Introduction
Gastric cancer (GC) is one of the most common
malignant diseases originating from the mucosal epithelium of the
stomach. GC has high morbidity and mortality in China, which
seriously affects the health of patients (1,2). GC is
more aggressive but is hard to find in early stage, so most GC
patients are diagnosed at an advanced stage (3). Although clinically significant progress
has been made in treatment, the clinical outcomes of patients with
advanced GC have not had a significant impact. Therefore, studies
to explore the underlying mechanisms of the GC development are
necessary, as they could provide novel therapeutic targets for GC
treatment (4).
Increasing evidence has been reported that microRNAs
(miRNAs) could function as tumor inhibitors or tumor promoters in
the GC development by targeting several mRNA genes, including
proliferation, migration and invasion (5,6). For
example, Ahn et al (7) showed
that miR-200 acted as an oncogene in modulating GC progression via
inhibiting CDH1. However, miR-22 was proved to suppress GC
metastasis and invasion via regulating MMP14 and Snail (8). So far, the miRNAs that were found to
participate in GC development are still relatively limited, and
their roles and potential mechanisms need to be further
studied.
Many previous studies showed that miR-16 is involved
in cell proliferation, invasion and metastasis of various cancers.
miR-16 was proven to function as a tumor suppressor in regulating
glioma cell proliferation, invasion and promoted apoptosis through
targeting Wip1 (9). A previous study
also showed that the effect of miR-127 on non-small cell lung
cancer proliferation was inhibition (10). In addition, one study stated that
miR-127 acted as a tumor promoter in regulating of the progression
of colorectal adenocarcinoma (11).
However, there are very few studies on the biological mechanism of
miR-127 in GC.
Sal-like protein 4 (SALL4) is a zincfinger
transcription factor encoded by a member of the SALLgene family
(12). Previous studies showed that
the role SALL4 played in early embryo development, organ formation
and the proliferation and pluripotency of embryonic stem cells was
very important (13–16). Recently, SALL4 was shown to be
involved in modulating various solid tumors. For instance, SALL4
expression was upregulated in liver, lung, breast and colorectal
cancer (17–20). Furthermore, SALL4 could promote the
migratory and invasive ability of breast cancer (21) and cell viability of endometrial cancer
(22). Therefore, to deeply
understand the mechanism of SALL4 in cancers would help researchers
to find a new target for cancer diagnosis and treatment (23). A study recently reported that SALL4
promoted GC progression as an oncogene (24,25).
However, the biological role of SALL4 in GC regulated by miR-16
remains unclear.
Our study examined miR-16 in GC development and its
biological mechanism in regulation of GC cell proliferation and
migration. We found that miR-16 showed inhibitory effect in GC.
miR-16 overexpression could suppress GC cell viability and
migration and make SALL4 expression lower, while knockdown of
miR-16 had the opposite effect. Furthermore, we demonstrated that
the relationship between miR-16 and SALL4 expression was negatively
correlated in GC tissues. Therefore, our results indicated that the
miR-16/SALL4 axis provided a therapeutic target for treating
GC.
Materials and methods
Samples and cell culture
Forty paired GC tissues and adjacent normal tissues
were obtained from GC patients who underwent surgery at the
China-Japan Union Hospital, Jilin University (Changchun, China).
All tissue specimens were confirmed by pathological diagnoses and
no patients received radiotherapy or chemotherapy before surgery.
All corrected tissues were immediately frozen in −80°C
refrigerator. All contents about this study were approved by the
Ethics Committee of China-Japan Union Hospital, Jilin University.
Each GC patient involved in this study signed the informed
consent.
The gastric epithelium cell line GES-1 and four GC
cell lines (SGC-7901, HGC-27, MKN45 and MGC-803) were obtained from
Shanghai Institute of Cell Biology of the Chinese Academy of
Sciences. The cells were cultured in RPMI-1640 medium (Invitrogen;
Thermo Fisher Scientific Inc., Waltham, MA, USA) containing 10%
fetal bovine serum, and then cultured in an incubator at 37°C under
5% CO2.
Cell transfection
miR-16 mimic and inhibitor were provided by the
company of GenePharma (Shanghai, China). miR-16 mimic and miR-16
inhibitor (50 nM) were transfected into SGC-7901 and HGC-27 cells
respectively in parallel to overexpress or suppress miR-16 and
SALL4 small interfering RNA (siRNA) to silence SALL4. All the cells
were plated in 24-well plates 24 h before transfection and the
transfections were performed using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific Inc.) the next day. The
transfected cells were divided into several groups.
RT-qPCR assays
TRIzol reagent (Invitrogen; Thermo Fisher Scientific
Inc.) was used to isolate total RNA from the GC tissues and cells.
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific,
Inc.), was used to quantify the RNA. The sequences of the primers
were: miR-16, TAGCAG CACGTAAATATTGGCG (forward) and TGCGTGTCGTG
GAGTC (reverse); for SALL4, TAGC CCTGCGTAGCCAGTTA (forward) and
TCATGCTTAGTCCACTGTCTGT (reverse); for U6, GCTTCGGCAGCACATATACTA
AAAT (forward) and CGCTTCACGAATTTGCGTGTCAT (reverse); for GAPDH,
AGAAGGCTGGGGCTCATTTG (forward) and AGGGGCCATCCACAGTCTTC (reverse).
