Introduction
Ovarian cancer is the second most common tumor of
the female reproductive system, with the highest mortality rate
(52.26%), United States in 2014 (1).
The American Cancer Society estimates that ~22,440 females in the
USA were diagnosed with ovarian cancer and ~14,080 succumbed to the
disease in 2017 (1). Epithelial
ovarian cancer (EOC), one of the three major gynecological
malignancy types, accounting for 85–90% of malignant ovarian tumors
and having the highest mortality rate (1). Due to the ovaries being deep in the
pelvic cavity and there being no notable clinical symptoms in the
early stages of ovarian cancer, the majority of patients with EOC
have an advanced disease stage or distant metastasis at the time of
diagnosis, resulting in limited treatment options and a 5-year
survival rate of <15% (2);
therefore, it is vital to improve the rate of early detection.
Currently, ovarian cancer is predominately diagnosed in clinical
practice through pelvic examination, laparoscopy, the detection of
serum cancer antigen 125 (CA125) (3),
human epididymis secretory protein 4 (4) and other biomarkers, and imaging
examination via computed tomography (CT), magnetic resonance
imagining (MRI) and dynamic contrast enhanced (DCE)-MRI (5); however, the low diagnostic sensitivity
and specificity of these methods has limited their efficacy in the
early diagnosis of EOC. Thus far, there have been numerous studies
regarding the diagnostic biomarkers of ovarian cancer (3,4,6–8); however,
there is not a suitable biomarker for the early diagnosis,
treatment efficacy monitoring and prognosis determination of
ovarian cancer. The study of Goff et al (6) demonstrated that the combination of
multiple indicators for the diagnosis of ovarian cancer can not
only improve the sensitivity and specificity of early diagnosis,
but can also determine the selection of an effective treatment and
prognosis.
In our preliminary study, surface-enhanced laser
desorption/ionization-time of flight-mass spectrometry
(SELDI-TOF-MS) technology was used to screen for the ovarian cancer
diagnosis significance of the serum antigens C-C motif chemokine
ligand 18 (CCL18) and C-X-C motif chemokine ligand 1 (CXCL1)
(7). Serological analysis of
recombinant cDNA expression library (SEREX) technology was
simultaneously used to screen for serum autoantibodies, including
C1D, transmembrane 4 L six family member 1 (TM4SF1), zinc finger
protein 675 (TIZ) and fragile X mental retardation 1 autosomal
homolog 1 (FXR1) (8). It was
determined that CCL18 and CXCL1 had a high sensitivity for EOC
diagnosis, but the specificity was not satisfactory; however, C1D,
TM4SF1, TIZ and FXR1 had a high specificity in EOC diagnosis,
compared with a combination of CCL18 and CXCL1. Therefore, it is
hypothesized that the combined use of these different types of
markers in the diagnosis of ovarian cancer may compensate for their
respective disadvantages.
In the present study, the expression of these six
genes/proteins in normal ovarian and EOC tissues were initially
analyzed using The Cancer Genome Atlas (TCGA) database. Secondly,
the association between these six genes/proteins and their
potential functions in EOC was analyzed. Finally, a mixed
suspension of fluorescent microspheres was applied to prepare a
liquid chip to combine these biomarkers for the differential
diagnosis of EOC. In the present study, the objective was to
combine the respective advantages of each set of biomarkers in
order to minimize the limitations of a singular biomarker, for the
early detection of EOC.
Materials and methods
Patients and samples
The present study was approved by the Ethics
Committee of the Affiliated Tumor Hospital of Guangxi Medical
University (Guangxi, China). All the patients provided written
informed consent prior to sample collection. Serum specimens were
collected from 60 patients diagnosed with early stage EOC,
confirmed by pathological examination by a pathologist in the
Affiliated Tumor Hospital of Guangxi Medical University, admitted
between September 2003 and December 2012 to the Department of
Gynecologic Oncology, Affiliated Tumor Hospital of Guangxi Medical
University. Additionally, samples from 30 patients with
gynecological benign tumors and 30 healthy females were collected
following routine physical examinations between June and September
2008 (Table I). Early stage EOC was
defined as stage I/II according to the International Federation of
Gynecology and Obstetrics (FIGO, 2013) (9). Serum specimens were also collected from
323 patients (Table II) diagnosed by
pathological examination admitted between September 2003 and
October 2009 to the Department of Gynecologic Oncology in the
Affiliated Tumor Hospital of Guangxi Medical University, including
119 patients with EOC with a median age of 48.5 years (range, 16–75
years) and 204 patients with benign pelvic tumors with a median age
of 43.6 years (range, 15–59 years), as well as 120 healthy females,
with samples collected from routine physical examinations in
between June and September 2008, with a median age of 41.5 years
(range, 21–70 years) (Table II). In
addition, serum specimens were collected from 40 female patients
with breast cancer (median age, 56.7 years; range, 20–75 years), 40
female patients with liver cancer (median age, 55.9 years; range,
38–87 years) and 40 female patients with lung cancer (median age,
54.3 years; range, 22–68 years). The Serum specimens were collected
from the Affiliated Tumor Hospital of Guangxi Medical University
between September 2003 and December 2012. Collected blood was
stored at 4°C for 2 h until coagulated, prior to being centrifuged
at 3,000 × g for 15 min at 4°C, and the serum was stored at −80°C
until further use.
 | Table I.Clinical pathological characteristics
of the 60 patients with EOC, 30 with gynecological benign tumors
and 30 healthy females. |
Table I.
Clinical pathological characteristics
of the 60 patients with EOC, 30 with gynecological benign tumors
and 30 healthy females.
| Variable | EOC (n=60) | Patients with
gynecological benign tumors (n=30) | Healthy females
(n=30) |
|---|
| Age, years |
|
|
|
| Mean | 46.7 | 32.4 | 34.4 |
| TNM stage (10) |
|
|
|
| I–II | 16 | 0 | 0 |
|
III–IV | 44 | 0 | 0 |
| OC type | Serous cancer
(n=42) | Ovarian lutein cysts
(n=3) |
|
|
| Mucinous carcinoma
(n=18) | Ovarian chocolate
cysts (n=13) |
|
|
|
| Ovarian mature
teratoma (n=12) |
|
|
|
| Ovarian corpus luteum
cyst (n=2) |
|
 | Table II.Clinical characteristics of serum
from patients. |
Table II.
Clinical characteristics of serum
from patients.
| Serum property | No. |
|---|
| Ovarian Cancer | 119 |
|
Epithelial tumor | 105 |
|
Non-epithelial tumors | 14 |
| FIGO staging
(10) |
|
| I | 15 |
| II | 22 |
|
III | 70 |
| IV | 12 |
| Pelvic benign
tumor | 204 |
|
Uterine-derived benign
tumors | 57 |
|
Tubal-derived benign
tumors | 15 |
|
Ovarian-derived benign
tumors | 111 |
| Other
sources of benign tumors | 21 |
| Healthy women | 120 |
| Breast cancer | 40 |
| Liver cancer | 40 |
| Lung cancer | 40 |
Non-ovarian cancer diseases were assessed in order
to detect whether the diagnostic method has higher specificity for
the diagnosis of ovarian cancer.
Bioinformatics analysis
The Human Protein Atlas (HPA; http://www.proteinatlas.org/) is an online tool with
the aim of recording the distribution of all human proteins through
the integration of various omics technologies, including
antibody-based imaging, mass spectrometry-based proteomics,
transcriptomics and systems biology. It consists of three separate
parts, each focusing on a particular aspect of the genome-wide
analysis of human proteins: The Tissue Atlas, which depicts the
distribution of the proteins across all major tissues and organs in
the human body; the Cell Atlas, which depicts the subcellular
localization of proteins in single cells; and the Pathology Atlas,
which depicts the impact of protein expression levels on the
survival of patients with cancer. HPA was used to analyze the
expression of C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ in normal
ovarian tissues and OC tissues, based on the clinical sample data
from TCGA database (https://cancergenome.nih.gov/). A
gene-gene/protein-gene/protein-protein interaction network was
generated by GeneMANIA (http://genemania.org/). The Coremine Medical online
tool (http://www.coremine.com/medical/) was used to annotate
the associated biological processes.
