Introduction
Osteosarcoma, an aggressive malignant neoplasm
arising from primitive transformed cells of mesenchymal origin, is
the most common type of human primary bone sarcoma and a leading
cause of cancer death in children and adolescents (1–5). The
treatment for osteosarcoma is unsatisfactory and new targets for
the treatment of osteosarcoma are urgently needed (6–9).
The mechanisms on the formation and development of
osteosarcoma have been studied for a long time (6,7,9). Recent evidence has shown that gene
expression is regulated by microRNAs (miRNAs/miRs), which are small
noncoding RNAs (about 22 nt in length) that play crucial roles in
regulating tumor growth by binding to the 3′ untranslated region
(UTR) of target mRNAs, which represses their translation (9–12).
Hundreds of target mRNAs have been associated with osteosarcoma,
but the underlying mechanisms are unclear (13–18).
One example is miR-2682-3p (19). Recently, we studied whether the
dysregulation of miR-2682-3p is involved in osteosarcoma. This
study is aimed at determining the role of miR-2682-3p in
osteosarcoma cell growth and the target genes for miR-2682-3p. In
the present study, miR-2682-3p was decreased in osteosarcoma
tissues and cell lines, and overexpression of miR-2682-3p inhibited
osteosarcoma cell proliferation. Moreover, in vitro
experiments proved that downregulation of miR-2682-3p promoted
tumor proliferation; these experiments also identified cyclin D1
(CCND)2, matrix metalloproteinase 8 (MMP8), and Myd88 as the
direct targets of miR-2682-3p in osteosarcoma cells. Our findings
suggest the involvement of miR-2682-3p in osteosarcoma cell
apoptosis induced by CCND2, MMP8, and Myd88.
Materials and methods
Ethics statement
All patients participated in the study provided
written informed consent, and the study was approved by the Ethics
Committee of Shanghai Tenth People's Hospital (20).
Tissues and cell culture
Twelve paired osteosarcoma tissues were obtained
from Shanghai Tenth People's Hospital. Human normal osteoblast
cells (hFOB) and osteosarcoma cell lines (Saos-2, MG-63, and U2OS)
were purchased from American Type Culture Collection (Manassas, VA,
USA) and cultured in Dulbecco's modified Eagle's medium at 37°C in
an atmosphere of 5% CO2 (9,17).
Cell proliferation assay
Cells were cultured in 96-well microplates at
1×104 per well for 3 days after transfection. CCK-8
(Dojindo, Kumamoto, Japan) were used to analyze the viability of
the cells (9). The viable cells were
counted by absorbance measurement at 450 nm using an
auto-microplate reader.
Colon formation assay
U2OS and MG63 cells were counted and diluted to 100
cells/ml. Aliquots (2 ml) of each suspension were added to wells of
six-well culture plates. The medium was refreshed every 3 days
until cell clones could be observed with the naked eye.
RNA extraction and quantitative
PCR
Total RNA was extracted from the cells or tissues
using the miRNA isolation kit (Ambion, Auston, TX, USA). Reverse
transcriptions were performed using an RNA PCR kit (Takara Bio,
Shiga, Japan) in accordance with manufacturer's instructions
(9). To quantify gene transcripts,
real-time PCR was performed using SYBR-Green Premix Ex Taq (Takara
Bio) on LightCycler 480 (Roche, Basel, Switzerland). U6 and
glycerladehyde-3-phosphate dehydrogenase (GAPDH) were used as the
normalizing controls for quantifying miRNA and mRNA, respectively
(9).
Oligonucleotide and transfection
miR-2682-3p mimics and scrambled miRNAs (Shanghai
GenePharma Co., Ltd., Shanghai, China) were transfected into cells
using DharmaFECT1 reagent (Dharmacon, Austin, TX, USA) according to
the manufacturer's instructions (9).
Western blot analysis
Tissues or cells were prepared using ice-cold lysis
buffer (50 mM Tris-HCl, pH 7.0, 1% w/v SDS, 10% glycerol), then
were centrifuged at 4°C. Proteins in each supernatant were
quantified. The proteins were separated by 10% SDS-PAGE and were
blotted to PVDF membranes (Amersham BioSciences, Buckinghamshire,
UK) (9). After blocking using 5%
nonfat milk for 1 h, the membranes were incubated with antibodies.
After using horseradish peroxidase-linked secondary antibodies
(Cell Signaling Technology, Inc., Beverley, MA, USA), proteins
bands were visualized (9).