U6 and GAPDH were used as internal controls. The 2−∆∆Cq
method was used to detect the relative expression of miR-16 and
SALL4 (26).
Cell proliferation assay
MTT assay was used to detect cell viability.
SGC-7901 and HGC-27 cells were seeded and RPMI-1640 medium was
subsequently added into 96-well plates and incubated for 24, 48,
and 72 h at 37°C with 5% CO2. MTT solution was added to
each well for incubation for 4 h. After centrifugation, at room
temperature, 1,000 × g for 10 min, the culture medium was removed
and DMSO (100 µl) was added into the plates to dissolve the
crystals. The absorbance value of each well was measured at the
OD490 nm using enzyme-linked immunoassay.
Cell migration assay
Cell migratory ability was performed using Transwell
assay. The Transwell chamber with 8 µm pore size polycarbonic
membrane (Costar; Corning Incorporated, Corning, NY, USA) was
placed into the 24-well plates to separate the top and the lower
chambers. GC cells (1×105) with different transfection
were seeded into the top chamber, and RPMI-1640 medium containing
20% fetal bovine serum was added into the lower chambers as an
attractant and then incubated for 24 h at 37°C. The cells in upper
chambers subsequently migrated into the lower chamber. Then the
migratory cells were stained with 0.1% crystal violet for 30 min.
Images of the migration cells were photographed under a microscope
(SZ61; Olympus Corporation, Tokyo, Japan).
Western blot analysis
Total protein was extracted from the GC cells or
tissues after transfection for 48 h, and the protein concentration
was measured. Then, 50 µg protein samples in each group were
subjected to 10% SDS-PAGE to separate the protein samples. Then,
they were electrophoretically transferred to NC membrane (EMD
Millipore, Billerica, MA, USA). Subsequently, skim milk (5–10%)
