Introduction
Colorectal cancer (CRC) is the third most common
type of cancer in men and the second most common type of cancer in
women worldwide (1). Despite advances
in the diagnosis and treatment of CRC in the last few years, the
overall prognosis remains poor. It has been reported that tumor
metastasis serves a function in the diagnosis and therapy of CRC
(2). Elucidation of the underlying
molecular mechanism of metastasis in CRC may lead to novel
treatment strategies.
Epithelial-mesenchymal transition (EMT) is an
important biological process for tumor polarized epithelial cells
to acquire mesenchymal cell phenotypes (3). In the progression of CRC, the typical
characteristics of EMT include the loss of epithelial (E-)cadherin,
which is usually accompanied by dysregulation of the Wnt signaling
pathway (4,5). In the canonical Wnt signaling pathway,
β-catenin regulates the expression of E-cadherin and the induction
of EMT (6,7). β-catenin and E-cadherin usually exist in
the form of an E-cadherin-β-catenin complex at the membrane
(8,9).
However, as tumors progress, this complex dissociates and is
translocated into the nucleus, thereby triggering the downstream
signaling pathways (8,9). Despite the known function of the
E-cadherin-β-catenin complex in cell-cell association, little is
understood concerning the molecular mechanisms underlying tumor
invasion and metastasis.
Increasing evidence suggests that Fusobacterium
nucleatum is involved in colorectal carcinogenesis. Previous
studies have indicated that F. nucleatum is present in human
colorectal adenomas and carcinomas (10,11).
Additionally, F. nucleatum is positively associated with
advanced tumor stage and poor prognosis in CRC (12–14).
Recent studies have identified that F. nucleatum adheres to
and invades epithelial cells mainly through the virulence factors,
including Fusobacterium adhesin A (FadA),
Fusobacterium autotransporter protein 2 and fusobacterial
outer membrane protein A (15–17).
Nevertheless, it is remains unclear whether F. nucleatum
triggers the colonic EMT process.
The aim of the present study was to identify the
molecular mechanism underlying F. nucleatum-mediated EMT
process, and to additionally elucidate the role of F.
nucleatum-mediated EMT in patients with CRC.
Materials and methods
Bacterial strain and culture
conditions
F. nucleatum strain was purchased from the
American Type Culture Collection (ATCC 25586; Manassas, VA, USA).
F. nucleatum was cultured in Columbia blood agar
supplemented with 5 µg/ml hemin, 5% defibrinated sheep blood (5%)
and 1 µg/ml vitamin K1 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) in an anaerobic glove box containing 85% N2,
10% H2 and 5% CO2 at 37°C. Escherichia
coli strain DH5α (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was propagated in Luria-Bertani medium (Difco; BD
Biosciences, Franklin Lakes, NJ, USA) aerobically at 37°C.
Cell culture
The human normal colon epithelial cell line NCM460
was obtained from Incell Corporation, LLC (San Antonio, TX, USA).
NCM460 cells were cultured in RPMI-1640 medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (Hyclone; GE Healthcare Life Sciences) at 37°C
in a humidified atmosphere containing 5% CO2.
Wound healing assays
NCM460 cells (1×106 cells/well) were
seeded into a 6-well plate to form a confluent monolayer and cells
were wounded using a pipette tip. Cells were incubated with F.
nucleatum or E. coli DH5α at a multiplicity of infection
(MOI) of 1,000:1. Images were captured at 6, 24, 48 and 72 h. The
wound size of each well was calculated as the distance between the
edges. The images were captured under a light microscope
(magnification, ×40; XDS-500D; Shanghai Caikon Optical Instrument
Co., Ltd., Shanghai, China).
Cell proliferation assays
NCM460 cells were seeded in 24-well plates at a
density of 1×104 cells/well with 1 ml complete RPMI-1640
medium (Hyclone; GE Healthcare Life Sciences). Cells were incubated
with F. nucleatum or E. coli DH5α at an MOI of
1,000:1. Cells treated with PBS were used as a negative control.
Cell numbers were calculated using a hemocytometer and cell
proliferation was analyzed using a Cell Counting Kit-8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan), according to the
manufacturer's protocol.
Cell cycle assays
NCM460 cells (~1×106) cells were trypsinized at room
temperature for 2 min, washed twice with PBS and fixed in 70%
ice-cold ethanol for 1 h at room temperature. The samples were
centrifuged at 300 × g for 5 min at 4°C, the ethanol was removed
and they were exposed to 100 mg/ml RNaseA (Sigma-Aldrich; Merck
KGaA) for 30 min at 37°C. Cellular DNA was stained with propidium
iodide (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
Cell-cycle distributions were determined by flow cytometry using a
BD FACSCalibur system (SKU#: 8044-30-1001, BD Biosciences, Franklin
Lakes, NJ, USA) and data was analyzed using the ModFit software
(version 4.1; Verity Software House, Inc., Topsham, ME, USA).
