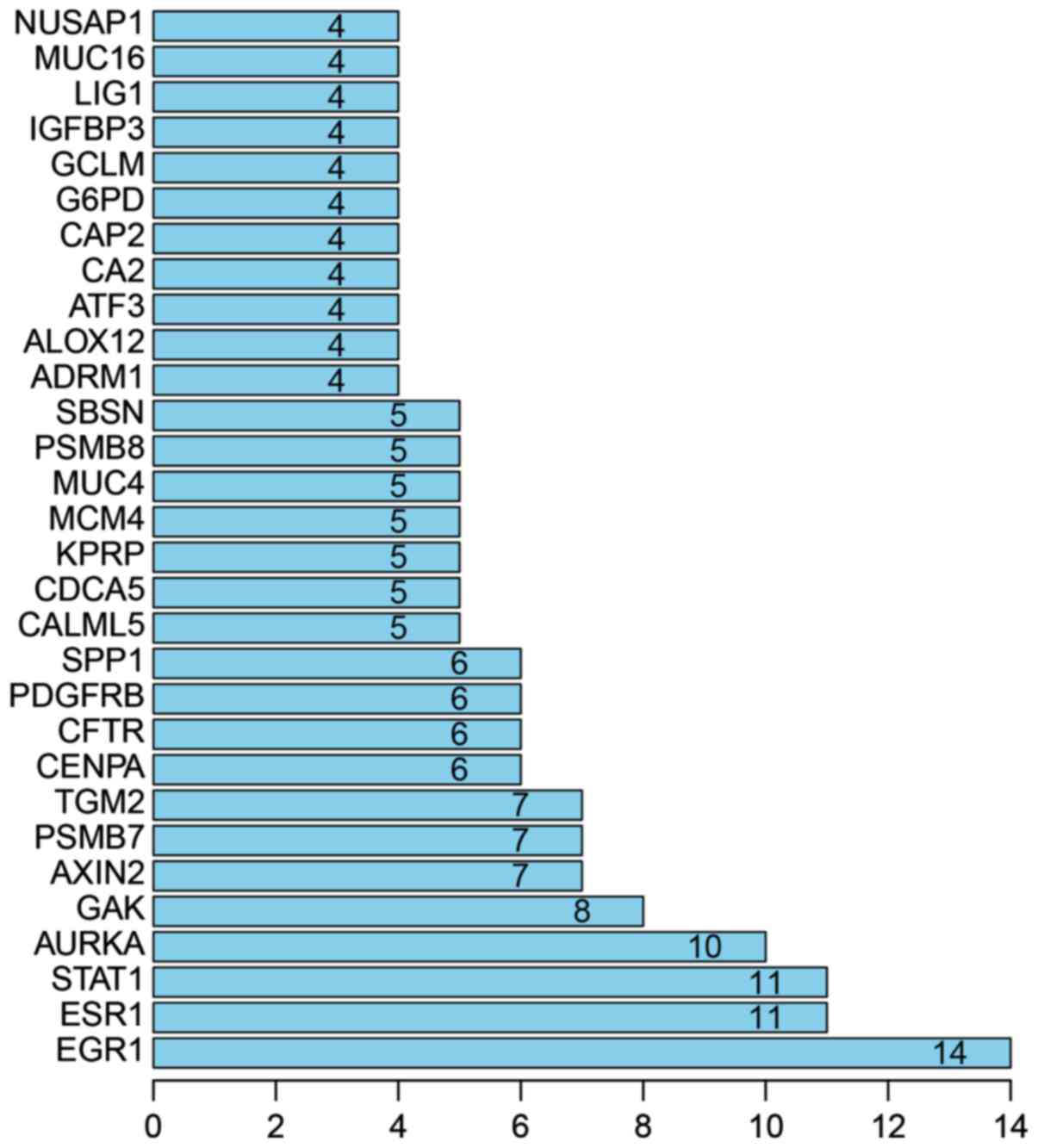
PPI analysis
With PPI analysis using STRING software, 30
prominent proteins were identified of which estrogen receptor α
(ESR1), STAT1, AURKA and GAK are the relatively important. EGR1 was
considered to be the most important protein and connected 14 nodes
(Figs. 3 and 4).
Discussion
Cervical cancer is a common malignant tumor in women
(10). In China, approximately 30,000
deaths and 100,000 new cases are reported annually (11). CIN, the precancerous lesion closely
related to cervical cancer, is considered to be of great
significance in studies of cervical cancer (12,13).
Transition from CIN to cervical cancer may take as long as 10 years
(14). Therefore, early diagnosis,
on-time follow-up and early treatment of CIN may effectively
inhibit the development of cervical cancer (15). The latest report of American Society
of Clinical Oncology (ASCO) showed that the patients with CIN were
becoming younger, especially for urban residents (16).