dissolved by 0.1% tris buffered saline with Tween-20 (TBST) was
added to block the membranes for 2 h at room temperature. Firstly,
the membranes were incubated with the primary antibody rabbit
monoclonal anti-SALL4 (cat. no. 5850S; 1:1,000, Cell Signaling
Technology, Inc., Danvers, MA, USA) at 4°C overnight, after washed
with 1×TBST (pH 7.4) three times later; the secondary antibodies
goat anti-rabbit IgG-HRP (cat. no. sc-2004; 1:3,000; Santa Cruz
Biotechnology, Inc. Santa Cruz, CA, USA) were added and incubated
at room temperature for 2 h. Finally, the enhanced
chemiluminescence kit (ECL; EMD Millipore, Billerica, MA, USA) was
used to detect the signals. GAPDH primary antibody (cat. no. 70699;
1:5,000; Abcam, Cambridge, MA, USA) was chosen as the internal
reference.
Dual-luciferase assay
The wild-type and mut-type miR-16 putative targets
on SALL4 3′UTR were synthesized and inserted into the pMIR-reporter
luciferase vector. We used Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific Inc.) to transfect HGC-27 cells with control
mimic and miR-16 mimic. The One-Glo luciferase assay instrument
(Promega Corporation, Madison, WI, USA) was then used to measure
the luciferase activity values.
Statistical analysis
All results are presented as the mean ± SD of three
experiments. Differences between groups were evaluated by Student's
t-test or Tukey's post hoc test after ANOVA in SPSS. The difference
between groups was significant at P-value <0.05. SPSS v.19.0
software (SPSS, Inc., Chicago, IL, USA) was used to perform
statistical analyses and GraphPad Prism 5.02 (GraphPad Software,
Inc., La Jolla, CA, USA) to complete graph presentation.
Results
Increase of miR-16 and decrease of
CRKL in GC
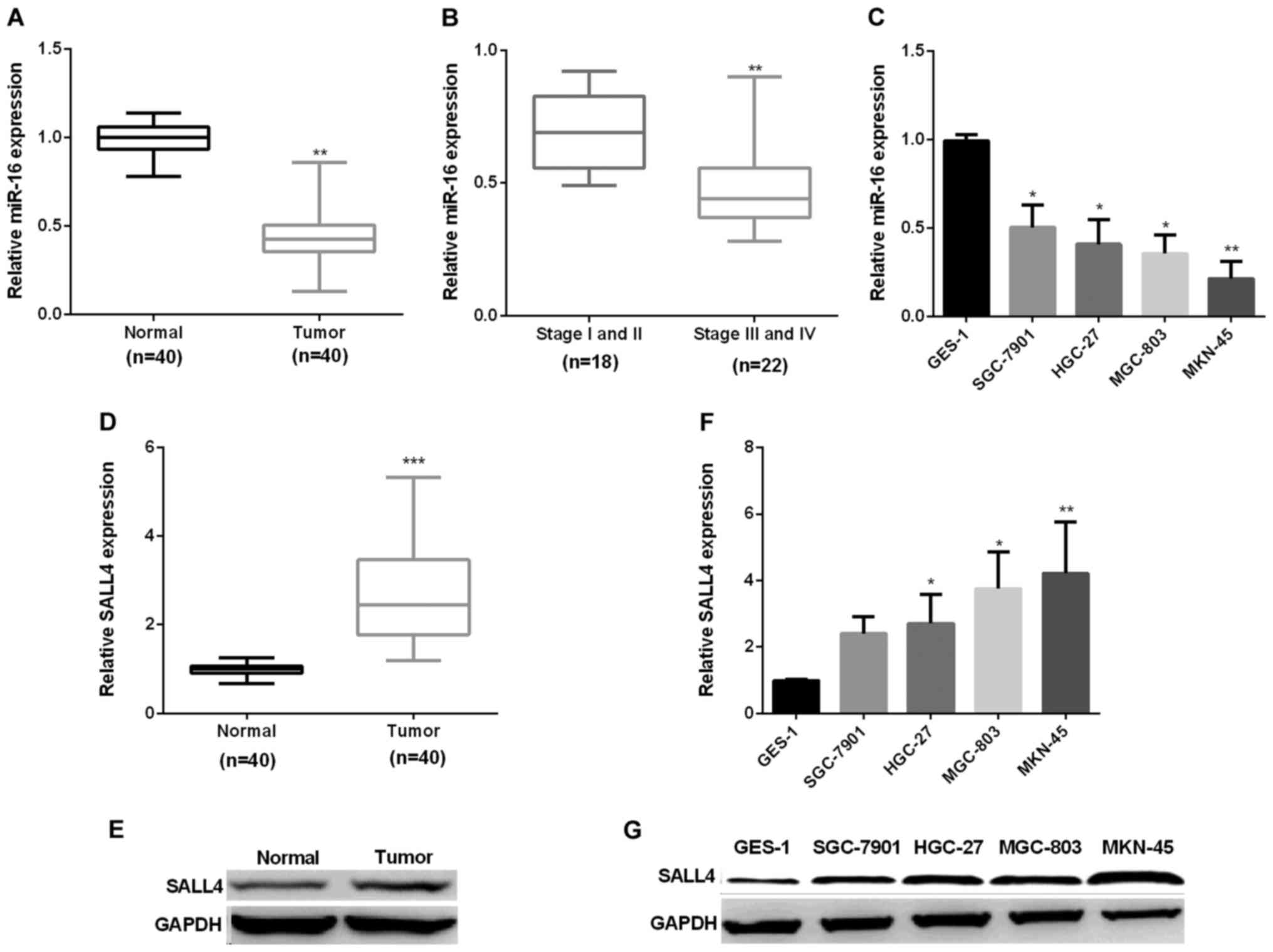
First, we examined miR-16 expression in forty pairs