Western blot analysis
NCM460 cells were lyzed using
radioimmunoprecipitation buffer (Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China). A BCA kit (Pierce; Thermo
Fisher Scientific, Inc.) was used to determine the protein
concentration. Equal quantities of protein (15 µg) were separated
by SDS-PAGE (10% gels). Electrophoresed proteins were transferred
onto nitrocellulose membranes. The membranes were then blocked with
5% fat-free milk suspended in TBS-T buffer for 2 h at room
temperature and incubated with the following primary antibodies at
4°C overnight: Anti-phospho-p65, a subunit of nuclear factor-κB
(NF-κB) (cat. no. ab86299, Abcam, Cambridge, MA, USA),
anti-interleukin (IL)-6 (cat. no. ab6672, Abcam), anti-IL-1β (cat.
no. ab156791, Abcam), anti-matrix metalloproteinase (MMP)-13 (cat.
no. ab39012, Abcam), anti-E-cadherin (cat. no. ab1416, Abcam),
anti-β-catenin (cat. no. ab32572, Abcam) and anti-GAPDH (cat. no.
ab8245, Abcam). Following several washes with TBST, the membranes
were incubated with HRP-conjugated goat anti-rabbit IgG (1:5,000;
cat. no. ZB-2306, Zhongshan Gold Bridge Biological Technology Co.,
Beijing, China) for 2 h at room temperature and then washed. The
proteins were detected using enhanced chemiluminescence, according
to the manufacturer's protocol (Merck KGaA). ImageJ 1.8.0 (National
Institutes of Health, Bethesda, MD, USA) was applied to quantify
the relative protein levels. GAPDH was used as an internal
control.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was extracted from NCM460 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The concentration and the
purity of the RNA samples were assayed by absorbent density
analysis using an optical density (OD) ratio of 260/280 nm. A total
of 2 µg RNA was reverse-transcribed using the TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR amplifications were performed in a 10-µl
reaction system containing 5 µl SYBR Green Supermix (Tarkara,
Japan) on Bio-Rad iQ5 Optical System (Bio-Rad Laboratories, Inc.,
Hercules, California, USA), 0.4 µl forward primer, 0.4 µl reverse
primer, 2.2 µl double-distilled water and 2 µl template cDNA. The
following thermal cycling conditions were used for the qPCR:
Initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 15
sec and a final extension at 60°C for 1 min. mRNA levels were
determined using the 2−ΔΔCq method (18). The data was analyzed using Bio-Rad CFX
Manager 3.1 (Bio-Rad Laboratories, Inc.). GAPDH was used as a
reference gene. The primers used in the current studies were as
follows: TNFα-forward: CTGGGCAGGTCTACTTTGGG; TNFα-reverse:
CTGGAGGCCCCAGTTTGAAT; IL6 foward: AGTGAGGAACAAGCCAGAGC; IL6
reverse: AGCTGCGCAGAATGAGATGA; IL-1β foward: CTTAAAGCCCGCCTGACAGA;
IL-1β reverse: ACACTGCTACTTCTTGCCCC; MMP-13 foward:
AGGCCATGGCATCCCTTTTT; MMP-13 reverse: AGCACCCTCCCCAAGTATCA; GAPDH
foward: GAGAAGGCTGGGGCTCATTT; GAPDH reverse:
AGTGATGGCATGGACTGTGG.
Transient transfection
Cells were transfected with small interfering RNA
(siRNA) targeting E-cadherin (si-E-cadherin) or β-catenin
(si-β-catenin), or with negative control siRNA (NC;
5′-ACUAGUCGAUCUAUGUGUGAUATT-3′) (Shanghai GenePharma Co., Ltd.,
Shanghai, China) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at the indicated concentrations,
according to the manufacturer's protocol. In brief, NCM460 cells
(1×106 cells/well) were seeded in a six well plate with
2 ml RPMI-1640 medium. At 60% confluence, 50 nM si-E-cadherin or 50
nM si-β-catenin or 50 nM NC was mixed with
Lipofectamine® 2000 at room temperature for 20 min.
Then, the mixture was added into each well at a final concentration
of 20 nM for 48 h. Then, the cells were collected for further
analysis.
Immunofluorescence
NCM460 cells were cultured in a 6-well plate with
glass coverslips and were fixed in 4% paraformaldehyde for 30 min
at room temperature. The samples were washed three times in PBS for
5 min. Following washing with PBS three times for 5 min, the slides
were blocked with 8% bovine serum albumin (Sigma-Aldrich; Merck
KGaA) at room temperature for 2 h The coverslips were incubated
with antibodies against E-cadherin (1:50; cat. no. ab1416, Abcam)
and β-catenin (1:50; cat. no. ab32572, Abcam) in a humidified
chamber overnight at 4°C. After that the slides were washed with
PBS for three times and incubated with
tetramethylrhodamine-conjugated anti-rabbit immunoglobulin G
(1:500; cat. no. ZDR5209, Zhongshan Gold Bridge Co.) and with DAPI
(1:1,000; cat. no. C0060, Solarbio Science & Technology Co.,
Ltd.) for 20 min at room temperature. Following incubation with the
secondary antibody, the slides were washed three times with PBS in
the dark and the coverslips were mounted with a mounting medium and
coated on glass slides. The slides were sealed at room temperature
for ~1 h in the dark and fluorescence intensity was examined using
a fluorescence microscope.
