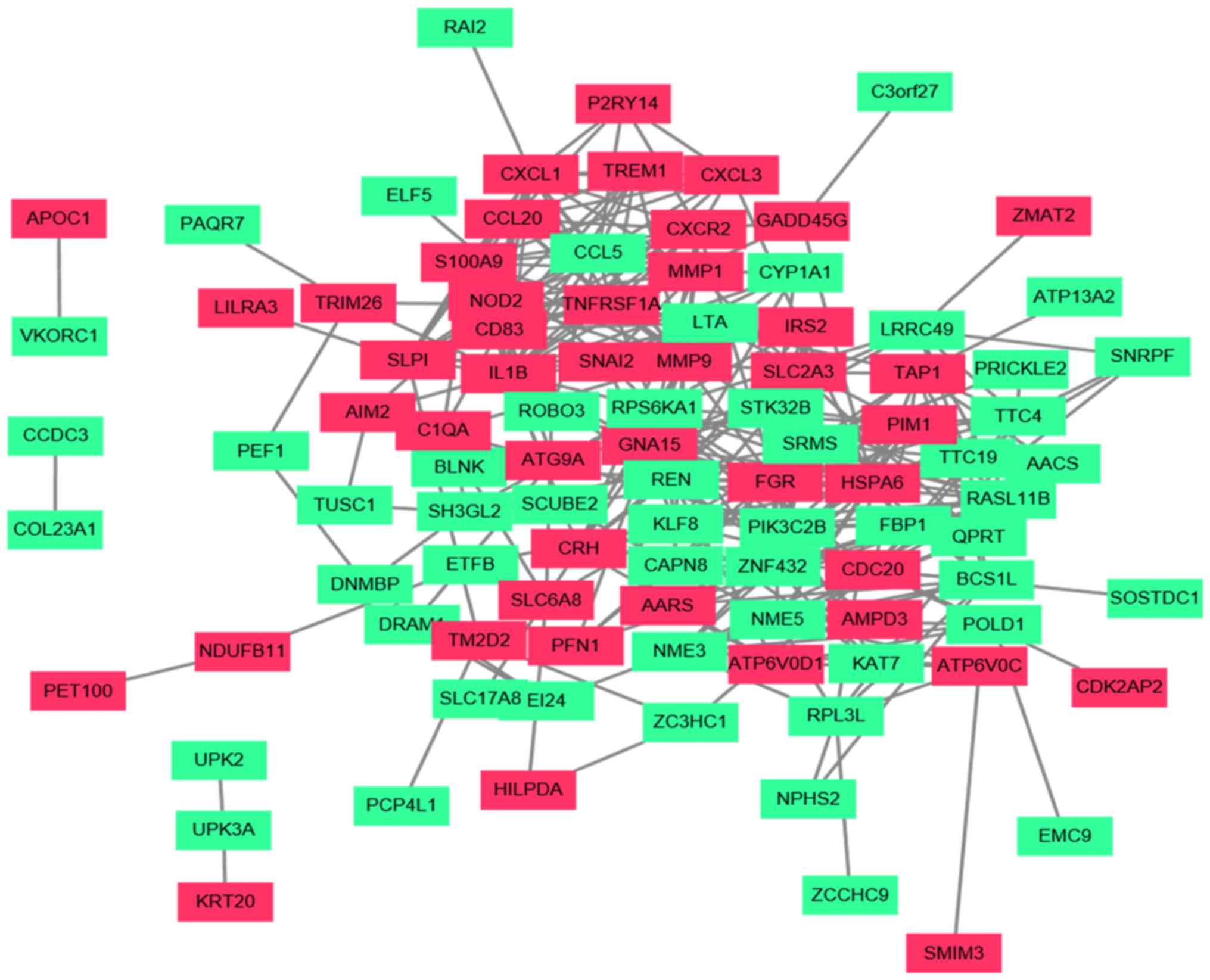
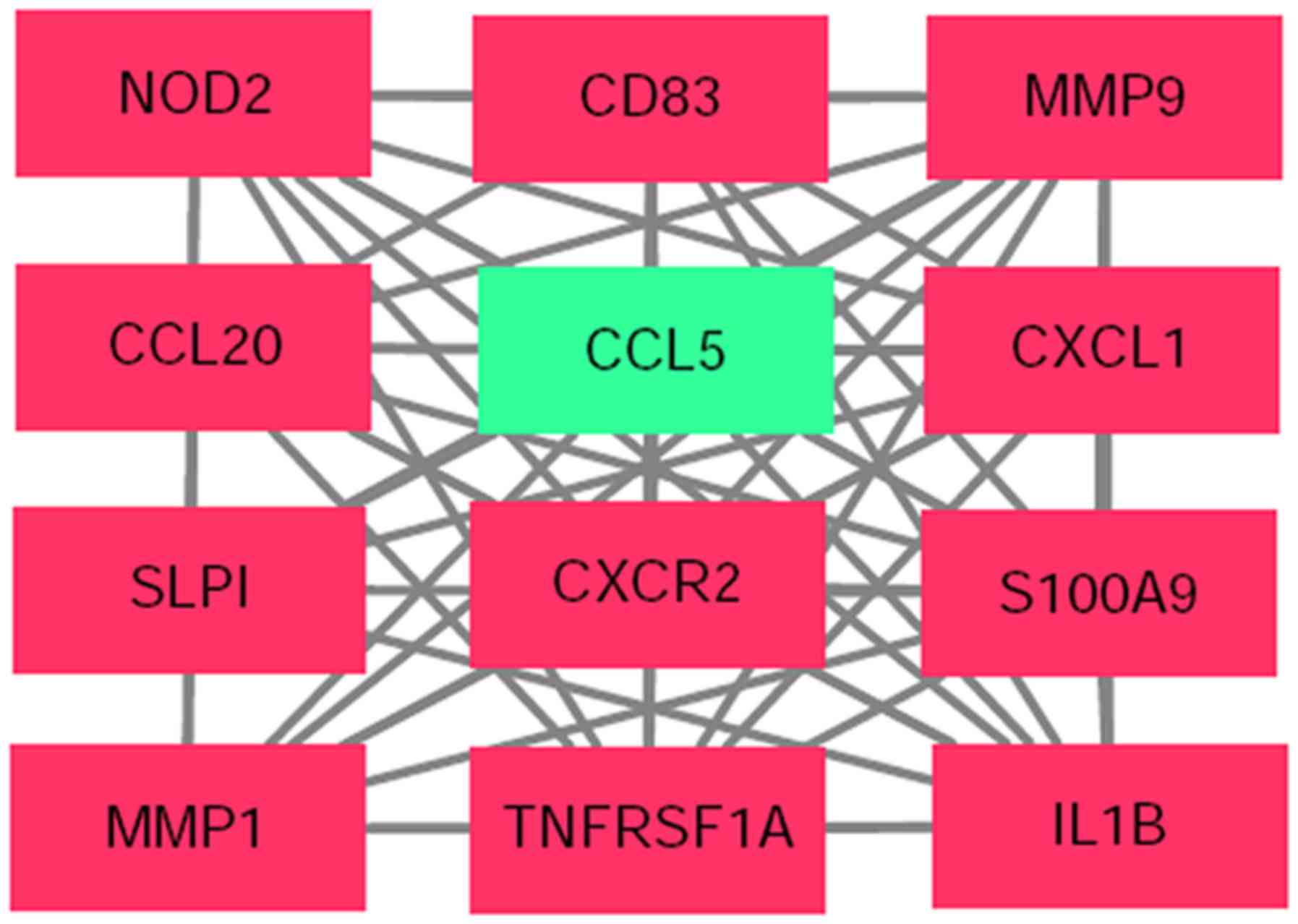
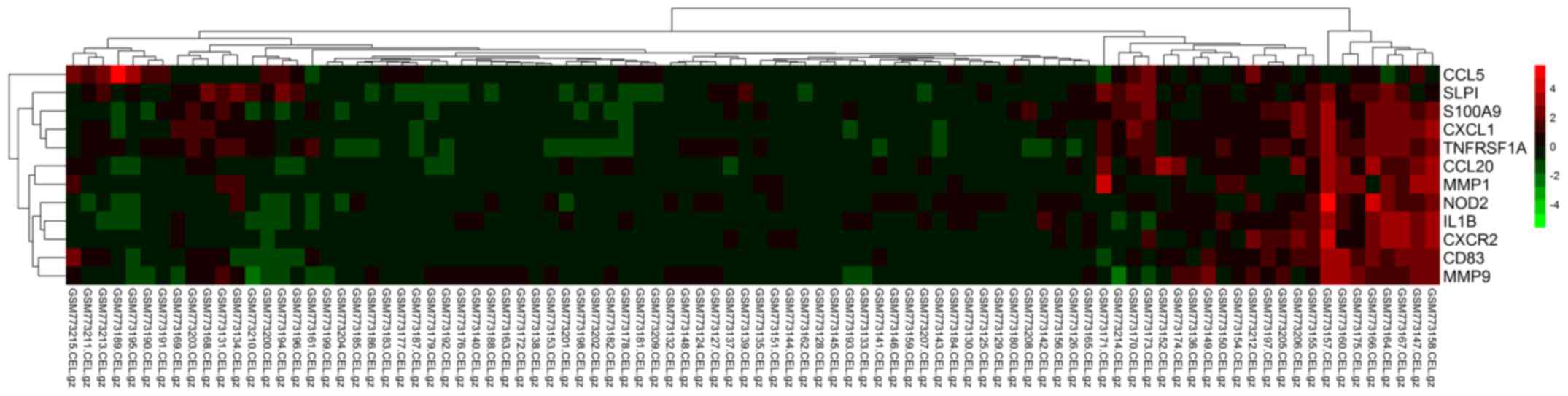