The Genotype-Tissue Expression (GTEx) project
collects and analyzes multiple human post mortem tissues. RNA-seq
data from 31 of their tissues having a corresponding tissue in
Human Protein Atlas were included to allow for comparisons between
the Human Protein Atlas data and GTEx data. The GTEx RNA-seq data
was mapped using the ensembl gene id available from GTEx, and the
RPKMs (number Reads Per Kilobase gene model and Million mapped
reads) for each gene were subsequently used to categorize the genes
using the same classification as described above but using 0.5 RPKM
as the threshold for detection.
Evaluating the diagnostic potential of
C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ in early stage EOC
As is subsequently described, C1D, CCL18, CXCL1,
TM4SF1, FXR1 and TIZ pET-SUMO prokaryotic expression vectors were
constructed in-house., and then purified high purity protein
was produced. A method for the combined detection of CCL18, CXCL1,
C1D, TM4SF1, FXR1 and TIZ was successfully developed using a
multi-analyte suspension array (MASA). MASA was used to examine the
C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ expression in patients with
EOC and gynecological benign tumors, and in healthy females.
The instrument used was a Bio-plex 200 suspension
chip system as described below.
Establishment of a liquid suspension
chip detection system
Subsequently, biotinylated antibody labeling was
performed using an EZ-Link® Sulfo-NHS-LC-Biotinylation
kit (cat. no. SF253102A; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and the antibodies were measured using the bicinchoninic
acid method (Pierce; Thermo Fisher Scientific, Inc.). In the second
step, each antigen or antibody [Anti-GRO-α antibody (cat. no.
ab89318; Abcam, Cambridge, UK), anti-macrophage inflammatory
protein 4 antibody (cat. no. ab89338; Abcam), goat anti-human
Immunoglobulin G Fc (cat. no. ab97221; Abcam) and natural human
Immunoglobulin G protein (cat. no. ab91102; Abcam) was applied to
carboxylated microspheres using the Bio-Plex® Amine
Coupling Kit (cat. no. 171-406001; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Finally, prepared samples of
1.25×106 microspheres were analyzed using the Bio-Plex
Manager™ software (version 6.1; cat. no. 171001010;
Bio-Rad Laboratories, Inc.) for MASA chip detection.
ELISA detection of C1D, TM4SF1, FXR1
and TIZ immunoglobulin G autoantibodies
The levels of serum antigens CCL18 and CXCL1, and
serum autoantibodies C1D, TM4SF1, FXR1 and TIZ were measured by an
ELISA. All procedures were performed as described by the
manufacturer's protocols for the ELISA kits. The ELISA test kits
used were the Quantikine® ELISA Human CCL18/PARC
Immunoassay kit (cat. no. DCL180B) and the Quantikine ELISA Human
CXCL1/GROα Immunoassay kit (cat. no. DGR00B) (both R&D Systems,
Inc., Minneapolis, MN, USA) and Rabbit anti-human IgG (H+L), Biotin
conjugated kit (cat. no. bsb-0297R) (Beijing Biosynthesis
Biotechnology Co. Ltd., China).
Comparison of the MASA and ELISA
methods
The accuracy, specificity and sensitivity between
the MASA and ELISA methods in combined multi-index detection were
compared. Firstly, the serum antigen and autoantibody contents were
detected by the liquid suspension chip and ELISA methods, and the
positive predictive value in the diagnosis of EOC was calculated by
the liquid suspension chip and ELISA through logistic regression
analysis and receiver operating characteristic (ROC) curves for the
six indicators. The point of maximum diagnostic value, based on the
Youden index, was defined as the diagnostic cut-off. Values above
this cut-off were defined as positive, and values below the cut-off
were defined as negative, If a value was equal to the cut-off, it
was defined as negative. The number of positive and negative cases
was counted, and the accuracy, specificity and sensitivity of
multi-index combined detection was compared with the four-table
method (Tables III and IV) (10).
 | Table III.Suspension liquid chip and ELISA
method ovarian cancer detection accuracy. |
Table III.
Suspension liquid chip and ELISA
method ovarian cancer detection accuracy.
|
| MASA |
| ELISA |
|---|
|
|
|
|
|
|---|
| Pathological
diagnosis | Positive | Negative | Total | Pathological
diagnosis | Positive | Negative | Total |
|---|
| Positive | 58 | 0 | 58 | Positive | 57 | 3 | 60 |
| Negative | 2 | 60 | 62 | Negative | 3 | 58 | 61 |
| Total | 60 | 60 | 120 | Total | 60 | 61 | 121 |
 | Table IV.Sensitivity and specificity of liquid
suspension chip and ELISA assay. |
Table IV.
Sensitivity and specificity of liquid
suspension chip and ELISA assay.
|
| MASA |
| ELISA |
|---|
|
|
|
|
|
|---|
| Pathological
diagnosis | Positive | Negative | Total | Pathological
diagnosis | Positive | Negative | Total |
|---|
| Positive | 58 | 0 | 58 | Positive | 57 | 3 | 60 |
| Negative | 2 | 60 | 62 | Negative | 3 | 58 | 61 |
| Total | 60 | 60 | 120 | Total | 60 | 61 | 121 |
Clinical evaluation of the multi-index
detection of EOC
In the present study, a system was successfully
established for the detection of the serum antigens CCL18 and
CXCL1, and autoantibodies C1D, TM4SF1, FXR1 and TIZ, by liquid
suspension microarray. The value and diagnostic performance of the
combined detection of six indicators or CA125 in the diagnosis of
early EOC was evaluated by testing the malignant and benign ovarian
clinical samples. The logistic regression model was validated by
detecting the levels of serum antigens CCL18 and CXCL1, and serum
autoantibodies C1D, TM4SF1, FXR1 and TIZ in the MASA and ELISA. ROC
curves were produced for the six indicators to determine their
positive predictive value.
The efficacy of the six markers in the diagnosis of
ovarian and non-ovarian malignancies was compared by measuring the
relative content of the serum antigens CCL18 and CXCL1, and
autoantibodies C1D, TM4SF1, FXR1 and TIZ, in the serum of 40
patients with breast cancer, 40 patients with liver cancer and 40
patients with lung cancer, using the liquid suspension chip method.
The diagnostic value of the markers for ovarian cancer was compared
with their value in other tumor types. ROC curves were produced for
the six indicators to determine their positive predictive
value.
Statistical analysis
SPSS statistical software (version 19.0; IBM Corp.,
Armonk, NY, USA) was used for all statistical analysis. Data are
presented as the mean ± standard deviation. Measurement data were
analyzed by Student's t-test and χ2 test. Logistic
regression analysis was used to draw the receiver operating curve
(ROC curve). P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression patterns of the six
potential biomarkers in ovarian cancer
The expression of the six proteins in normal and
tumor tissues was compared using the TCGA data. Their expression is
associated with multiple tumor types, including ovarian cancer, and
expression was notably lower in normal ovarian tissues, compared
with ovarian cancer tissues (P<0.05; Figs. 1 and 2).