Statistical analysis
Data are presented as the mean + standard deviation
for the indicated number of separate experiments. A total of three
independent experiments were performed for each experiment.
Differences in the quantitative data between two groups were
determined using the unpaired Student's t-test. Comparisons of
means among multiple groups were determined using one-way analysis
of variance. All statistical analyses were performed using GraphPad
Software (version 6.0; GraphPad Software, Inc., La Jolla, CA, USA).
All P-values were two-tailed. P<0.05 was considered to indicate
a statistically significant difference.
Results
F. nucleatum enhances the
proliferation and invasion of NCM460 cells
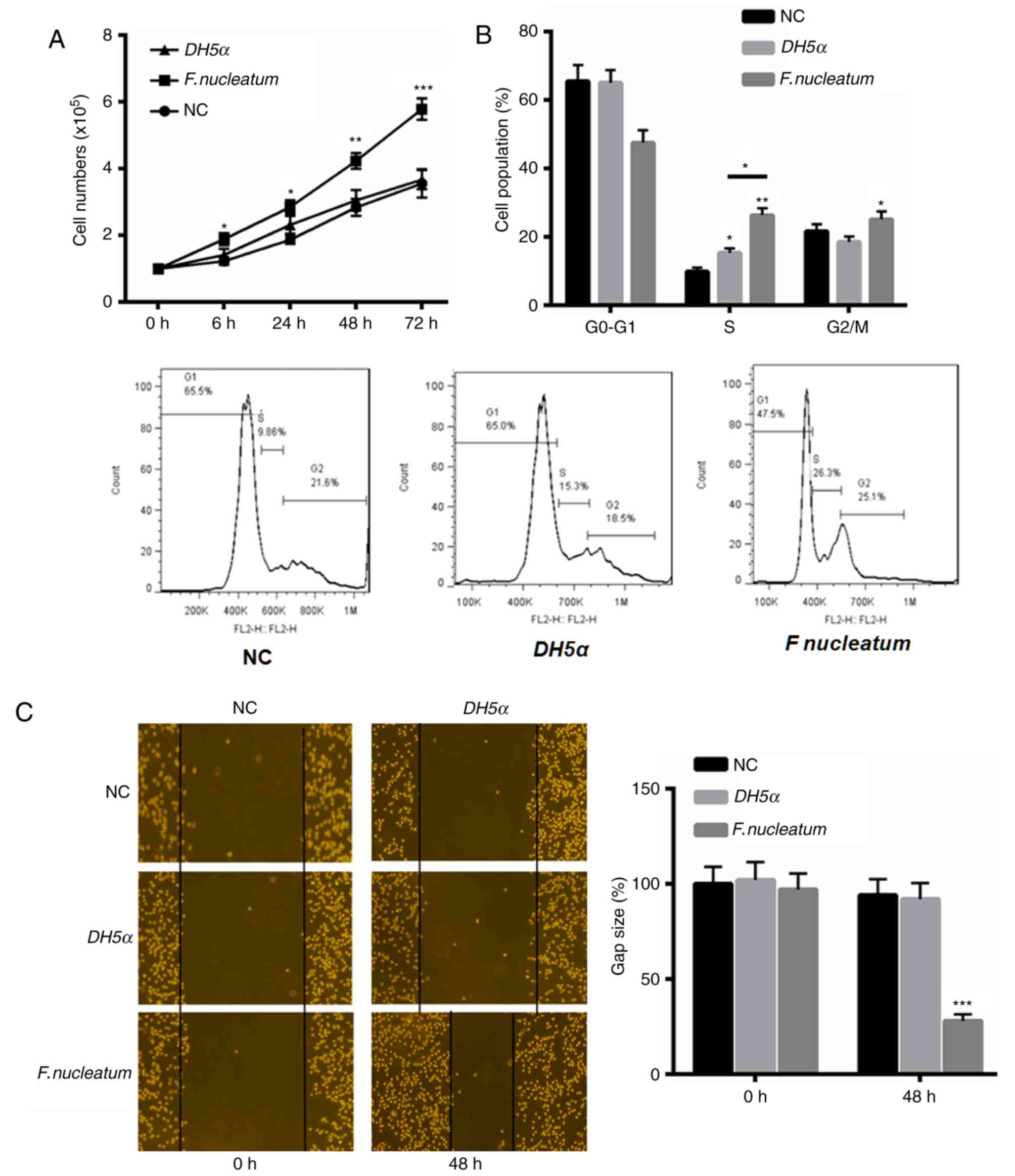
First, the function of F. nucleatum in the
proliferation and invasion of NCM460 cells was investigated. The
results indicated that F. nucleatum significantly enhanced
the proliferation of NCM460 cells at 24, 48 and 72 h, compared with
the control and E. coli DH5α groups (Fig. 1A). Additionally, flow cytometric
analysis of the cell cycle revealed an increased proportion of
cells at S and G2/M phase in response to F.
nucleatum treatment (Fig. 1B).