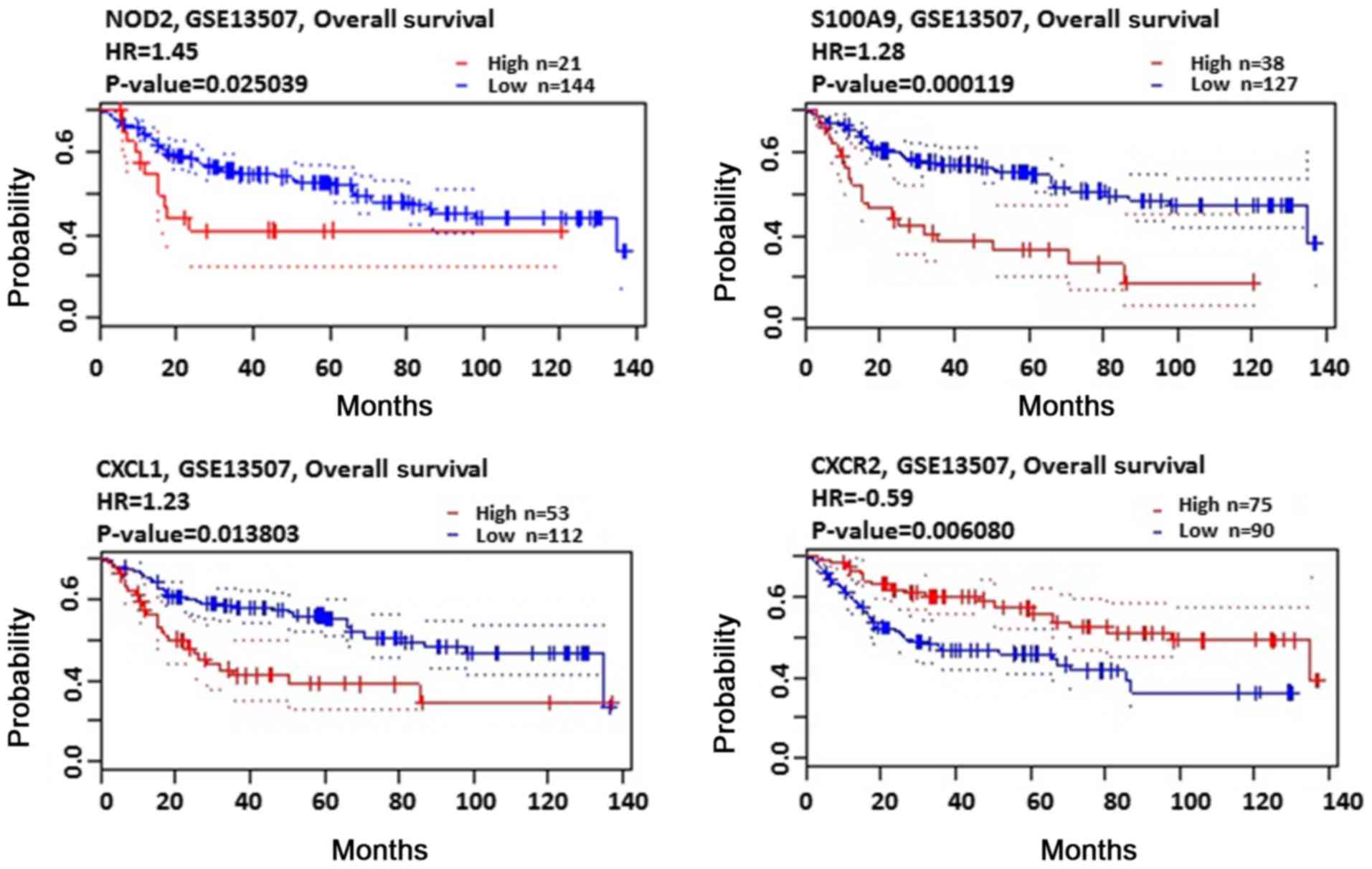
|
1
|
Hao L, Zhao Y, Li ZG, He HG, Liang Q,
Zhang ZG, Shi ZD, Zhang PY and Han CH: Tumor necrosis
factor-related apoptosis-inducing ligand inhibits proliferation and
induces apoptosis of prostate and bladder cancer cells. Oncol Lett.