of GC tissues. RT-qPCR showed that miR-16 average expression was
markedly decreased in GC tissues (Fig.
1A). Next, we assessed the correlation of miR-16 expression
level and the stage of cancer. The results indicated that miR-16
showed higher expression in stage I/II (early stage) GC tissues
than in stage III/IV (late stage) (Fig.
1B). Subsequently, we detected miR-16 mRNA expression in GC
cell lines. Compared with the normal GES-1 cells, miR-16 expression
was reduced significantly in GC cell lines (Fig. 1C). Secondly, we examined SALL4
expression in forty pairs of GC tissues and cells by RT-qPCR and
immunoblotting respectively. The results showed that SALL4
expression was markedly increased in GC in comparison with normal
(Fig. 1D and E), similar results were
seen in GC cell lines (Fig. 1F and
G).
Inhibition effect of miR-16 on GC cell
proliferation and migration
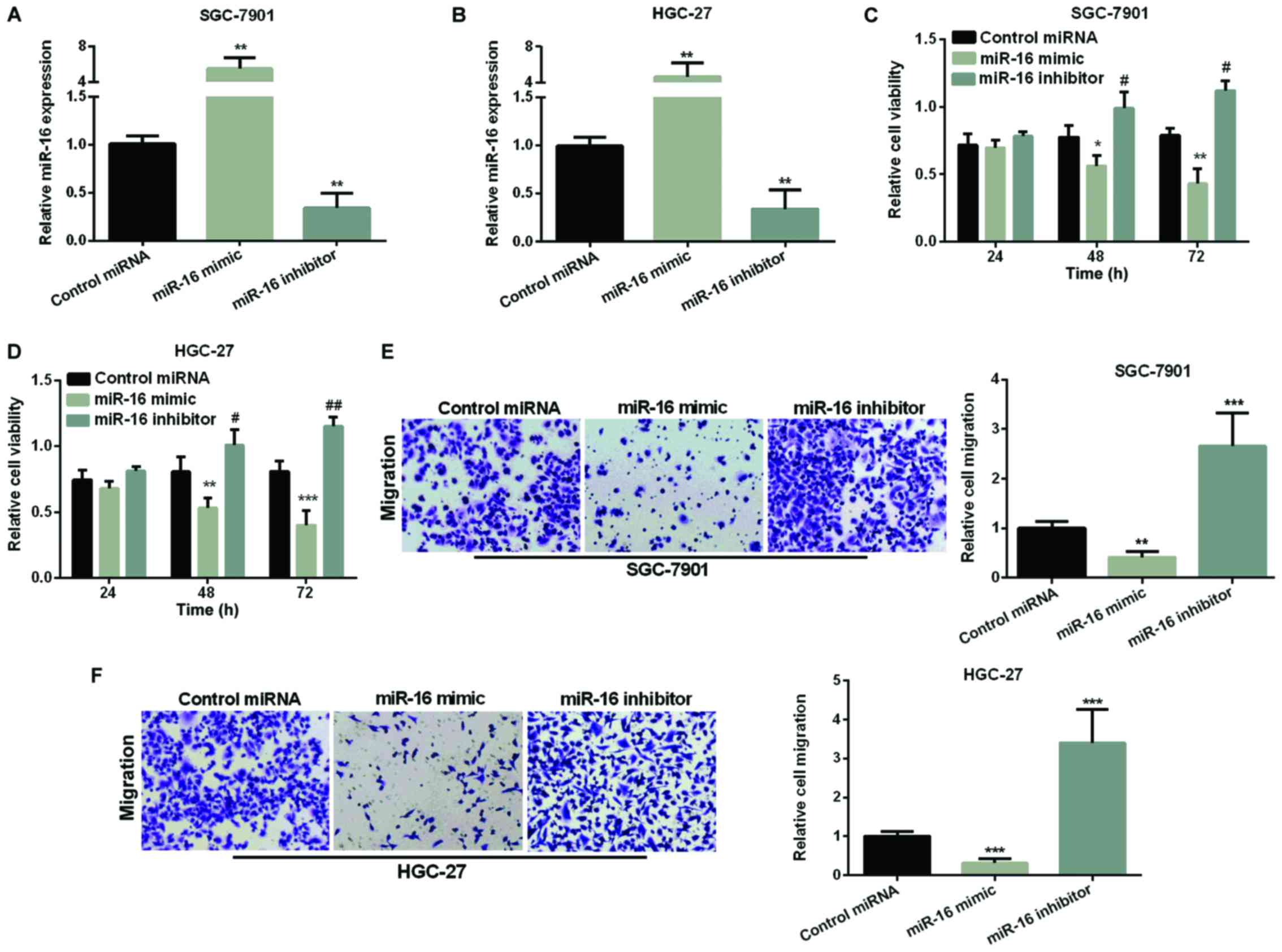
We overexpressed or silenced miR-16 by transfection
of miR-16 mimic or inhibitor into SGC-7901 and HGC-27 cells. The
efficiency of the miR-16 transfection was assessed by RT-qPCR and
found that miR-16 expression was obviously higher in both GC cells
after overexpression of miR-16 but was decreased after silencing
miR-16 compared with the control (Fig. 2A
and B). We used MTT assay to measure miR-16 effect on GC cell
proliferation. As Fig. 2C and D show,
re-expression of miR-16 made cell viability reduced in both GC cell
lines, while, inhibiting miR-16 significantly raised cell
viability. Next, we used Transwell assay to examine miR-16's effect
on GC cell migration. As seen in Fig. 2E
and F, miR-16 re-expression significantly reduced the migration
cells in both GC cell lines, whereas, miR-16 silencing increased
the migration of cells remarkably.
SALL4 silencing inhibits GC cell
viability and migration