F. nucleatum also increased the invasive ability of NCM460
cells at 48 h compared with the control and E. coli DH5α
groups (Fig. 1C).
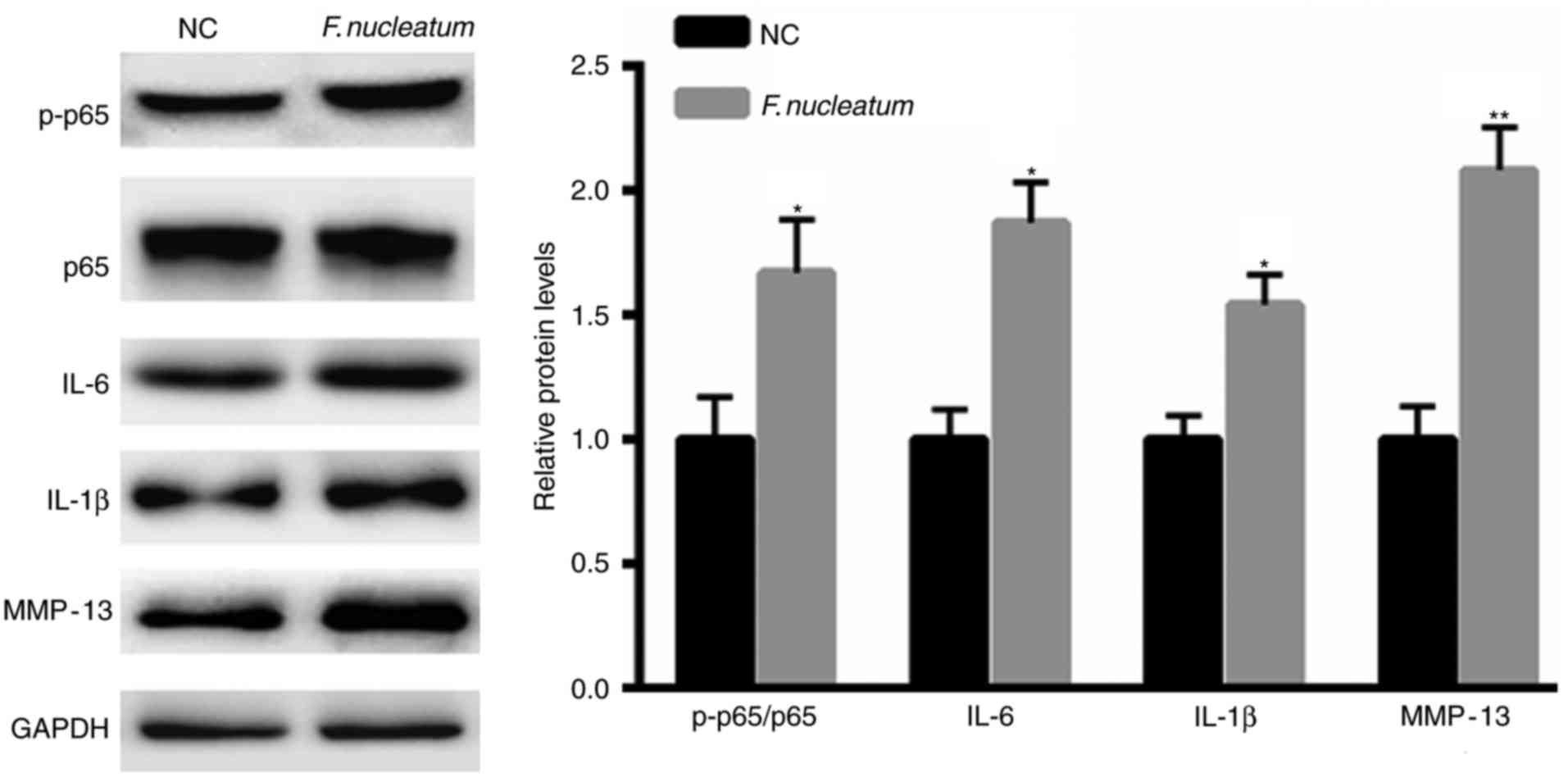
F. nucleatum promotes the activation
of NF-κB signaling
Increasing evidence has indicated the important
function of pro-inflammatory factors during tumorigenesis (19,20).
Therefore, whether F. nucleatum infection regulates
signaling was also evaluated. F. nucleatum infection
significantly enhanced the phosphorylation levels of p65 and the
expression of IL-6, IL-1β and MMP-13 (Fig. 2).