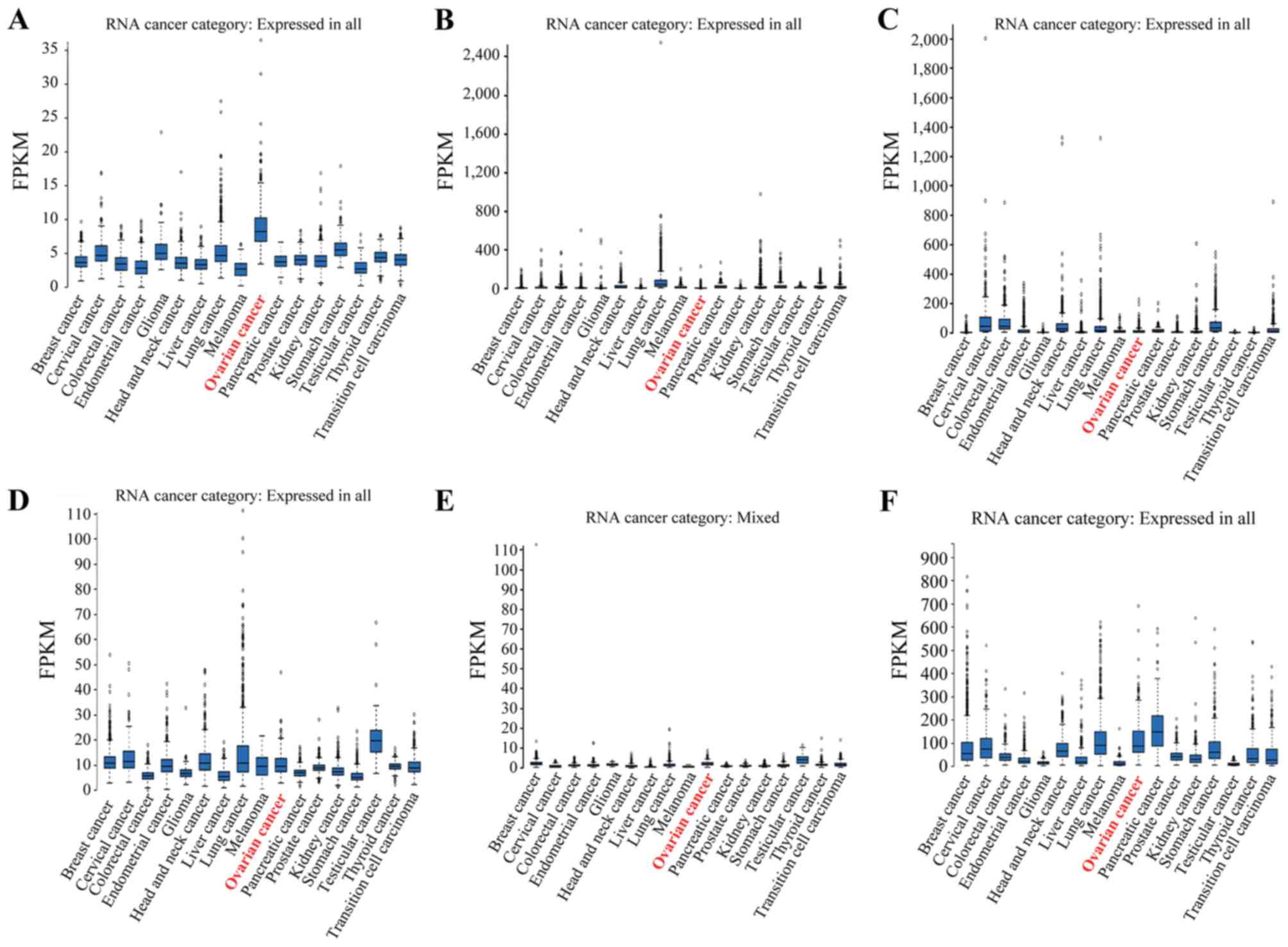
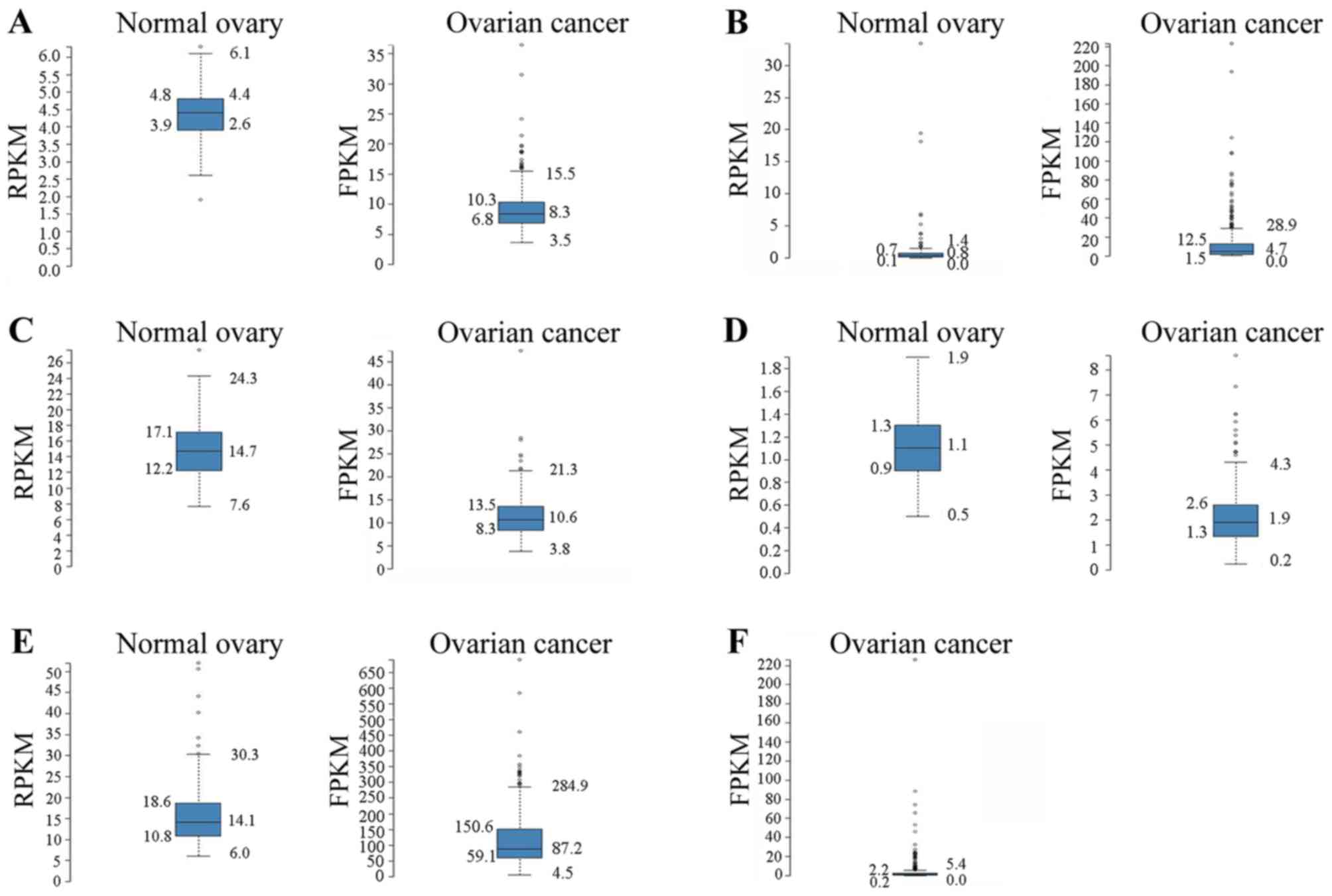 | Figure 2.Ovarian RNA-seq data from the GTEx
project 7 samples) and ovarian cancer RNA-seq data from TCGA (373
samples). The RNA-seq data are reported as the median number of
RPKM for the GTEx data and as number of FPKM for TCGA data. The
normalized distribution across the dataset is visualized with box
plots, including the median, and 25th and 75th percentiles. Points
are displayed as outliers if they are 1.5-fold above or below the
interquartile range. RPKM/FPKM values of the individual samples are
presented next to the box plot. (A) Left panel, C1D expression in
ovarian tissue based on GTEx RNA-seq data (mean RPKM, 4.4); and
right panel, C1D expression in ovarian cancer tissue based on TCGA
RNA-seq data (mean FPKM, 8.9). (B) Left panel, CXCL1 expression in
ovarian tissue based on GTEx RNA-seq data (mean RPKM, 1.5); and
right panel, CXCL1 expression in ovarian cancer based on TCGA
RNA-seq data (mean FPKM, 12.4). (C) Left panel, FXR1 expression in
ovarian tissue based on GTEx RNA-seq data (mean RPKM, 14.8); and
right panel, FXR1 expression in ovarian cancer tissue based on TCGA
RNA-seq data (mean FPKM, 11.2). (D) Left panel, TIZ expression in
ovarian tissue based on GTEx RNA-seq data (mean RPKM, 1.1); and
right panel, TIZ expression in ovarian cancer tissue based on TCGA
RNA-seq data (mean FPKM, 2.0). (E) Left panel, TM4SF1 expression in
ovary tissue based on GTEx RNA-seq data (mean RPKM, 16.7); and
right panel, TM4SF1 expression in ovarian cancer based on TCGA
RNA-seq data (mean FPKM, 114.7). (F) CCL18 expression in ovarian
cancer tissue based on TCGA RNA-seq data (mean FPKM, 3.7). No GTEx
RNA-seq data was identified for CCL18 in normal ovarian tissue.