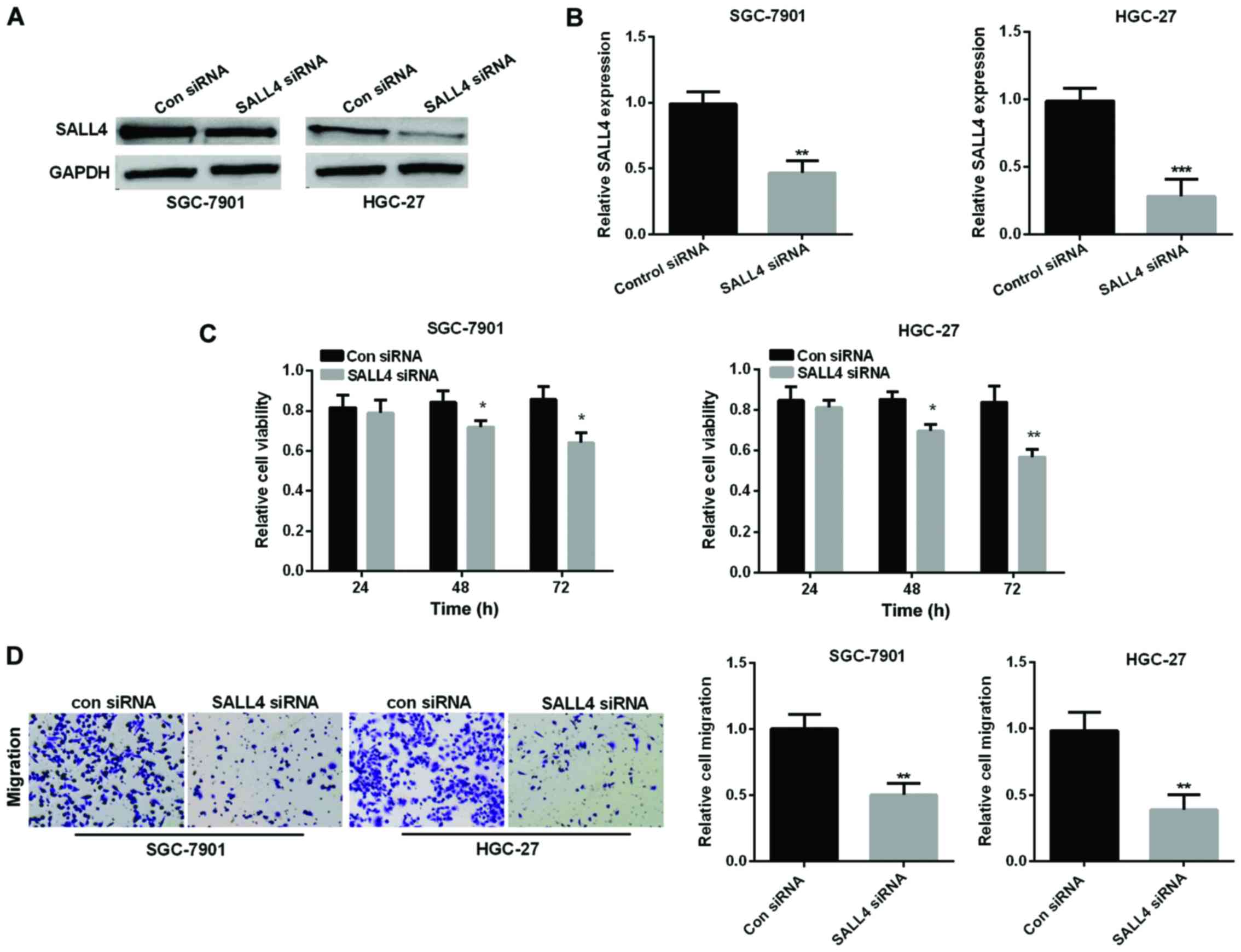
SALL4 siRNA was performed to knock down SALL4
expression to examine SALL4 function in GC progression. Relative
SALL4 expression was detected by western blot analysis and RT-qPCR
in SGC-7901 and HGC-27 cell lines after transfected with small
interfering RNA, respectively, as shown in Fig. 3A and B, the corresponding SALL4
protein expression and mRNA expression was significantly reduced by
downregulation of SALL4 in SGC-7901 and HGC-27 cell lines. Then, we
used MTT assay to investigate the cell viability in GC cell lines
to explore SALL4 effect on GC cell proliferation. We found that
si-SALL4 suppressed cell viability in both GC cell lines (Fig. 3C). Transwell assay revealed that
si-SALL4 curbed cell migration also in the GC cell lines (Fig. 3D).
SALL4 is a specific target of miR-16
in GC development
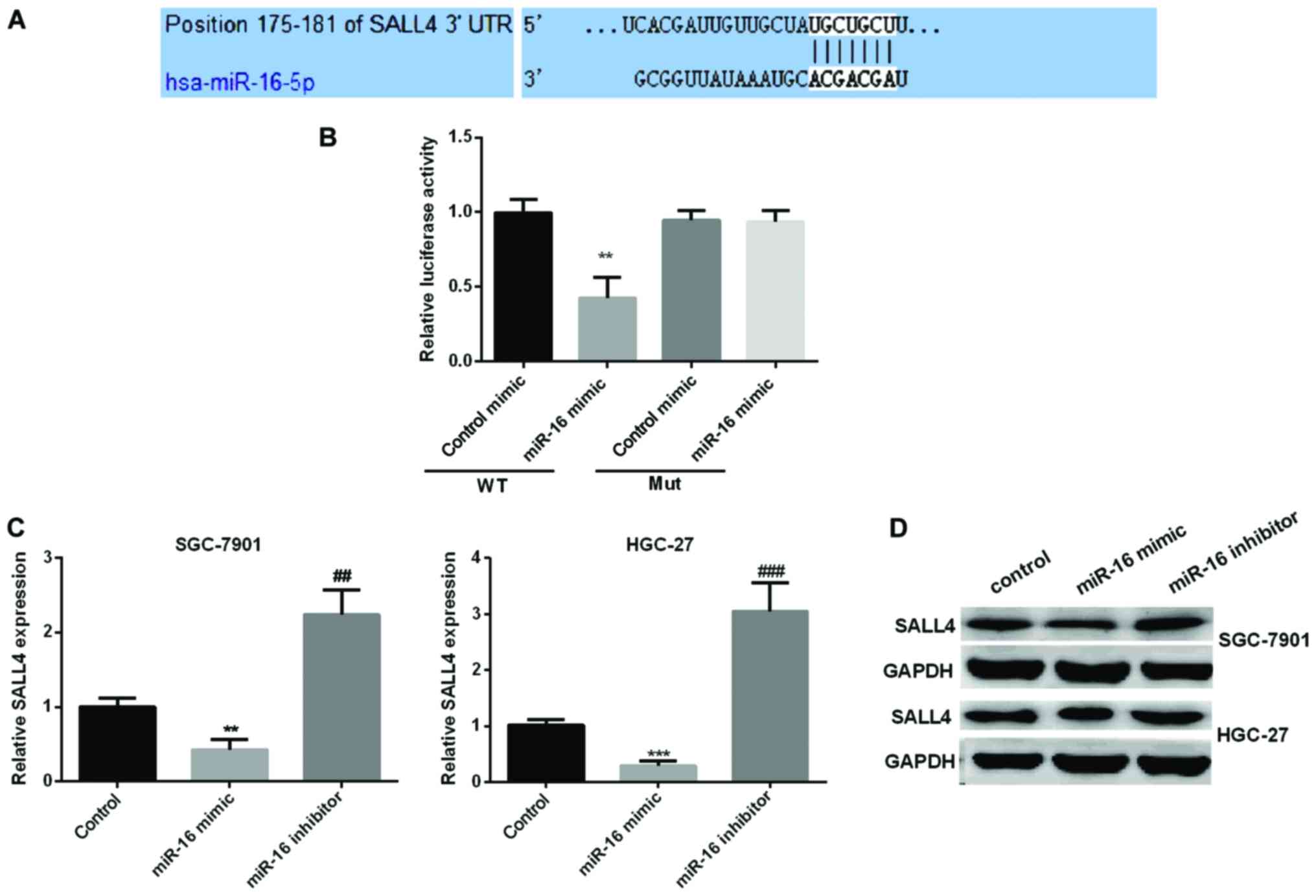
We used TargetScan algorithms to look for possible
targets of miR-16. Based on its important role in the process of
cell proliferation and migration, we selected SALL4 for further
study. To corroborate the hypothesis that SALL4 was a novel target
of miR-16 in GC progression, dual-luciferase reporter assay was
carried out to check the luciferase activity of HGC-27 cells
treated with miR-16 mimic. The results indicated that miR-16 mimic
significantly reduced the relative SALL4 luciferase activity in
wild-type, however, there were no changed in mut-type (Fig. 4A and B). We then explored the
connection between miR-16 and SALL4 expression. RT-qPCR and
immunoblotting were carried out to detect SALL4 expression in the
GC cell lines by re-expression or knockdown of miR-16. As seen in
Fig. 4C and D, the relative SALL4
mRNA and protein expression was reduced observably in miR-16 mimic
group, while increased in miR-16 inhibitor group.