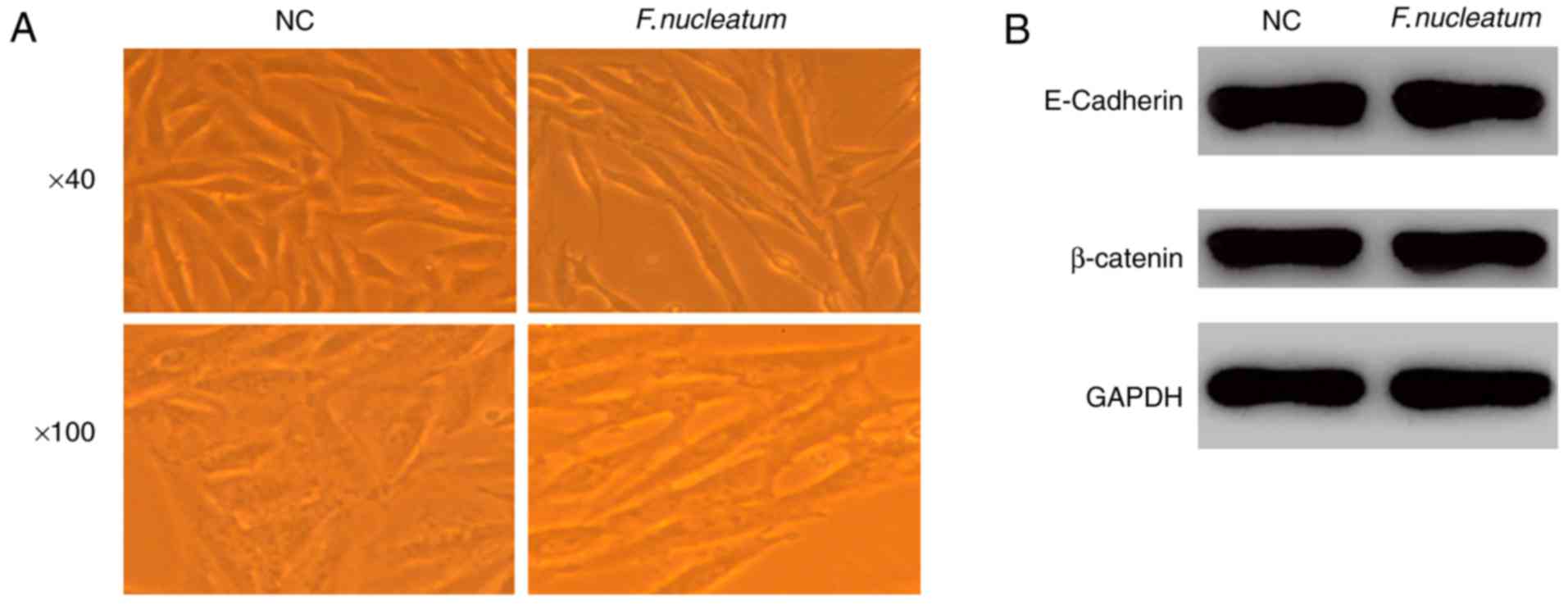
F. nucleatum promotes EMT without
affecting the expression of E-cadherin and
Attachment and invasion are hallmarks of F.
nucleatum. Therefore, the effects of F. nucleatum
infection on the induction of EMT were also assessed. At 48 h
post-infection, epithelial cells transdifferentiated into
mesenchymal-like cells as observed using microscopy (Fig. 3A). Furthermore, F. nucleatum
infection did not alter the protein levels of E-cadherin and
β-catenin as demonstrated using western blot analysis (Fig. 3B), indicating that F. nucleatum
infection induced EMT without affecting the expression of
E-cadherin.
F. nucleatum promotes malignancy of
NCM460 cells by interacting with E-cadherin
To further elucidate the underlying molecular
mechanism of F. nucleatum in malignant transformation of
NCM460 cells, E-cadherin or β-catenin expression was silenced in
NCM460 cells (Fig. 4A and B). An
immunofluorescence assay revealed decreased expression of
E-cadherin or β-catenin in NCM460 cells following transfection with
si-E-cadherin or si-β-catenin for 48 h (Fig. 4C). qPCR was performed to evaluate the
expression levels of inflammatory factors, including tumor necrosis
factor (TNF)-α, IL-6, IL-1β and MMP-13. The results indicated that
F. nucleatum did not increase the mRNA levels of TNF-α,
IL-6, IL-1β and MMP-13 in NCM460 cells when E-cadherin was silenced
(Fig. 4D). However, increased
expression of TNF-α, IL-6, IL-1β and MMP-13 was identified in
NCM460 cells when transfected with si-β-catenin (Fig. 4E). Additionally, F. nucleatum
infection did not increase the proportion of cells at S phase when
E-cadherin was silenced, but it increased the number of cells at S
phase when NCM460 cells were transfected with si-β-catenin
(Fig. 4F). These results indicated
that although E-cadherin or β-catenin are required for tumor growth
and inflammatory responses, E-cadherin and β-catenin are
differentially regulated by F. nucleatum infection.