In order to investigate the molecular pathogenesis
of CIN, we analyzed the differential expression between CIN
patients and healthy controls using gene expression profiling data
with multiple bioinformatic methods including enrichment analysis
and PPI analysis. In the present study, strict inclusion criteria
were followed to select the most reliable chips from microarray
candidates. The reliability of the results was secured by the use
of microarray data from multiple samples, which can reduce the
error rate.
Based on GEO public database, we analyzed and
integrated the chip data using software package, and the resulting
287 differential genes were further treated for PPI analysis with
STRING. PPI analysis with differential genes showed that
EGR1 and ESR1 genes are important factors affecting
CIN. EGR1 protein is regarded as the protein with the most
significant impact on CIN. Van den Brandt et al (17) suggested that EGR1 was closely
associated with the development of myopia and non-small cell lung
cancer in human. EGR1 is also an important factor affecting the
cell dysplasia, dedifferentiation, and synthesis of nucleoli and
ribosomes (18). The activation of
EGR1 is related to the normal growth and differentiation of cells.
However, cell dysplasia and dedifferentiation play an important
role in the development of CIN. Therefore, EGR1 can serve as a
marker for the diagnosis of CIN. Schiavon et al (19) suggested that ESR1 is a potential risk
factor for breast cancer and can be used as a tumor marker for
targeted therapy of breast cancer. The biological effects of ESR1
mainly affect estrogen-targeted organs. ESR1 is mainly expressed in
cytoplasm of cervical cancer cells, but not in the nucleus,
possibly due to the blocked protein translation and modification.
However, with the progression of CIN, the expression of ESR1 in
epithelial cells is gradually declining, indicating that ESR1 can
be used as marker for the early diagnosis of CIN. But further
studies are needed to confirm these conclusions. GO enrichment
analysis found that differential expression between CIN cancer
cells and normal cells was mainly observed in epithelial cell
differentiation, epithelial cell development, and epidermal
development. Nagaoka et al (20) concluded that the biological processes
of ‘epithelial cell differentiation’ and ‘development’ played an
equally dominant role in the pathogenesis of breast and lung
cancer, making the two the focus of study on the biological process
of lung cancer. The specific maintenance of differentiation ability
in squamous epithelial cells is an important feature of CIN,
indicating the important impact of EGR1 and ESR1 on CIN. KEGG
pathway analysis revealed the dominant role of glutathione
metabolism and arachidonic acid metabolism pathways in CIN. A
previous study carried out by Liu et al (21) suggested that glutathione metabolism
was involved in varying aspects of the development of cancers by
affecting the rate of cancer progression. Glutathione, which can be
found in every cell in the body, plays importance roles in the
maintenance of normal immune system function. Glutathione has been
used widely as basic ingredient in functional foods due to its
function in improving resistance and inhibiting tumorigenesis.
Arachidonic acid in the human body can be synthesized by linoleic
acid. The metabolism of arachidonic acid can affect cell
proliferation rate, which is related to cell dysplasia of CIN.
These four signal pathways also play pivotal roles in the
progression of other tumors, but the correlation with CIN still has
not been reported. The specific mechanism remains to be further
explored.
Chip data used in this study are relatively old and
sample size was relatively small. Considering that CIN-relevant
genes may change with contributing factors or demographic reason
(region, and ethnicity), occult genetic difference may exist. We
have reduced the avoidable human error to the lowest possible
level. Tumor development is difficult to predict. Further studies
should focus on the gene and pathway candidates to elucidate the
mechanism.
The long-term analysis led to identification that,
CIN was closely related to EGR1 and ESR1 genes,
epithelial cell differentiation and glutathione metabolism.
However, the internal connection of the three factors remains to be
explored with further studies. Considering the fact that only few
studies on CIN-relevant genes have been reported, better
understanding of CIN at the genetic level may significantly benefit
the diagnosis, treatment and prognosis of CIN.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SY conceived the design of this study and wrote the
manuscript. TL helped with pretreatment of raw data, identification
of differential genes, and preparation of a heat map and KEGG
pathway analysis. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luo Q, Lei B, Liu S, Chen Y, Sheng W, Lin
P, Li W, Zhu H and Shen H: Expression of PBK/TOPK in cervical
cancer and cervical intraepithelial neoplasia. Int J Clin Exp
Pathol. 7:8059–8064. 2014.PubMed/NCBI
|
|
2
|
Ni L, Zheng RS, Zhang SW, Zhou XN, Zeng HM
and Chen WQ: An analysis of incidence and mortality of cervical
cancer in China 2003–2007. China Cancer. 21:801–804. 2012.
|
|
3
|
Shi YH, Wang BW, Tuokan T, Li QZ and Zhang
YJ: Association between micronucleus frequency and cervical
intraepithelial neoplasia grade in Thinprep cytological test and
its significance. Int J Clin Exp Pathol. 8:8426–8432.