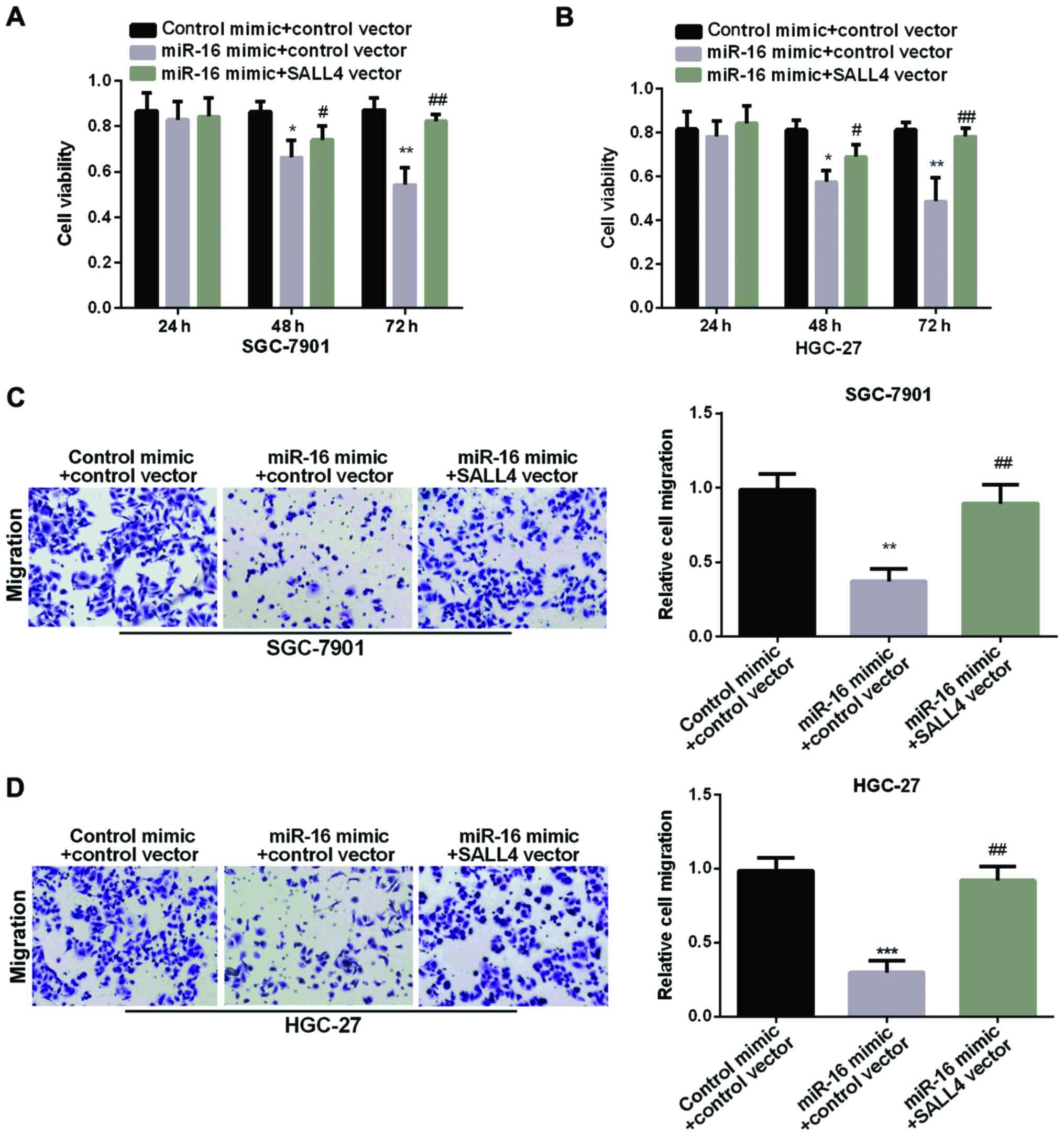
The reversal of SALL4 in miR-16
suppression effect in GC
We carried out MTT and Transwell assay to examine
SALL4 function in GC cell proliferation and migration regulated by
miR-16. As proved above, the miR-16 mimic group showed decreased
cell viability. However, re-expression of both miR-16 and SALL4
showed higher cell viability than cell overexpression of miR-16
alone (Fig. 5A and B), suggesting
that SALL4 attenuated the inhibition effect of miR-16 on GC cell
proliferation. In addition, Fig. 5C and
D results showed that the relative cell migration in GC cells
was decreased in miR-16 mimic group. However, re-expression of both
miR-16 and SALL4 showed higher migration than cell overexpression
of miR-16 alone, suggesting that SALL4 attenuated miR-16 inhibition
effect on GC cell migration. In conclusion, miR-16 could inhibit GC
cell proliferation and migration by targeting SALL4.