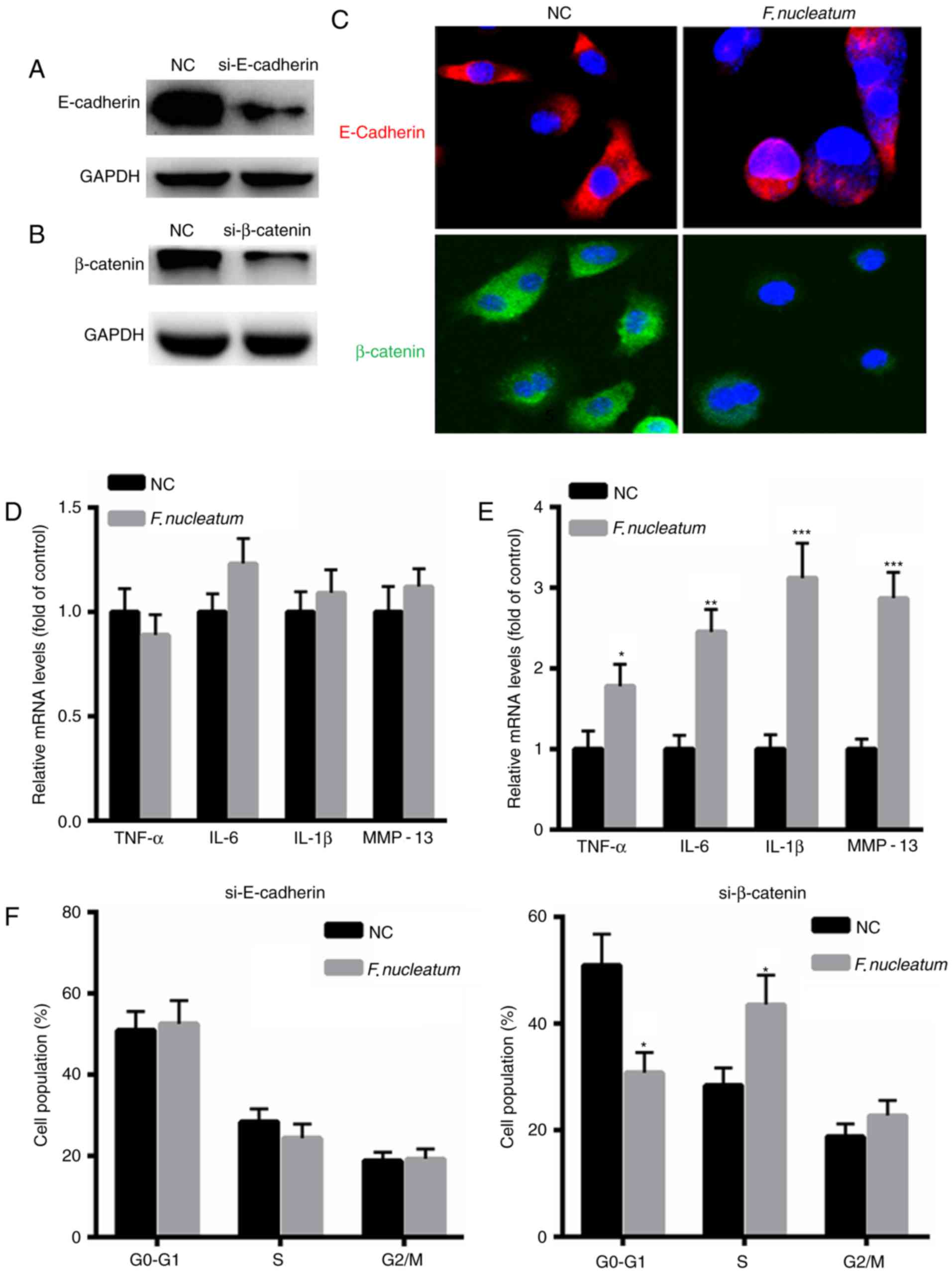 | Figure 4.F. nucleatum promotes the
malignant phenotype of NCM460 cells by interacting with E-cadherin.
Western blot analysis of (A) E-cadherin or (B) β-catenin in NCM460
cells transfected with si-E-cadherin or si-β-catenin for 48 h. (C)
Immunofluorescence assay revealed decreased expression of
E-cadherin or β-catenin following transfection with si-E-cadherin
(Red) or si-β-catenin (Green) for 48 h in NCM460 cells infected
with F. nucleatum or control siRNA. Magnification, ×400. The
nucleus was stained DAPI). (D) F. nucleatum did not increase
the mRNA levels of TNF-α, IL-6, IL-1β and MMP-13 in NCM460 cells
when E-cadherin was silenced. (E) Increased expression of TNF-α,
IL-6, IL-1β and MMP-13 in NCM460 cells when transfected with a
si-β-catenin. (F) Flow cytometric analysis of the cell cycle
following transfection with si-E-cadherin (Red) or si-β-catenin
(Green) for 48 h in NCM460 cells (106/well) infected
with F. nucleatum or control siRNA. *P<0.05, **P<0.01,
***P<0.001 vs. control. NC, negative control group; IL,
interleukin; MMP, matrix metalloproteinase; TNF, tumor necrosis
factor; si, siRNA. |
Discussion
It is estimated that >1,000,000 new cases of CRC
are diagnosed every year (21,22).