2015.PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller JW, Hanson V, Johnson GD, Royalty
JE and Richardson LC: From cancer screening to treatment: Service
delivery and referral in the National Breast and Cervical Cancer
Early Detection Program. Cancer. 120:2549–2556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bueno CT, da Silva Dornelles CM, Barcellos
RB, da Silva J, Dos Santos CR, Menezes JE, Menezes HS and Rossetti
ML: Association between cervical lesion grade and micronucleus
frequency in the Papanicolaou test. Genet Mol Biol. 37:496–499.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Q, Liu S, Liu H, Zhang J, Guo S and
Wang L: Significance and implication on changes of serum squamous
cell carcinoma antigen in the diagnosis of recurrence squamous cell
carcinoma of cervix. Zhonghua Fu Chan Ke Za Zhi. 50:131–136.
2015.(In Chinese). PubMed/NCBI
|
|
8
|
Rebolj M, Helmerhorst T, Habbema D, Looman
C, Boer R, van Rosmalen J and van Ballegooijen M: Risk of cervical
cancer after completed post-treatment follow-up of cervical
intraepithelial neoplasia: Population based cohort study. BMJ.
345:e68552012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luesley D and Leeson S: Colposcopy and
programme management: Guidelines for the NHS Cervical Screening
Programme2nd edition. SBA Questions for the MRCOG. Jones A: NHSCSP
Publication No. 20. Cambridge University Press; Cambridge: pp.
1262010
|
|
10
|
Leinonen M, Nieminen P, Kotaniemi-Talonen
L, Malila N, Tarkkanen J, Laurila P and Anttila A: Age-specific
evaluation of primary human papillomavirus screening vs
conventional cytology in a randomized setting. J Natl Cancer Inst.
101:1612–1623. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McCredie MR, Sharples KJ, Paul C, Baranyai
J, Medley G, Jones RW and Skegg DC: Natural history of cervical
neoplasia and risk of invasive cancer in women with cervical
intraepithelial neoplasia 3: A retrospective cohort study. Lancet
Oncol. 9:425–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hakama M, Miller AB and Day NE: Screening
for cancer of the uterine cervix. Oxford University Press; UK:
2012
|
|
13
|
Cuzick J, Arbyn M, Sankaranarayanan R, Tsu
V, Ronco G, Mayrand MH, Dillner J and Meijer CJ: Overview of human
papillomavirus-based and other novel options for cervical cancer
screening in developed and developing countries. Vaccine. 26:29–41.
2008. View Article : Google Scholar
|
|
14
|
Bulkmans NW, Berkhof J, Rozendaal L, van
Kemenade FJ, Boeke AJ, Bulk S, Voorhorst FJ, Verheijen RH, van
Groningen K, Boon ME, et al: Human papillomavirus DNA testing for
the detection of cervical intraepithelial neoplasia grade 3 and
cancer: 5-year follow-up of a randomised controlled implementation
trial. Lancet. 370:1764–1772. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ronco G, Sideri MG and Ciatto S: Cervical
intraepithelial neoplasia and higher long term risk of cancer. BMJ.
335:1053–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Werkgroep Oncologische Gynaecologie, .
Cervical intraepithelial neoplasia (CIN), nationwide guidelines.
1.1. 2011.(in Dutch).
|
|
17
|
Van den Brandt PA, Schouten LJ, Goldbohm
RA, Dorant E and Hunen PM: Development of a record linkage protocol
for use in the Dutch Cancer Registry for Epidemiological Research.
Int J Epidemiol. 19:553–558. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Van den Akker-van Marle ME, van
Ballegooijen M and Habbema JD: Low risk of cervical cancer during a
long period after negative screening in the Netherlands. Br J
Cancer. 88:1054–1057. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schiavon G, Hrebien S, Garcia-Murillas I,
Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I,
Lopez-Knowles E, Ribas R, et al: Analysis of ESR1 mutation in
circulating tumor DNA demonstrates evolution during therapy for
metastatic breast cancer. Sci Transl Med. 7:313ra1822015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nagaoka K: Zhang H, Watanabe G and Taya K:
Epithelial cell differentiation regulated by McroRNA-200a in
mammary glands. PLoS One. 8:e651272013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Y, Hyde AS, Simpson MA and Barycki JJ:
Emerging regulatory paradigms in glutathione metabolism. Adv Cancer
Res. 122:69–101. 2014. View Article : Google Scholar : PubMed/NCBI
|