Discussion
Previous studies have shown that expression of
several miRNAs was abnormal in gastric cancer (GC) which in turn
induced the changed cell proliferation, invasion and apoptosis
(27). Thus, these miRNAs could be
used as biomarkers to predict the prognosis of GC and search for a
specific miRNA and its target gene is critical.
miR-16 had been proved to be expressed abnormally in
a variety of human cancers. It was reported that miR-16 was
obviously reduced in pituitary tumors (28). Moreover, miR-16 was reported to be
decreased in chronic lymphocytic leukemia cells and targeted Bcl-2
to induce cell apoptosis (29).
Recently a study showed that miR-16 was expressed abnormally in GC
development and progression (3,30). Our
study stated an observably reduced miR-16 expression in GC, and
miR-16 mimic suppressed GC cell proliferation and migration, while
miR-127 inhibitor facilitated it. It was in line with the recent
studies that miR-16 was downregulated in GC and it could inhibit GC
cell progression (31,32).
SALL4 is well known to be involved in progression of
many human cancers, including colorectal cancer, breast cancer,
liver cancer and lung cancer (20,33–35), by
regulating cell growth, metastasis and invasion. SALL4 expression
detected in our experiment was obviously higher in GC consistent
with reports that SALL4 was upregulated in GC (24,36). A
previous study also showed that SALL4 promoted cell proliferation
and metastasis regulated by the miR-33b, and miR-33b exhibited
significant inverse correlation with SALL4 in hepatocellular
carcinoma cells (37). Zhou et
al (38) found that SALL4
expression was directly regulated by miR-16 in glioma cell
proliferation, migration and invasion. Our present study indicated
that SALL4 expression increased in GC and silencing SALL4 could
inhibit GC cell viability and migratory ability.
Colletively, miR-16 expression was downregulated
while SALL4 was upregulated in GC. The relationship between miR-16
and SALL4 expression was negatively correlated. We first proved
that SALL4 was a directly target of miR-16 in regulation of the
progress of GC and SALL4 could partially reverse the suppression
effect of miR-16 in GC, indicating miR-16/SALL4 axis to have a
potential application vlue in GC diagnosis and therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XJ collected and analyzed the data, interpreted the
data and drafted the manuscript. ZW conceived and designed this
study, finally revised and approved the manuscript. Both authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
China-Japan Union Hospital, Jilin University (Changchun, China).
Signed informed consents were obtained from the patients or the
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng L, Jiao W, Song H, Qu H, Li D, Mei
H, Chen Y, Yang F, Li H, Huang K, et al: miRNA-558 promotes gastric
cancer progression through attenuating Smad4-mediated repression of
heparanase expression. Cell Death Dis. 7:e23822016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma
Y, Chen Y, Pan F, Wang K, Ni J, et al: Circulating miR-16-5p and
miR-19b-3p as two novel potential biomarkers to indicate
progression of gastric cancer. Theranostics. 5:733–745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi
HC, Lin YH, Lu PH and Lin KH: Potential diagnostic, prognostic and
therapeutic targets of microRNAs in human gastric cancer. Int J Mol
Sci. 17:9452016. View Article : Google Scholar
|
|
5
|
An Y, Zhang Z, Shang Y, Jiang X, Dong J,
Yu P, Nie Y and Zhao Q: miR-23b-3p regulates the chemoresistance of
gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.
6:e17662015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hurst DR, Edmonds MD and Welch DR:
Metastamir: The field of metastasis-regulatory microRNA is
spreading. Cancer Res. 69:7495–7498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ahn SM, Cha JY, Kim J, Kim D, Trang HT,
Kim YM, Cho YH, Park D and Hong S: Smad3 regulates E-cadherin via
miRNA-200 pathway. Oncogene. 31:3051–3059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhan XH, Xu QY, Tian R, Yan H, Zhang M, Wu
J, Wang W and He J: MicroRNA16 regulates glioma cell proliferation,
apoptosis and invasion by targeting Wip1-ATM-p53 feedback loop.
Oncotarget. 8:54788–54798. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Chen J, Dai J, Zhang B, Wang F and
Sun Y: MicroRNA-16-1 inhibits tumor cell proliferation and induces
apoptosis in A549 non-small cell lung carcinoma cells. Oncol Res.
24:345–351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diamantopoulos MA, Kontos CK, Kerimis D,
Papadopoulos IN and Scorilas A: Upregulated miR-16 expression is an
independent indicator of relapse and poor overall survival of
colorectal adenocarcinoma patients. Clin Chem Lab Med. 55:737–747.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tatetsu H, Kong NR, Chong G, Amabile G,
Tenen DG and Chai L: SALL4, the missing link between stem cells,
development and cancer. Gene. 584:111–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warren M, Wang W, Spiden S, Chen-Murchie
D, Tannahill D, Steel KP and Bradley A: A Sall4 mutant mouse model
useful for studying the role of Sall4 in early embryonic
development and organogenesis. Genesis. 45:51–58. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Rao S, Chu J, Shen X, Levasseur
DN, Theunissen TW and Orkin SH: A protein interaction network for
pluripotency of embryonic stem cells. Nature. 444:364–368. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY,
Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al: Sall4 modulates
embryonic stem cell pluripotency and early embryonic development by
the transcriptional regulation of Pou5f1. Nat Cell Biol.