13:3638–3640. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Ma X, Tang L, Wang B, Chen L, Zhang
F and Zhang X: Prognostic value of neutrophil-to-lymphocyte ratio
in urothelial carcinoma of the upper urinary tract and bladder: A
systematic review and meta-analys. Oncotarget. 8:62681–62692.
2016.PubMed/NCBI
|
|
3
|
Peng M, Huang Y, Tao T, Peng CY, Su Q, Xu
W, Darko KO, Tao X and Yang X: Metformin and gefitinib cooperate to
inhibit bladder cancer growth via both AMPK and EGFR pathways
joining at Akt and Erk. Sci Rep. 6:286112016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO,
Kim KH, Lee JK, Kim SW and Kim YT: The long noncoding RNA HOXA11
antisense induces tumor progression and stemness maintenance in
cervical cancer. Oncotarget. 7:83001–83016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo G, Wang M, Wu X, Tao D, Xiao X, Wang
L, Min F, Zeng F and Jiang G: Long non-coding RNA meg3 inhibits
cell proliferation and induces apoptosis in prostate cancer. Cell
Physiol Biochem. 37:2209–2220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cui X, Jing X, Long C, Yi Q, Tian J and
Zhu J: Accuracy of the urine UCA1 for diagnosis of bladder cancer:
A meta-analysis. Oncotarget. 8:35222–35233. 2017.PubMed/NCBI
|
|
8
|
Cui X, Jing X and Wu X: The prognostic
value of long non coding RNAs in cervical cancer: A meta-analysis.
Oncotarget. 8:62470–62477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing X, Liang H, Cui X, Han C, Hao C and
Huo K: Long noncoding RNA CCAT2 can predict metastasis and a poor
prognosis: A meta-analysis. Clin Chim Acta. 468:159–165. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui X, Jing X, Yi Q, Long C, Tan B, Li X,
Chen X, Huang Y, Xiang Z, Tian J and Zhu J: Systematic analysis of
gene expression alterations and clinical outcomes of STAT3 in
cancer. Oncotarget. 9:3198–3213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui X, Jing X, Yi Q, Long C, Tian J and
Zhu J: Clinicopathological and prognostic significance of SDC1
overexpression in breast cancer. Oncotarget. 8:111444–111455. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo Y, Sheng Q, Li J, Ye F, Samuels DC and
Shyr Y: Large scale comparison of gene expression levels by
microarrays and RNAseq using TCGA data. PLoS One. 8:e714622013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Senchenko VN, Kisseljova NP, Ivanova TA,
Dmitriev AA, Krasnov GS, Kudryavtseva AV, Panasenko GV, Tsitrin EB,
Lerman MI, Kisseljov FL, et al: Novel tumor suppressor candidates
on chromosome 3 revealed by NotI-microarrays in cervical cancer.
Epigenetics. 8:409–420. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen G, Li Y, Su Y, Zhou L, Zhang H, Shen
Q, Du C, Li H, Wen Z, Xia Y and Tang W: Identification of candidate
genes for necrotizing enterocolitis based on microarray data. Gene.