Luciferase reporter assay
Primers were designed in accordance with the
pGL3-CCND2, and MMP8, and Myd88 gene mRNA sequence. MG-63 cells
were cultured in 96-well plates for 48 h at 1×104 cells
per well, and then were transfected with miR-2682-3p mimics or
scramble, 10 ng pGL3, and pGL3-CCND2, MMP8 and Myd88-3′UTR or
pGL3-CCND2, and MMP8 and Myd88-3′UTR Mut plasmid per well using
Lipofectamine 3000 (9). The
Dual-Luciferase Reporter Assay System (Promega Corporation,
Madison, WI, USA) was used to calculate relative luciferase
activity of the cells after 48 h. Normalized firefly luciferase
activity for each construct was compared with that of the pmirGLO
Vector no insert (NO) control (9).
Statistical analysis
Data are presented as the mean ± SD from three
separate experiments. Student's t-test was used to compare the
differences between the two groups, and ordinary one-way ANOVA
followed by Dunnett's multiple comparisons test was used to analyze
the differences in multigroups. The Pearson's correlation analysis
was used to verify the relevance and the log-rank test was used to
compare the statistical significance difference. A P-value <0.05
was considered statistically significant. GraphPad Prism 6
(GraphPad Software, Inc., La Jolla, CA, USA) was used for
statistical analyses.
Results
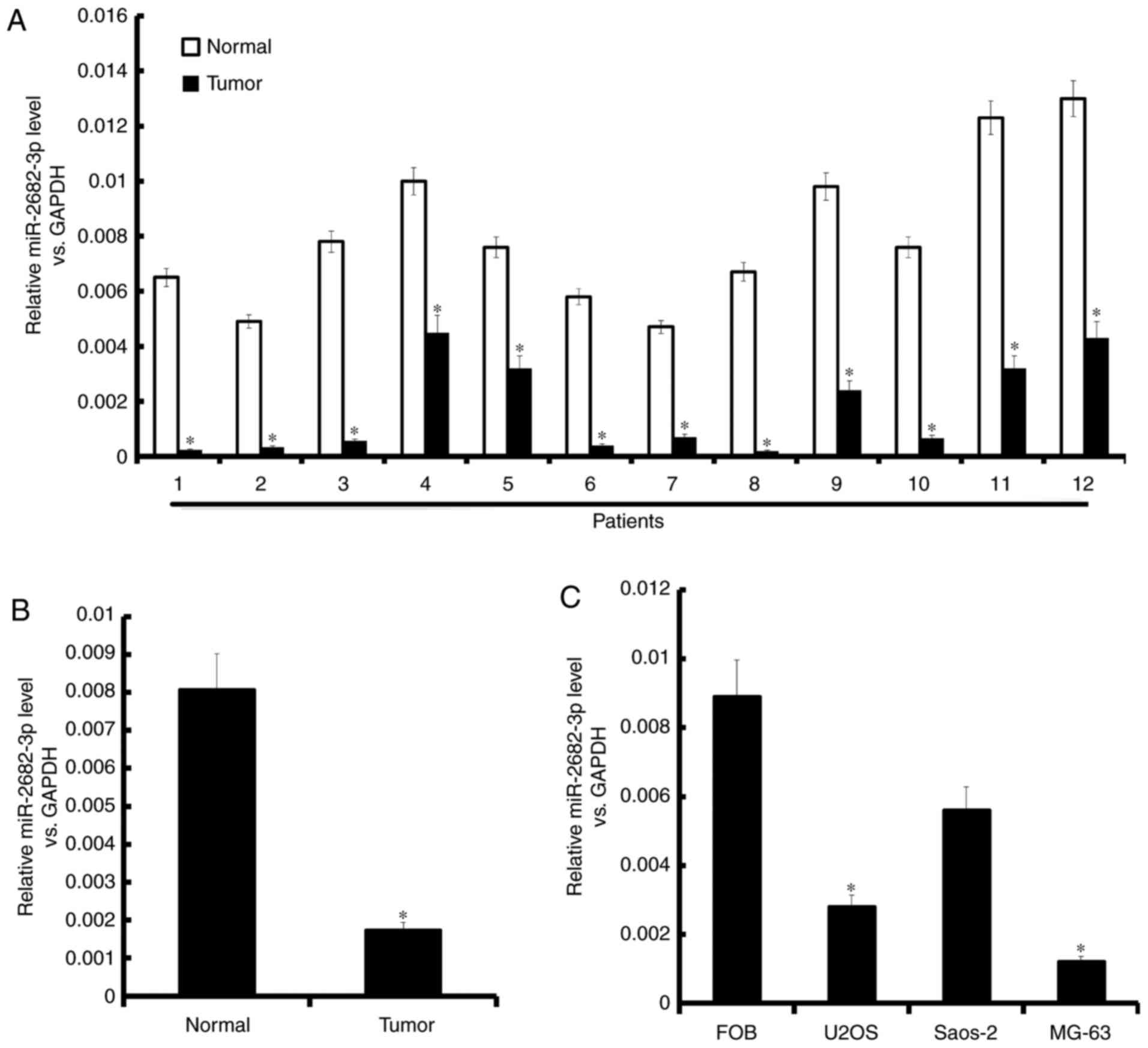
miR-2682-3p expression is decreased in
osteosarcoma tissues and cell lines
miR-2682-3p expression was lower in tumor
tissues than in normal tissues (Fig.