Therefore, it is important to understand the underlying molecular
mechanisms in CRC and identify potential risk factors. Infectious
agents have attracted attention since they are associated with
several human malignancies (23). In
the present study, the function of F. nucleatum in CRC
progression through modulating the expression of
E-cadherin/β-catenin signaling was investigated.
Previous studies have demonstrated the significant
overabundance of F. nucleatum in colorectal adenomas and
cancer. However, it remains unclear how F. nucleatum is
involved in the CRC microenvironment (24,25). In
the present study, F. nucleatum enhanced the growth and
proliferation of NCM460 cells, suggesting that F. nucleatum
may contribute to the development of CRC. The results of the
present study also suggested that F. nucleatum is associated
with intratumoral immune cell responses and promotes tumorigenesis
by regulating the tumor microenvironment (10,26). F.
nucleatum invades epithelial cells through the virulence factor
FadA which then triggers pro-oncogenic signaling and promotes CRC
development (17). The present study
demonstrated that F. nucleatum infection activated NF-κB
signaling in NCM460 cells, thereby enhancing downstream signaling
pathways.
E-cadherin is a transmembrane glycoprotein, which is
located at the adherens junctions and mediates calcium-dependent
cell-cell adhesion (17). At the
C-terminus, E-cadherin is linked to α-catenin and the actin
cytoskeleton through interaction with β-catenin. E-cadherin,
α-catenin and β-catenin form a complex, thus creating tight
cell-cell interactions and restraining cell mobility. Dysregulation
of the E-cadherin-β-catenin complex leads to the invasion and
metastasis of CRC cells (13,14). Thus, the function of F.
nucleatum in the E-cadherin-β-catenin complex in invasion and
metastasis of CRC cells was examined. The results indicated that
infection by F. nucleatum induced EMT and did not affect the
expression levels of E-cadherin and β-catenin.
The present study sought to investigate the
molecular mechanism underlying F. nucleatum-mediated EMT.
Loss of E-cadherin-mediated cell adhesion is associated with tumor
invasion. Decreased β-catenin expression is associated with the
loss or decrease of E-cadherin expression in invasive tumor cells
(27,28). Additionally, membrane
β-catenin/cytosolic expression level is decreased in primary
tumors, indicating that low β-catenin expression level may be a
potential marker for the metastasis and a worse outcome in breast
cancer (29). The results of the
present study demonstrated that F. nucleatum may only
increase the inflammatory responses when β-catenin expression is
knocked down in NCM460 cells, whereas no changes were identified
when E-cadherin expression is knocked down. These results indicated
that F. nucleatum may promote the malignant phenotype of CRC
by interacting with E-cadherin.
In conclusion, to the best of our knowledge, the
present study is the first to demonstrate that F. nucleatum
infection may interact with E-cadherin, which in turn induces the
malignant phenotype of CRC cells. The results of the present study
may provide insights that lead to the development of novel
antimicrobial targets for the treatment of CRC.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lugli A, Zlobec I, Minoo P, Baker K,
Tornillo L, Terracciano L and Jass JR: Prognostic significance of
the wnt signalling pathway molecules APC, beta-catenin and
E-cadherin in colorectal cancer: A tissue microarray-based
analysis. Histopathology. 50:453–464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaller M and Hermeking H: Interplay
between transcription factors and MicroRNAs regulating
epithelial-mesenchymal transitions in colorectal cancer. Adv Exp
Med Biol. 937:71–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
The importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fiering S, Ang LH, Lacoste J, Smith TD and
Griner E: Reproducibility Project: Cancer Biology: Registered
report: Biomechanical remodeling of the microenvironment by stromal
caveolin-1 favors tumor invasion and metastasis. Elife.