8:1114–1123. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Q, Chen X, Zhang J, Loh YH, Low TY,
Zhang W, Zhang W, Sze SK, Lim B and Ng HH: Sall4 interacts with
Nanog and co-occupies Nanog genomic sites in embryonic stem cells.
J Biol Chem. 281:24090–24094. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oikawa T, Kamiya A, Zeniya M, Chikada H,
Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et
al: Sal-like protein 4 (SALL4), a stem cell biomarker in liver
cancers. Hepatology. 57:1469–1483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
19
|
Kobayashi D, Kuribayshi K, Tanaka M and
Watanabe N: SALL4 is essential for cancer cell proliferation and is
overexpressed at early clinical stages in breast cancer. Int J
Oncol. 38:933–939. 2011.PubMed/NCBI
|
|
20
|
Khales Ardalan S, Abbaszadegan MR,
Abdollahi A, Raeisossadati R, Tousi MF and Forghanifard MM: SALL4
as a new biomarker for early colorectal cancers. J Cancer Res Clin
Oncol. 141:229–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Itou J, Matsumoto Y, Yoshikawa K and Toi
M: Sal-like 4 (SALL4) suppresses CDH1 expression and maintains cell
dispersion in basal-like breast cancer. FEBS Lett. 587:3115–3121.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan
B, Srivastava S, Lim GS, Tang P, Yang H, et al: SALL4 is a new
target in endometrial cancer. Oncogene. 34:63–72. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Yuan X, Zhu W, Qian H and Xu W:
SALL4: An emerging cancer biomarker and target. Cancer Lett.
357:55–62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang
M, Zhang X, Yang T, Cai J, Yan Y, et al: SALL4, a novel marker for
human gastric carcinogenesis and metastasis. Oncogene.
33:5491–5500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan X, Zhang X, Zhang W, Liang W, Zhang
P, Shi H, Zhang B, Shao M, Yan Y, Qian H, et al: SALL4 promotes
gastric cancer progression through activating CD44 expression.
Oncogenesis. 5:e2682016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ishiguro H, Kimura M and Takeyama H: Role
of microRNAs in gastric cancer. World J Gastroenterol.
20:5694–5699. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amaral FC, Torres N, Saggioro F, Neder L,
Machado HR, Silva WA Jr, Moreira AC and Castro M: MicroRNAs
differentially expressed in ACTH-secreting pituitary tumors. J Clin
Endocrinol Metab. 94:320–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Venturutti L, Russo Cordo RI, Rivas MA,
Mercogliano MF, Izzo F, Oakley RH, Pereyra MG, De Martino M,
Proietti CJ, Yankilevich P, et al: MiR-16 mediates trastuzumab and
lapatinib response in ErbB-2-positive breast and gastric cancer via
its novel targets CCNJ and FUBP1. Oncogene. 35:6189–6202. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang W, Tong JH, Lung RW, Dong Y, Zhao J,
Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al: Targeting of YAP1
by microRNA-15a and microRNA-16-1 exerts tumor suppressor function
in gastric adenocarcinoma. Mol Cancer. 14:522015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang T, Hou J, Li Z, Zheng Z, Wei J, Song
D, Hu T, Wu Q, Yang JY and Cai JC: miR-15a-3p and miR-16-1-3p
negatively regulate Twist1 to repress gastric cancer cell invasion
and metastasis. Int J Biol Sci. 13:122–134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gautam AK, Wang C, Zeng J, Wang J, Lu J,
Wei J, Huang G and Mo B, Luo M and Mo B: Expression and clinical
significance of SALL4 and LGR5 in patients with lung cancer. Oncol
Lett. 10:3629–3634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tanaka Y, Aishima S, Kohashi K, Okumura Y,
Wang H, Hida T, Kotoh K, Shirabe K, Maehara Y, Takayanagi R, et al:
Spalt-like transcription factor 4 immunopositivity is associated
with epithelial cell adhesion molecule expression in combined
hepatocellular carcinoma and cholangiocarcinoma. Histopathology.
68:693–701. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dirican E and Akkiprik M: Functional and
clinical significance of SALL4 in breast cancer. Tumour Biol.
37:11701–11709. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu J, Wang L, Yang A, Jiang P and Wang M:
Up-regulation of SALL4 associated with poor prognosis in gastric
cancer. Hepatogastroenterology. 61:1459–1464. 2014.PubMed/NCBI
|
|
37
|
Tian Q, Xiao Y, Wu Y, Liu Y, Song Z, Gao
W, Zhang J, Yang J, Zhang Y, Guo T, et al: MicroRNA-33b suppresses
the proliferation and metastasis of hepatocellular carcinoma cells
through the inhibition of Sal-like protein 4 expression. Int J Mol
Med. 38:1587–1595. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Liu Y, Hu C and Jiang Y:
MicroRNA-16 inhibits the proliferation, migration and invasion of
glioma cells by targeting Sal-like protein 4. Int J Mol Med.
38:1768–1776. 2016. View Article : Google Scholar : PubMed/NCBI
|