GTEx, Genotype-Tissue Expression; TCGA, The Cancer Genome Atlas;
RKPM, reads per kb of exon per million reads; FPKM, fragments per
kb of exon per million reads; CXCL1, C-X-C motif chemokine ligand
1; FXR1, fragile X mental retardation 1 autosomal homolog 1; TIZ,
zinc finger protein 675; TM4SF1, transmembrane 4 L six family
member 1; CCL18, C-C motif chemokine ligand 18. |
Prediction and analysis of function
based on gene/protein-gene/protein interactions
The interaction networks between C1D, CCL18, CXCL1,
TM4SF1, FXR1 and TIZ and diagnosis-associated genes in EOC were
analyzed using the GeneMANIA tool. The results demonstrated that
C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ interacted with 26 genes in
total (Fig. 3).
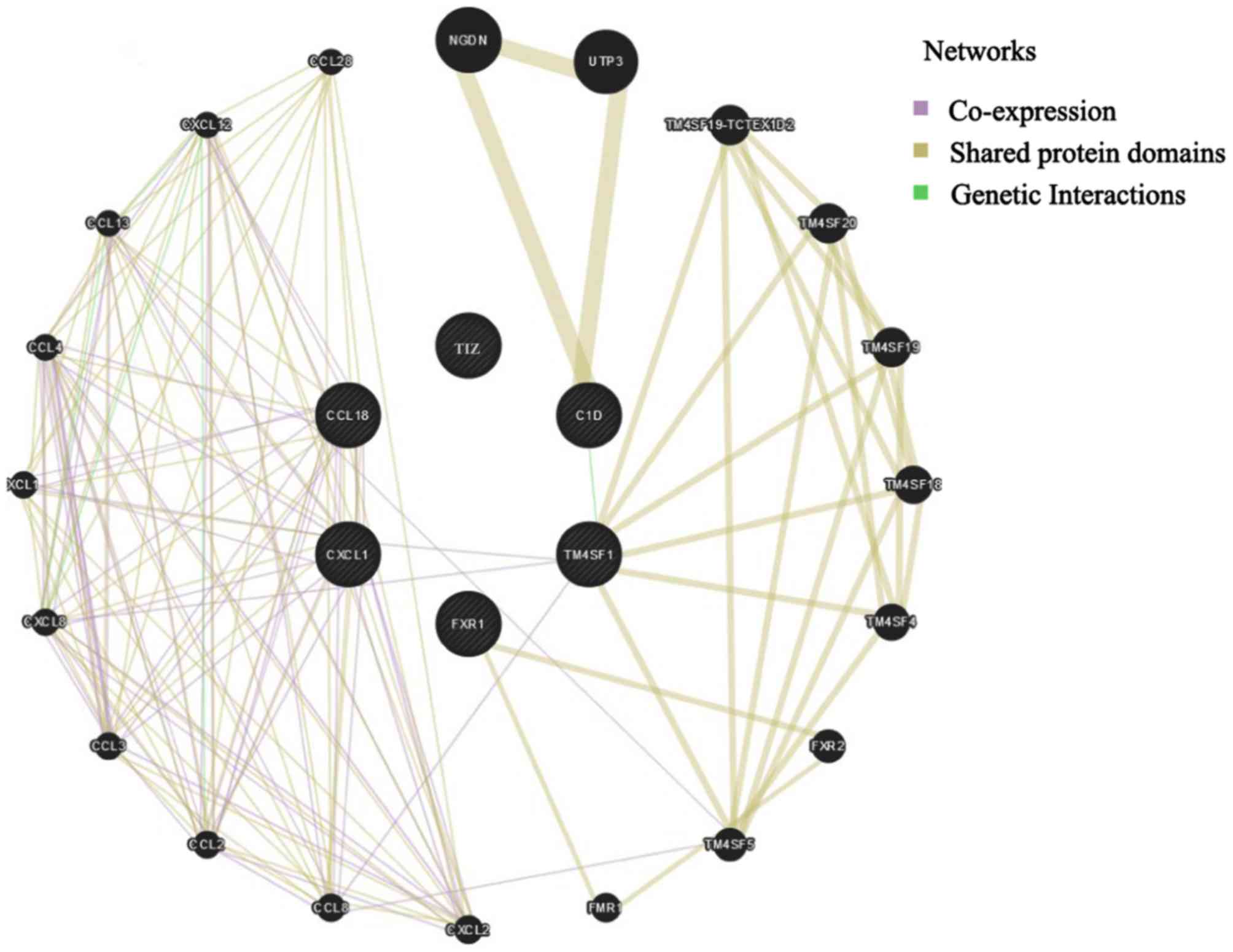 | Figure 3.Gene/protein interaction networks for
C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ, based on the GeneMANIA
online tool. CXCL1, C-X-C motif chemokine ligand 1; FXR1, fragile X
mental retardation 1 autosomal homolog 1; TIZ, zinc finger protein
675; TM4SF1, transmembrane 4 L six family member 1; CCL18, C-C
motif chemokine ligand 18. |
Functional prediction based on the
annotated biological processes
Coremine Medical is an online tool to identify the
terms relevant to the biological function and importance in disease
of genes and proteins. This tool was used with a P-threshold of
P<1.000, in order to associate CCL18, CXCL1, C1D, TM4SF1, TIZ
and FXR1 with biological processes. As depicted in Fig. 4, 22 biological processes associated
with early stage ovarian cancer and the expression levels of CCL18,
CXCL1, C1D, TM4SF1, TIZ and FXR1 were annotated, indicating their
mutual suitability for the early diagnosis of EOC. Among these, the
association between the six proteins and early EOC is in accordance
with the previous observation that CCL18, CXCL1, C1D, TM4SF1, TIZ
and FXR1 were suitable biological markers for the early diagnosis
of EOC.
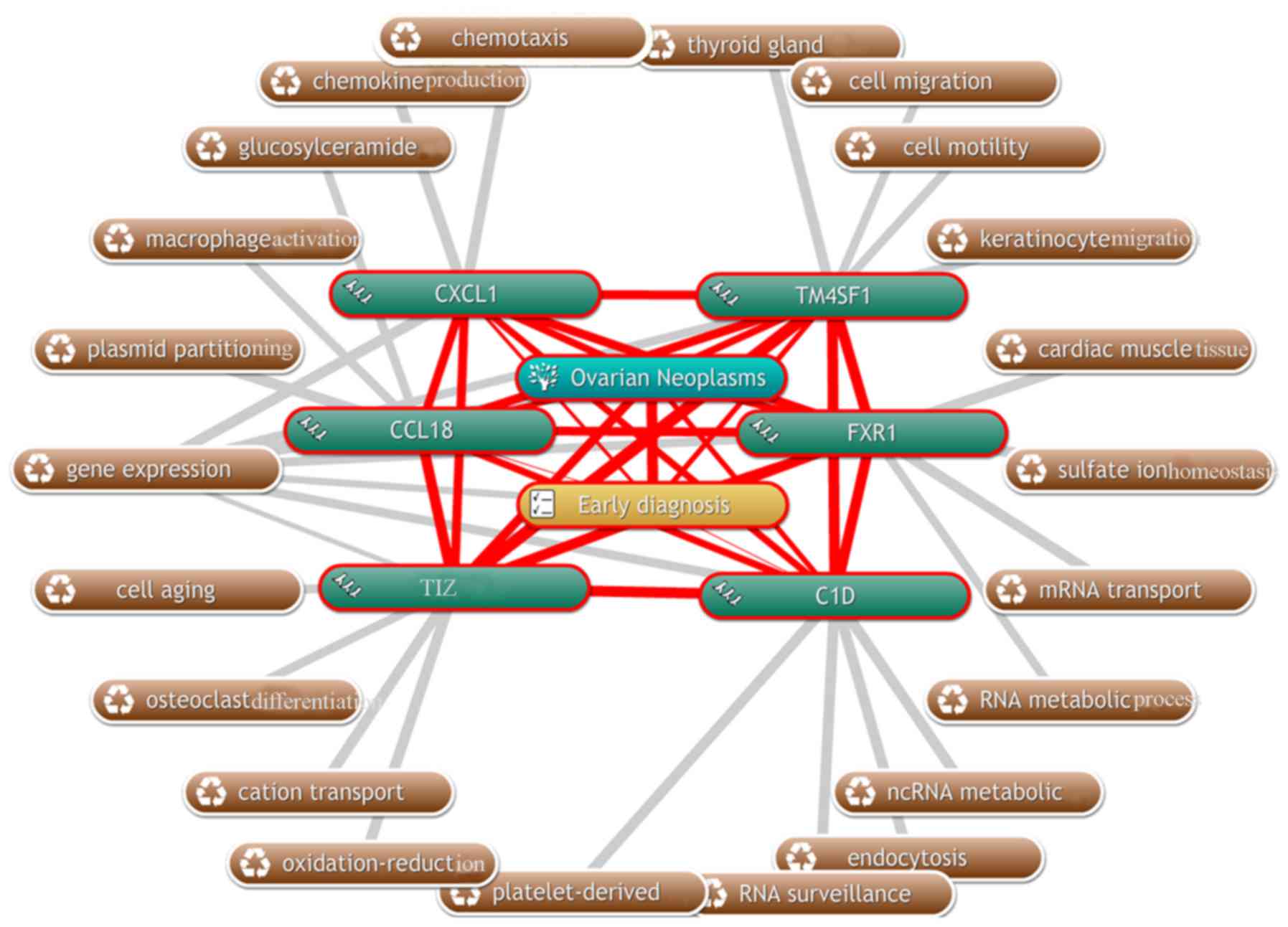 | Figure 4.Diagram of the biological processes