661:152–159. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shu B, Fang Y, He W, Yang J and Dai C:
Identification of macrophage-related candidate genes in lupus
nephritis using bioinformatics analysis. Cell Signal. 46:43–51.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shao K, Shen LS, Li HH, Huang S and Zhang
Y: Systematic-analysis of mRNA expression profiles in skeletal
muscle of patients with type II diabetes: The glucocorticoid was
central in pathogenesis. J Cell Physiol. 233:4068–4076. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Ding J, Wang Z, Zhu J, Wang X and
Du J: Identification of downstream metastasis-associated target
genes regulated by LSD1 in colon cancer cells. Oncotarget.
8:19609–19630. 2017.PubMed/NCBI
|
|
18
|
Urquidi V, Goodison S, Cai Y, Sun Y and
Rosser CJ: A candidate molecular biomarker panel for the detection
of bladder cancer. Cancer Epidemiol Biomarkers Prev. 21:2149–2158.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gene Ontology Consortium, . The Gene
Ontology (GO) project in 2006. Nucleic Acids Res. 34:(Database
Issue). D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The Gene Ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:(Database Issue). D258–D261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36:(Database Issue). D480–D484.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wishart DS, Knox C, Guo AC, Shrivastava S,
Hassanali M, Stothard P, Chang Z and Woolsey J: DrugBank: A
comprehensive resource for in silico drug discovery and
exploration. Nucleic Acids Res. 34:(Database Issue). D668–D672.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui X, Jing X, Long C, Tian J and Zhu J:
Long noncoding RNA MEG3, a potential novel biomarker to predict the
clinical outcome of cancer patients: A meta-analysis. Oncotarget.
8:19049–19056. 2017.PubMed/NCBI
|
|
32
|
Gnesin S, Mitsakis P, Cicone F, Deshayes
E, Dunet V, Gallino AF, Kosinski M, Baechler S, Buchegger F, Viertl
D and Prior JO: First in-human radiation dosimetry of
68Ga-NODAGA-RGDyK. EJNMMI Res. 7:432017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cohen T, Ricchiuti D and Memo M: Bladder
cancer that metastasized to the skin: A unique presentation that
signifies poor prognosis. Rev Urol. 19:67–71. 2017.PubMed/NCBI
|
|
34
|
Zhang Y, Fang L, Zang Y and Xu Z:
Identification of core genes and key pathways via integrated
analysis of gene expression and DNA methylation profiles in bladder
cancer. Med Sci Monit. 24:3024–3033. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, He C, Xu Q, Xing C and Yuan Y: NOD2
polymorphisms associated with cancer risk: A meta-analysis. PLoS
One. 9:e893402014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang X, Yang C, Liao X, Han C, Yu T, Huang
K, Yu L, Qin W, Zhu G, Su H, et al: NLRC and NLRX gene family mRNA
expression and prognostic value in hepatocellular carcinoma. Cancer
Med. 6:2660–2672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Duan L, Wu R, Ye L, Wang H, Yang X, Zhang
Y, Chen X, Zuo G, Zhang Y, Weng Y, et al: S100A8 and S100A9 are
associated with colorectal carcinoma progression and contribute to
colorectal carcinoma cell survival and migration via Wnt/β-catenin
pathway. PLoS One. 8:e620922013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Källberg E, Vogl T, Liberg D, Olsson A,
Björk P, Wikström P, Bergh A, Roth J, Ivars F and Leanderson T:
S100A9 interaction with TLR4 promotes tumor growth. PLoS One.