4:e047962015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goetz JG, Minguet S, Navarro-Lerida I,
Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibáñez T,
Pellinen T, Echarri A, et al: Biomechanical remodeling of the
microenvironment by stromal caveolin-1 favors tumor invasion and
metastasis. Cell. 146:148–163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen HN, Yuan K, Xie N, Wang K, Huang Z,
Chen Y, Dou Q, Wu M, Nice EC, Zhou ZG and Huang C: PDLIM1
stabilizes the E-cadherin/β-Catenin complex to prevent
epithelial-mesenchymal transition and metastatic potential of
colorectal cancer cells. Cancer Res. 76:1122–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang X, Xu X, Wang F, Chen X, Li N, Wang
C and He J: E-cadherin knockdown increases β-catenin reducing
colorectal cancer chemosensitivity only in three-dimensional
cultures. Int J Oncol. 47:1517–1527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kostic AD, Chun E, Robertson L, Glickman
JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold
GL, et al: Fusobacterium nucleatum potentiates intestinal
tumorigenesis and modulates the tumor-immune microenvironment. Cell
Host Microbe. 14:207–215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ito M, Kanno S, Nosho K, Sukawa Y,
Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M,
Takahashi H, et al: Association of Fusobacterium nucleatum
with clinical and molecular features in colorectal serrated
pathway. Int J Cancer. 137:1258–1268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mima K, Nishihara R, Qian ZR, Cao Y,
Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al:
Fusobacterium nucleatum in colorectal carcinoma tissue and
patient prognosis. Gut. 65:1973–1980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tahara T, Shibata T, Kawamura T, Okubo M,
Ichikawa Y, Sumi K, Miyata M, Ishizuka T, Nakamura M, Nagasaka M,
et al: Fusobacterium detected in colonic biopsy and
clinicopathological features of ulcerative colitis in Japan. Dig
Dis Sci. 60:205–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tahara T, Yamamoto E, Suzuki H, Maruyama
R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al:
Fusobacterium in colonic flora and molecular features of colorectal
carcinoma. Cancer Res. 74:1311–1318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gur C, Ibrahim Y, Isaacson B, Yamin R,
Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N,
Coppenhagen-Glazer S, et al: Binding of the Fap2 protein of
Fusobacterium nucleatum to human inhibitory receptor TIGIT
protects tumors from immune cell attack. Immunity. 42:344–355.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abed J, Emgard JE, Zamir G, Faroja M,
Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al:
Fap2 mediates Fusobacterium nucleatum colorectal
adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc.
Cell Host Microbe. 20:215–225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G
and Han YW: Fusobacterium nucleatum promotes colorectal
carcinogenesis by modulating E-cadherin/β-catenin signaling via its
FadA adhesin. Cell Host Microbe. 14:195–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ellah Abd NH, Taylor L, Ayres N, Elmahdy
MM, Fetih GN, Jones HN, Ibrahim EA and Pauletti GM: NF-κB decoy
polyplexes decrease P-glycoprotein-mediated multidrug resistance in
colorectal cancer cells. Cancer Gene Ther. 23:149–155. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren H, Wang Z, Zhang S, Ma H, Wang Y, Jia
L and Li Y: IL-17A promotes the migration and invasiveness of
colorectal cancer cells through NF-κB-Mediated MMP expression.
Oncol Res. 23:249–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L,
Li Q, Wu J and Fu X: Invasive Fusobacterium nucleatum
activates beta-catenin signaling in colorectal cancer via a
TLR4/P-PAK1 cascade. Oncotarget. 8:31802–31814. 2017.PubMed/NCBI
|
|
22
|
Mehta RS, Nishihara R, Cao Y, Song M, Mima
K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y, et al:
Association of dietary patterns with risk of colorectal cancer
subtypes classified by Fusobacterium nucleatum in tumor
tissue. JAMA Oncol. 3:921–927. 2017.PubMed/NCBI
|
|
23
|
Nosho K, Sukawa Y, Adachi Y, Ito M,
Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi
H, et al: Association of Fusobacterium nucleatum with
immunity and molecular alterations in colorectal cancer. World J
Gastroenterol. 22:557–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nie S, Tian B, Wang X, Pincus DH, Welker
M, Gilhuley K, Lu X, Han YW and Tang YW: Fusobacterium
nucleatum subspecies identification by matrix-assisted laser
desorption ionization-time of flight mass spectrometry. J Clin
Microbiol. 53:1399–1402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han YW: Fusobacterium nucleatum: A
commensal-turned pathogen. Curr Opin Microbiol. 23:141–147. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mima K, Sukawa Y, Nishihara R, Qian ZR,
Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al:
Fusobacterium nucleatum and T cells in colorectal carcinoma.
JAMA Oncol. 1:653–661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ismail AF, Halacli Oskay S, Babteen N, De
Piano M, Martin TA, Jiang WG, Khan MS, Dasgupta P and Wells CM:
PAK5 mediates cell: Cell adhesion integrity via interaction with
E-cadherin in bladder cancer cells. Biochem J. 474:1333–1346. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qu J, Jiang Y, Liu H, Deng H, Yu J, Qi X,
Ge W and Li G: Prognostic value of E-cadherin-, CD44-, and
MSH2-associated nomograms in patients with stage II and III
colorectal cancer. Transl Oncol. 10:121–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Jin L, Ye F, Ma Q, Yang Z, Liu D,
Yang J, Ma D and Gao Q: Isoform expression patterns of EPHA10
protein mediate breast cancer progression by regulating the
E-Cadherin and β-catenin complex. Oncotarget. 8:30344–30356.
2017.PubMed/NCBI
|