associated with C1D, CCL18, CXCL1, TM4SF1, FXR1 and TIZ (grey
lines), and their association with early state ovarian cancer (red
lines) using the Coremine Medical online tool. CXCL1, C-X-C motif
chemokine ligand 1; FXR1, fragile X mental retardation 1 autosomal
homolog 1; TIZ, zinc finger protein 675; TM4SF1, transmembrane 4 L
six family member 1; CCL18, C-C motif chemokine ligand 18. |
Comparison of the accuracy of the
liquid suspension chip and ELISA methods in multi-index combined
detection
The areas under the ROC curves for the liquid
suspension chip and the ELISA method detection of the six indexes
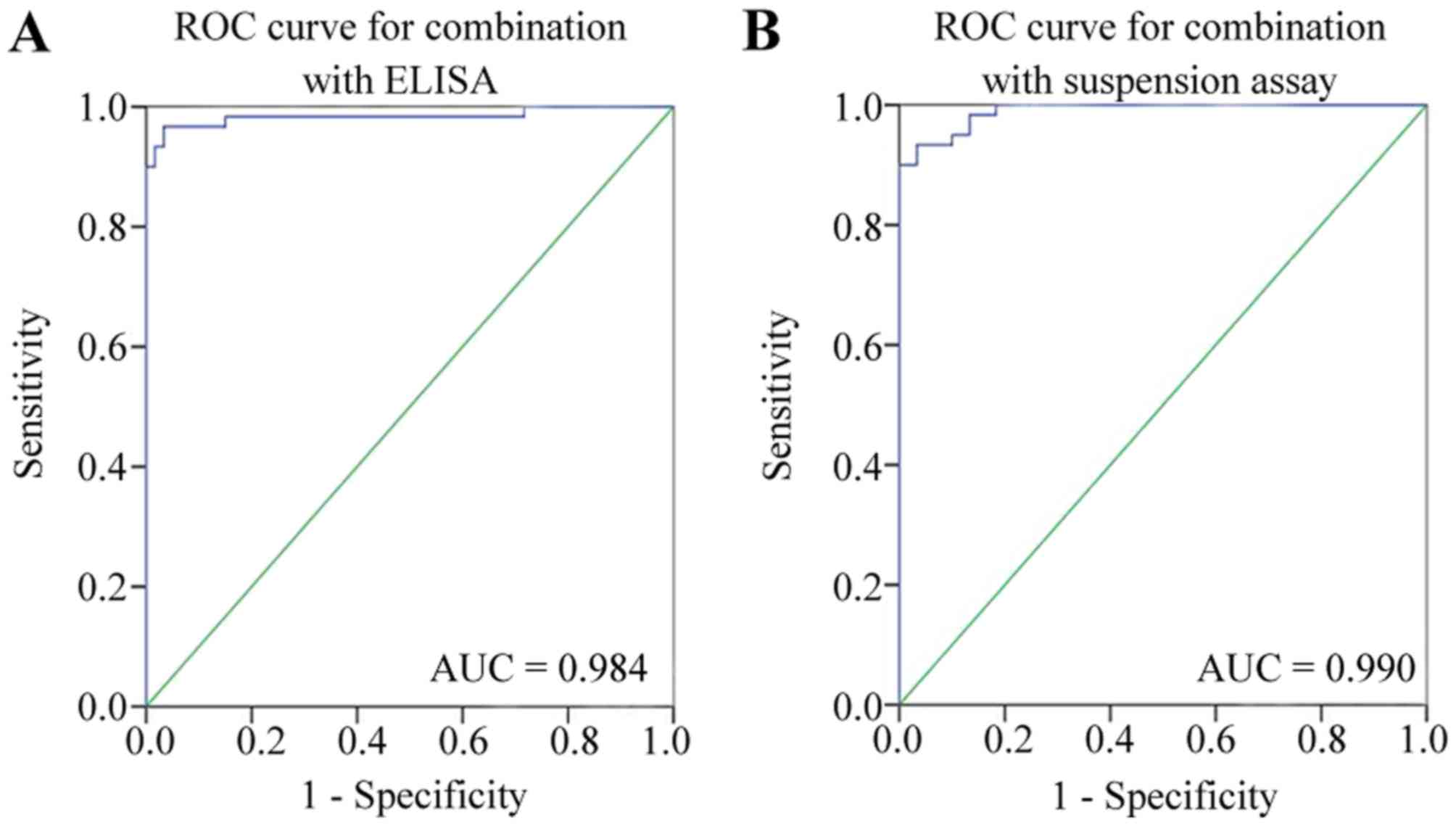
for EOC diagnosis were 0.990 and 0.984, respectively (Fig. 5). The accuracy of diagnosis using the
liquid suspension chip and the ELISA method was 98.33 and 95.33%,
respectively (Table III). The
differences between the methods was statistically significant
(P=0.02). It was concluded that the diagnostic performance of the
combined detected serum antigens CCL18 and CXCL1, and
autoantibodies C1D, TM4SF1, FXR1 and TIZ was improved in terms of
accuracy in the liquid suspension chip, compared with the ELISA
method.
Comparison of sensitivity and
specificity between the liquid suspension chip and ELISA
methods
The sensitivity and specificity for the detection of
the combination of the six genes by ELISA were 96.1 and 95.1%,
respectively, in the diagnosis of EOC (Table IV). The positive and negative
likelihood ratio and predictive value, and the false positive and
false negative rates for the liquid chip and ELISA methods are
displayed in Table V. The diagnosis
of EOC was improved in terms of sensitivity and specificity when
using the suspension liquid chip, compared with the ELISA
method.
 | Table V.Diagnostic performance of the
analysis of six proteins by liquid suspension assay and ELISA. |
Table V.
Diagnostic performance of the
analysis of six proteins by liquid suspension assay and ELISA.