7:e342072012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
De Veirman K, De Beule N, Maes K, Menu E,
De Bruyne E, De Raeve H, Fostier K, Moreaux J, Kassambara A, Hose
D, et al: Extracellular S100A9 protein in bone marrow supports
multiple myeloma survival by stimulating angiogenesis and cytokine
secretion. Cancer Immunol Res. 5:839–846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yasar O, Akcay T, Obek C and Turegun FA:
Significance of S100A8, S100A9 and calprotectin levels in bladder
cancer. Scand J Clin Lab Invest. 77:437–441. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu P, Quan H, Kang J, He J, Luo S, Xie C,
Xu J, Tang Y and Zhao S: Downregulation of calcium-binding protein
S100A9 inhibits hypopharyngeal cancer cell proliferation and
invasion ability through inactivation of NF-κB signaling. Oncol
Res. 25:1479–1488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Q, Li X, Chen H, Cao Y, Xiao Q, He Y,
Wei J and Zhou J: IRF7 regulates the development of granulocytic
myeloid-derived suppressor cells through S100A9 transrepression in
cancer. Oncogene. 36:2969–2980. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lim SY, Yuzhalin AE, Gordon-Weeks AN and
Muschel RJ: Tumor-infiltrating monocytes/macrophages promote tumor
invasion and migration by upregulating S100A8 and S100A9 expression
in cancer cells. Oncogene. 35:5735–5745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang D, Yang W, Du J, Devalaraja MN, Liang
P, Matsumoto K, Tsubakimoto K, Endo T and Richmond A:
MGSA/GRO-mediated melanocyte transformation involves induction of
Ras expression. Oncogene. 19:4647–4659. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Verbeke H, Struyf S, Laureys G and Van
Damme J: The expression and role of CXC chemokines in colorectal
cancer. Cytokine Growth Factor Rev. 22:345–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyake M, Lawton A, Goodison S, Urquidi V,
Gomes-Giacoia E, Zhang G, Ross S, Kim J and Rosser CJ: Chemokine
(C-X-C) ligand 1 (CXCL1) protein expression is increased in
aggressive bladder cancers. BMC Cancer. 13:3222013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han KQ, Han H, He XQ, Wang L, Guo XD,
Zhang XM, Chen J, Zhu QG, Nian H, Zhai XF and Jiang MW: Chemokine
CXCL1 may serve as a potential molecular target for hepatocellular
carcinoma. Cancer Med. 5:2861–2871. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Z, Wang Z, Li G, Wu H, Sun K, Chen J,
Feng Y, Chen C, Cai S, Xu J and He Y: CXCL1 from tumor-associated
lymphatic endothelial cells drives gastric cancer cell into
lymphatic system via activating integrin β1/FAK/AKT signaling.
Cancer Lett. 385:28–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kawanishi H, Matsui Y, Ito M, Watanabe J,
Takahashi T, Nishizawa K, Nishiyama H, Kamoto T, Mikami Y, Tanaka
Y, et al: Secreted CXCL1 is a potential mediator and marker of the
tumor invasion of bladder cancer. Clin Cancer Res. 14:2579–2587.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cong L, Qiu ZY, Zhao Y, Wang WB, Wang CX,
Shen HC and Han JQ: Loss of β-arrestin-2 and activation of CXCR2
correlate with lymph node metastasis in non-small cell lung cancer.
J Cancer. 8:2785–2792. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Devapatla B, Sharma A and Woo S: CXCR2
inhibition combined with sorafenib improved antitumor and
antiangiogenic response in preclinical models of ovarian cancer.
PLoS One. 10:e01392372015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang T, Tseng C, Zhang Y, Sirin O, Corn
PG, Li-Ning-Tapia EM, Troncoso P, Davis J, Pettaway C, Ward J, et
al: CXCL1 mediates obesity-associated adipose stromal cell
trafficking and function in the tumour microenvironment. Nat
Commun. 7:116742016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ignacio RM, Kabir SM, Lee ES, Adunyah SE
and Son DS: NF-κB-mediated CCL20 reigns dominantly in CXCR2-driven
ovarian cancer progression. PLoS One. 11:e01641892016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhao J, Ou B, Feng H, Wang P, Yin S, Zhu
C, Wang S, Chen C, Zheng M, Zong Y, et al: Overexpression of CXCR2
predicts poor prognosis in patients with colorectal cancer.
Oncotarget. 8:28442–28454. 2017.PubMed/NCBI
|
|
55
|
Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo
L, Shao Z and Li X: miR-940 suppresses tumor cell invasion and
migration via regulation of CXCR2 in hepatocellular carcinoma.
Biomed Res Int. 2016:76183422016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu H, Lin F, Wang Z, Yang L, Meng J, Ou Z,
Shao Z, Di G and Yang G: CXCR2 promotes breast cancer metastasis
and chemoresistance via suppression of AKT1 and activation of COX2.
Cancer Lett. 412:69–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Singh JK, Farnie G, Bundred NJ, Simões BM,
Shergill A, Landberg G, Howell SJ and Clarke RB: Targeting CXCR1/2
significantly reduces breast cancer stem cell activity and
increases the efficacy of inhibiting HER2 via HER2-dependent and
-independent mechanisms. Clin Cancer Res. 19:643–656. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan
J, Wang X and Li L: CXCL5/CXCR2 axis promotes bladder cancer cell
migration and invasion by activating PI3K/AKT-induced upregulation
of MMP2/MMP9. Int J Oncol. 47:690–700. 2015. View Article : Google Scholar : PubMed/NCBI
|