| Detection
methods | Accuracy (%) | Sensitivity
(%) | Specificity
(%) | LR+ | LR- | Positive predictive
value (%) | Negative predictive
value (%) | Rate of missed
diagnosis (%) | Rate of
misdiagnosis (%) |
|---|
| MASA | 98.33 | 96.67 | 100.00 | – | 0.03 | 100.00 | 96.77 | 3.33 | 0.00 |
| ELISA | 95.04 | 95.00 | 95.08 | 19.32 | 0.05 | 95.00 | 95.08 | 5.00 | 4.92 |
Comparison of diagnostic value of the
combination of the six proteins and CA125 in the diagnosis of
EOC
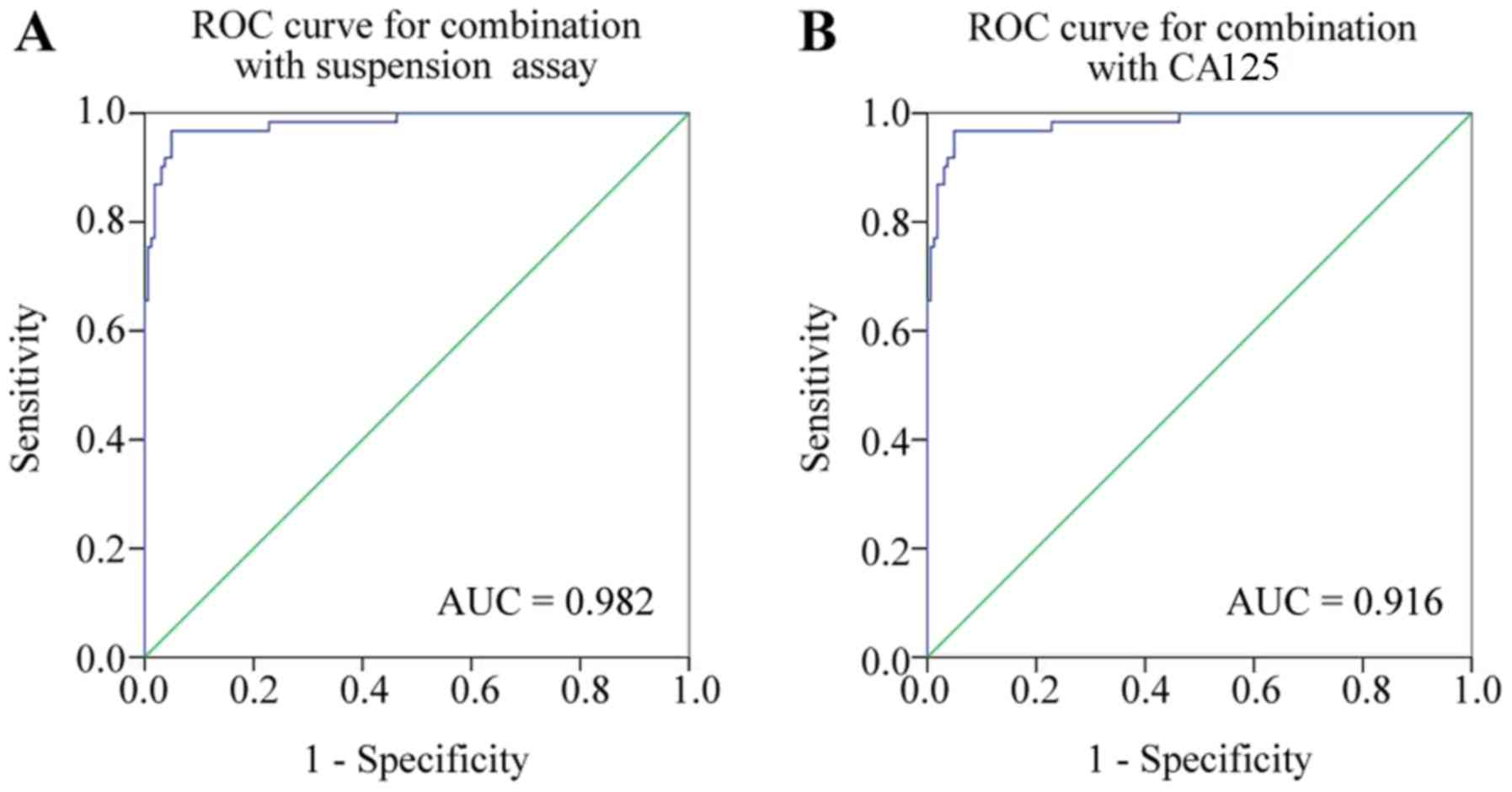
Using a combination of the six indicators or CA125
to detect EOC resulted in an area under the ROC curve of 0.982 and
0.916, respectively (Fig. 6). The
positive and negative likelihood ratio and predictive value, and
the false positive and false negative rates for the liquid chip
combined detection and CA125 are depicted in Table VI. The diagnosis of EOC was improved
when using the liquid chip, compared with CA125. It was concluded
that the diagnostic performance of the combination of the serum
antigens CCL18 and CXCL1, and autoantibodies C1D, TM4SF1, FXR1 and
TIZ was improved when using the liquid suspension chip, compared
with CA125.
 | Table VI.Diagnostic performance of the
analysis of six proteins by liquid suspension assay and CA125. |
Table VI.
Diagnostic performance of the
analysis of six proteins by liquid suspension assay and CA125.
| Methods | Accuracy (%) | Sensitivity
(%) | Specificity
(%) | LR+ | LR- | Positive predictive
value (%) | Negative predictive
value (%) | Rate of missed
diagnosis (%) | Rate of
misdiagnosis (%) |
|---|
| MASA | 96.38 | 90.48 | 98.11 | 47.95 | 0.10 | 95.00 | 96.30 | 9.52 | 1.89 |
| CA125 | 81.08 | 60.47 | 94.12 | 10.28 | 0.42 | 86.67 | 79.01 | 39.53 | 5.88 |
Comparison of the efficacy of the six
markers in the diagnosis of ovarian and non-ovarian
malignancies
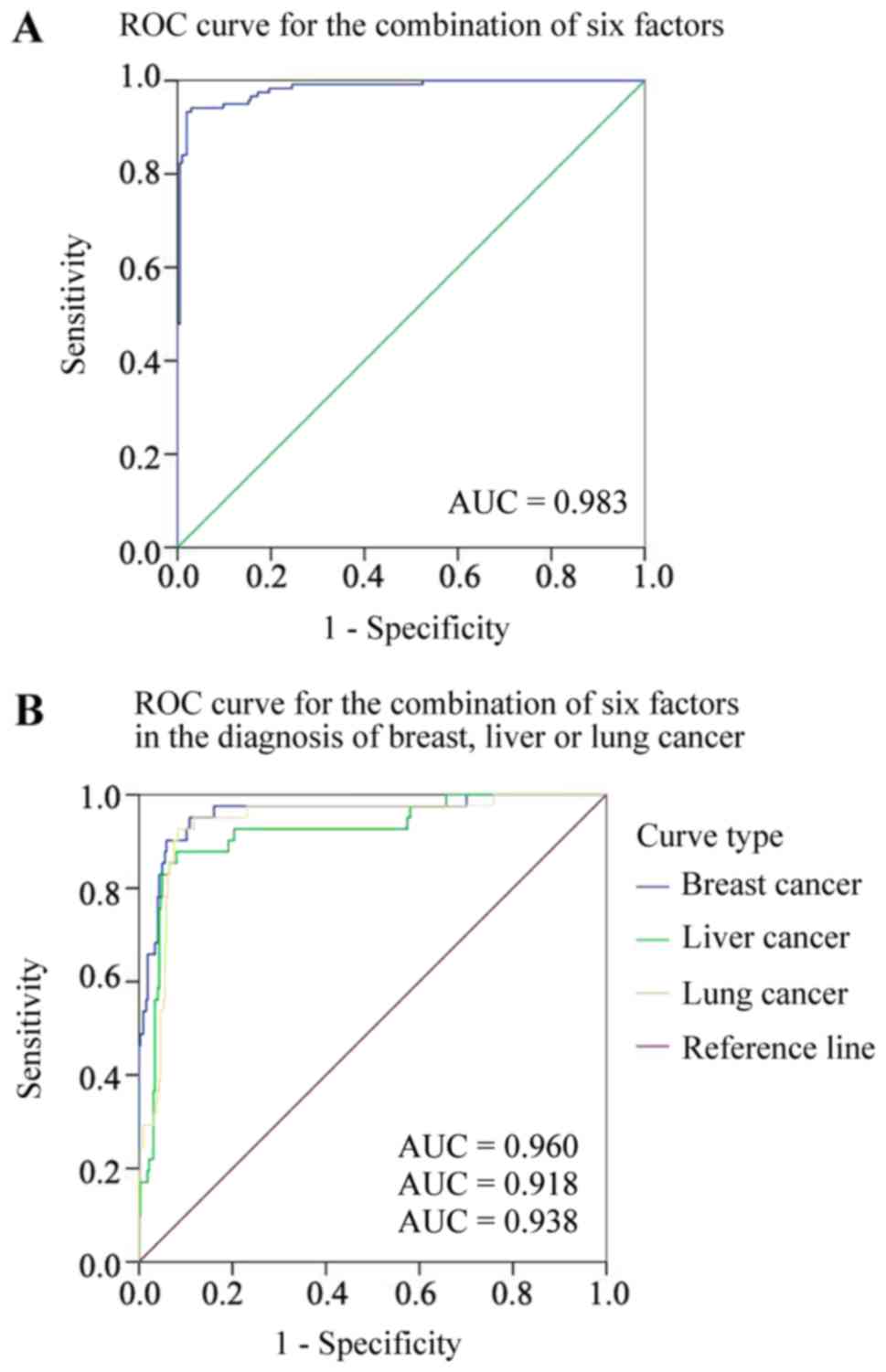
The positive rates for breast cancer, liver cancer,
lung cancer and ovarian cancer based on the combined detection of
the six indicators were 82.5, 77.5, 72.5 and 94.9%, respectively
(Table VII), with an area under the
ROC curve of 0.960, 0.918, 0.938 and 0.983, respectively (Fig. 7). The positive and negative likelihood
ratio and predictive value, and the false positive and false
negative rates for the diagnosis of these malignancies are
displayed in Table VIII. It was
concluded that the combined detection of the six indicators had a
greater probability of indicating EOC, compared with the other
malignancy types.
 | Table VII.Positive rates for other malignant
tumor types detected by a suspension array combining six
factors. |
Table VII.
Positive rates for other malignant
tumor types detected by a suspension array combining six
factors.
| Sample
category | n | Positive case | Positive rate
(%) |
|---|
| Ovarian cancer | 119 | 113 | 94.90 |
| Breast cancer | 40 | 33 | 82.50 |
| Liver cancer | 40 | 31 | 77.50 |
| Lung cancer | 40 | 29 | 72.50 |
 | Table VIII.Diagnostic performance of a
suspension array combining six factors for other malignant tumor
types. |
Table VIII.
Diagnostic performance of a
suspension array combining six factors for other malignant tumor
types.
| MASA | Accuracy (%) | Sensitivity
(%) | Specificity
(%) | Negative predictive
value (%) | LR+ | LR- | Rate of missed
diagnosis (%) | Rate of
misdiagnosis (%) |
|---|
| Ovarian cancer | 56.51 | 73.14 | 94.95 | 73.14 | 2.10 | 0.59 | 43.5 | 2.46 |
| Breast cancer | 30.84 | 68.51 | 82.50 | 68.51 | 0.98 | 1.01 | 69.15 | 4.16 |
| Liver cancer | 26.05 | 68.34 | 77.50 | 68.34 | 0.82 | 1.08 | 74.94 | 4.52 |
| Lung cancer | 25.89 | 68.67 | 72.50 | 68.67 | 0.83 | 1.08 | 74.10 | 5.69 |
Discussion
The high mortality rate for ovarian cancer is
primarily due to the predominately asymptomatic nature of the
disease in its early stages; therefore, surgery and chemotherapy
are the primary treatment choices for patients with ovarian cancer.
Additionally, it is estimated that 70–75% of all females with
ovarian cancer will experience recurrence, with a 5-year survival
rate of ~30% (11). When the disease
can be detected in stage I and limited to the ovaries, up to 90% of
patients can be successfully treated with the currently available
surgical and chemotherapeutic methods. Early stage (I and II)
ovarian cancer is only detected in a limited number of patients by
conventional examination; therefore, a thorough understanding of
the biomarkers for ovarian cancer identification and early
detection may improve the 5-year survival rate (12).
TM4SF1 is overexpressed in EOC and regulates cancer
cell motility and invasion. Additionally, it is associated with
metastasis and regulates the development of angiogenesis, making it
a potential target for anti-angiogenesis and antitumor therapies
(13). TM4SF1 has been identified in
human liver cancer (14), breast
cancer (15), colorectal cancer
(16) and ovarian cancer. Liu et
al (17) identified that the
positive expression rate of TM4SF1 protein in EOC tissue was higher
than that in benign ovarian tumors or normal ovarian epithelial
tissues, and may be associated with the abnormal proliferation of
ovarian epithelial cells, malignant transformation and other
clinical characteristics. TM4SF1 protein is associated with the
occurrence and progression of EOC, but the underlying mechanism of
this requires further study.
C1D is a small, 16-kDa mammalian nuclear matrix
protein involved in higher-order chromatin folding and tight DNA
binding. A recent study indicated that the C1D protein may be
involved in the maintenance of genomic integrity by regulating the
activity of double stranded DNA break repair proteins (18). C1D induces the production of
autoantibodies in the serum of patients with EOC, possibly due to
the stimulation of the immune system by the ectopic expression of
C1D (19); however, regarding the
role of C1D in the occurrence, development and apoptosis resistance
of EOC, further research is required.
The FXR1 gene encodes an RNA-binding protein that is
a critical regulator of post-transcriptional gene expression in
differentiation, development and immunity. Due to FXR1 coordinating
RNA-protein and protein-protein interaction networks, the altered
function of FXR1 is expected to contribute toward the progression
of cancer (20). In addition to its
amplification in lung cancer, breast cancer, head and neck cancer
and ovarian cancer (21,22), FXR1 has been demonstrated to be
downregulated in other human pathology types, including
facioscapulohumeral muscular dystrophy. The overexpression of FXR1
is associated with cell growth, migration and invasion, indicating
that FXR1 has oncogenic activities. Anti-FXR1 autoantibodies can be
detected in the serum of patients with EOC, but their function in
the development of EOC requires further investigation (23).
TIZ belongs to the C2H2-type zinc finger protein
family, which can interact with tumor necrosis factor
receptor-associated factor 6, in order to serve a role in the
regulation of the spread and migration of tumors (24). According to a previous clinical study,
the expression level of TIZ protein in the serum of patients with
malignant ovarian tumors was significantly higher, compared with
patients with benign ovarian tumor and healthy individuals, which
indicates a notable association between the expression level of TIZ
and ovarian cancer (25).
CXCL1 belongs to the CXC chemokine family and is
associated with cellular transformation, tumor growth and an
increase in invasive potential (26).
A previous study demonstrated that CXCL1 is associated with the
occurrence, growth and metastasis of malignant tumors, and the
overexpression of CXCL1 in EOC cells promoted proliferation,
whereas CXCL1-silencing inhibited proliferation (27). The detection of serum CXCL1 provides a
novel direction for the early diagnosis of EOC, but its clinical
application requires further validation.
CCL18 is predominately produced by tumor-associated
macrophages, and promotes the migration and invasion of breast
cancer cells (28). It is expressed
at higher levels in ovarian cancer ascites, compared with
non-ovarian carcinomas. Its increased expression in OC tissue is
associated with metastasis (29). The
study of Schutyser et al (30)
determined that patients with EOC and ascites had higher CCL18
expression levels than ovarian benign tumor patients with high
immune staining in interstitial areas and a limited number of CCL18
antibodies. EOC cells exhibited positive CCL18 expression, and the
relative expression level was higher than that in benign ovarian
tumors and normal ovarian epithelial cells (31). CCL18 protein expression has potential
as a novel tumor marker for EOC.
The detection of CCL18 and CXCL1 antigens has
greater sensitivity for the diagnosis of EOC compared with CA125,
but its specificity was not satisfactory, as the detection of CCL18
and CXCLL1 antigens had a poor diagnostic specificity for EOC;
however, autoantibodies, including TM4SF1, have a high specificity
for the diagnosis of EOC, but the diagnostic sensitivity was
reduced, compared with CCL18. Liquid chips can contain multiple
types of microspheres with different fluorescent dyes, combined
with the corresponding serum antigens and antibodies in a covalent
cross-linking manner. The analytes are mixed with the fluorescently
encoded microspheres to form immune complexes, which can be
detected through the two-laser detection of fluorescent signals, to
simultaneously detect a number of different molecules in the same
sample (32). The serum markers of
EOC were screened by SELDI-TOF-MS and SEREX techniques and used to
produce a liquid microarray. By combining these two different types
of markers for the early diagnosis and differential diagnosis of
ovarian cancer, their respective advantages can make up for the
drawbacks of one marker type alone. The high-throughput,
high-content liquid suspension chip method is highly applicable in
clinical practice, representing the current direction of the
development of serum molecular diagnostic technology. It also
provides a method for the simultaneous detection of multiple types
of markers.
In conclusion, based on a comprehensive
bioinformatics analyses and MASA, the six indicators were
identified to have an improved performance, compared with CA125,
for the early diagnosis of EOC. The genes identified in the present
study have the potential to improve the early diagnosis of EOC,
although these possibilities require further research, including
further validation with an increased number of clinical
samples.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Development Plan of Guangxi, China (grant no.
1140003A-33) and Science and Technology Development Plan of
Guangxi, China (grant no. 1140003A-34).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
Study development and design of methodology was the
responsibility of LL, experiments were conducted and analyzed by YZ
and LL. Application of statistical, mathematical, computation and
presentation of the published work and revision and approval of the
final manuscript was undertaken by YZ and LL.
Ethics approval and consent to
participate
The present study was endorsed by the Ethics
Committee of the Affiliated Tumor Hospital of Guangxi Medical
University (Guangxi, China). All the patients provided written
informed consent prior to sample collection.
Patient consent for publication
All study participants involved in this experiment
provided consent to publish any relevant data or images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sopik V, Iqbal J, Rosen B and Narod SA:
Why have ovarian cancer mortality rates declined? Part II.
Case-fatality. Gynecol Oncol. 138:750–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abu Hassan SO, Nielsen DL, Tuxen MK,
Petersen PH and Sölétormos G: Performance of seven criteria to
assess CA125 increments among ovarian cancer patients monitored
during first-line chemotherapy and the post-therapy follow-up
period. Future Sci OA. 3:FSO2162017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Capriglione S, Luvero D, Plotti F,
Terranova C, Montera R, Scaletta G, Schirò T, Rossini G, Panici
Benedetti P and Angioli R: Ovarian cancer recurrence and early
detection: May HE4 play a key role in this open challenge? A
systematic review of literature. Med Oncol. 34:1642017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan SJ, Qiao TK, Qiang JW, Cai SQ and Li
RK: The value of DCE-MRI in assessing histopathological and
molecular biological features in induced rat epithelial ovarian
carcinomas. J Ovarian Res. 10:652017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goff BA, Agnew K, Neradilek MB, Gray HJ,
Liao JB and Urban RR: Combining a symptom index, CA125 and HE4
(triple screen) to detect ovarian cancer in women with a pelvic
mass. Gynecol Oncol. 147:291–295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Q, Zhang W, Li DR and Li L:
Identification of two potencial serum biomarkers for ovarian cancer
and clinical validation thereof. Zhonghua Yi Xue Za Zhi.
88:1012–1016. 2008.(In Chinese). PubMed/NCBI
|
|
8
|
Yang ZJ, Yang G, Jiang YM, Ran YL, Yang
ZH, Zhang W, Zhang JQ, Pan ZM and Li L: Screening and
sero-immunoscreening of ovarian epithelial cancer associative
antigens. Zhonghua Fu Chan Ke Za Zhi. 42:834–839. 2007.PubMed/NCBI
|
|
9
|
Kandukuri SR and Rao J: FIGO 2013 staging
system for ovarian cancer: What is new in comparison to the 1988
staging system? Curr Opin Obstet Gynecol. 27:48–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Y: Consistency test of the paired
fourfold table. J Mathe Med. 23:386–387. 2010.
|
|
11
|
Han XR, Wen X, Li YY, Fan SH, Zhang ZF, Li
H, Sun XF, Geng GQ, Sun S, Huang SQ, et al: Effect of different
anesthetic methods on cellular immune functioning and the prognosis
of patients with ovarian cancer undergoing oophorectomy. Biosci
Rep. 37:pii: BSR20170915. 2017. View Article : Google Scholar
|
|
12
|
Teng C and Zheng H: Low expression of
microRNA-1908 predicts a poor prognosis for patients with ovarian
cancer. Oncol Lett. 14:4277–4281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao J, Yang JC, Ramachandran V, Arumugam
T, Deng DF, Li ZS, Xu LM and Logsdon CD: TM4SF1 regulates
pancreatic cancer migration and invasion in vitro and in vivo. Cell
Physiol Biochem. 39:740–750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang YK, Fan XG and Qiu F: TM4SF1
promotes proliferation, invasion, and metastasis in human liver
cancer cells. Int J Mol Sci. 17:pii: E661. 2016. View Article : Google Scholar
|
|
15
|
Sun Y, Xu Y, Xu J, Lu D and Wang J: Role
of TM4SF1 in regulating breast cancer cell migration and apoptosis
through PI3K/AKT/mTOR pathway. Int J Clin Exp Pathol. 8:9081–9088.
2015.PubMed/NCBI
|
|
16
|
Park YR, Lee ST, Kim SL, Liu YC, Lee MR,
Shin JH, Seo SY, Kim SH, Kim IH, Lee SO and Kim SW: MicroRNA-9
suppresses cell migration and invasion through downregulation of
TM4SF1 in colorectal cancer. Int J Oncol. 48:2135–2143. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Yan Y, Gao C and Yang Z: Expression
and clinical significance of TM4SF1 protein in epithelial ovarian
cancer. Chin J Oncol Prev Treat. 6:20–24. 2014.(In Chinese).
|
|
18
|
Jackson RA, Wu JS and Chen ES: C1D family
proteins in coordinating RNA processing, chromosome condensation
and DNA damage response. Cell Div. 11:22016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li G, Liu J, Abu-Asab M, Masabumi S and
Maru Y: XPB induces C1D expression to counteract UV-induced
apoptosis. Mol Cancer Res. 8:885–895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raheja R and Gandhi R: FXR1: Linking
cellular quiescence, immune genes and cancer. Cell Cycle.
15:2695–2696. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Majumder M, House R, Palanisamy N, Qie S,
Day TA, Neskey D, Diehl JA and Palanisamy V: RNA-binding protein
FXR1 regulates p21 and TERC RNA to bypass p53-mediated cellular
senescence in OSCC. PLoS Genet. 12:e10063062016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian J, Hassanein M, Hoeksema MD, Harris
BK, Zou Y, Chen H, Lu P, Eisenberg R, Wang J, Espinosa A, et al:
The RNA binding protein FXR1 is a new driver in the 3q26-29
amplicon and predicts poor prognosis in human cancers. Proc Natl
Acad Sci USA. 112:3469–3474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jo YS, Kim SS, Kim MS, Yoo NJ and Lee SH:
Frameshift mutation of FXR1 encoding a RNA-binding protein in
gastric and colorectal cancers with microsatellite instability.
Pathol Oncol Res. 23:453–454. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bing-bing Z, Wei Z, Qi W, Zhi-jun Y and Li
L: The effect of TIZ gene overexpression on biological
characteristics of epithelial ovarian cancer cells. Tumor.
32:2012.
|
|
25
|
Zheng HY, Zheng HY, Zhou YT, Liu EL, Li J
and Zhang YM: Changes of TIZ expression in epithelial ovarian
cancer cells. Asian Pac J Trop Med. 8:157–161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wani N, Nasser MW, Ahirwar DK, Zhao H,
Miao Z, Shilo K and Ganju RK: C-X-C motif chemokine 12/C-X-C
chemokine receptor type 7 signaling regulates breast cancer growth
and metastasis by modulating the tumor microenvironment. Breast
Cancer Res. 16:R542014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sahingur SE and Yeudall WA: Chemokine
function in periodontal disease and oral cavity cancer. Front
Immunol. 6:2142015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li HY, Cui XY, Wu W, Yu FY, Yao HR, Liu Q,
Song EW and Chen JQ: Pyk2 and Src mediate signaling to
CCL18-induced breast cancer metastasis. J Cell Biochem.
115:596–603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Tang Y, Yu H, Yin Q, Li M, Shi L,
Zhang W, Li D and Li L: CCL18 from tumor-cells promotes epithelial
ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog.
55:1688–1699. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schutyser E, Struyf S, Proost P,
Opdenakker G, Laureys G, Verhasselt B, Peperstraete L, Van de Putte
I, Saccani A, Allavena P, et al: Identification of biologically
active chemokine isoforms from ascitic fluid and elevated levels of
CCL18/pulmonary and activation-regulated chemokine in ovarian
carcinoma. J Biol Chem. 277:24584–24593. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei Z, Ying-Zhu Y, Li L, Zhong-Mian P,
Zhi-Jun Y and Qi W: Study on the localization and quantification of
CCL18 protein in epithelial cells of ovarian cancer. J Guangxi Med
Univ. 9–13. 2014.
|
|
32
|
Lin A, Salvador A and Carter JM:
Multiplexed microsphere suspension array-based immunoassays.
Methods Mol Biol. 1318:107–118. 2015. View Article : Google Scholar : PubMed/NCBI
|