1). miR-2682-3p expression was decreased in the
osteosarcoma tissues and cell lines of the patients (Fig. 1A and B). Moreover, miR-2682-3p
expression was lower in the three osteosarcoma cell lines (Saos-2,
MG-63, and U2OS) than in normal osteoblast cells (FOB) (Fig. 1C).
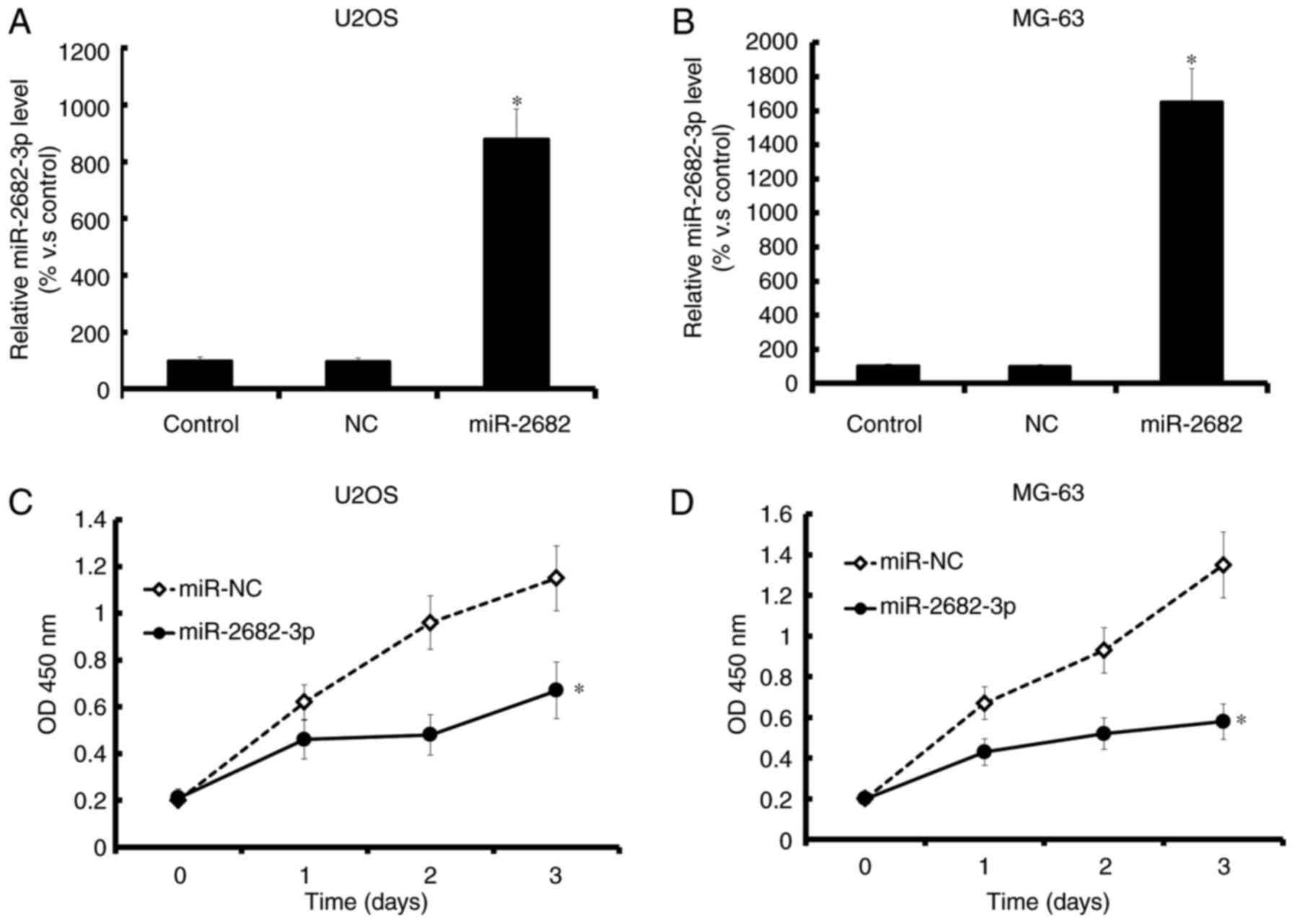
Overexpression of miR-2682-3p
inhibited osteosarcoma cell proliferation
Increased miR-2682-3p expression was
confirmed by qRT-PCR (Fig. 2A and B);
overexpression of miR-2682-3p decreased the proliferation of
tumor cells (MG-63, and U2OS) (Fig. 2C
and D).
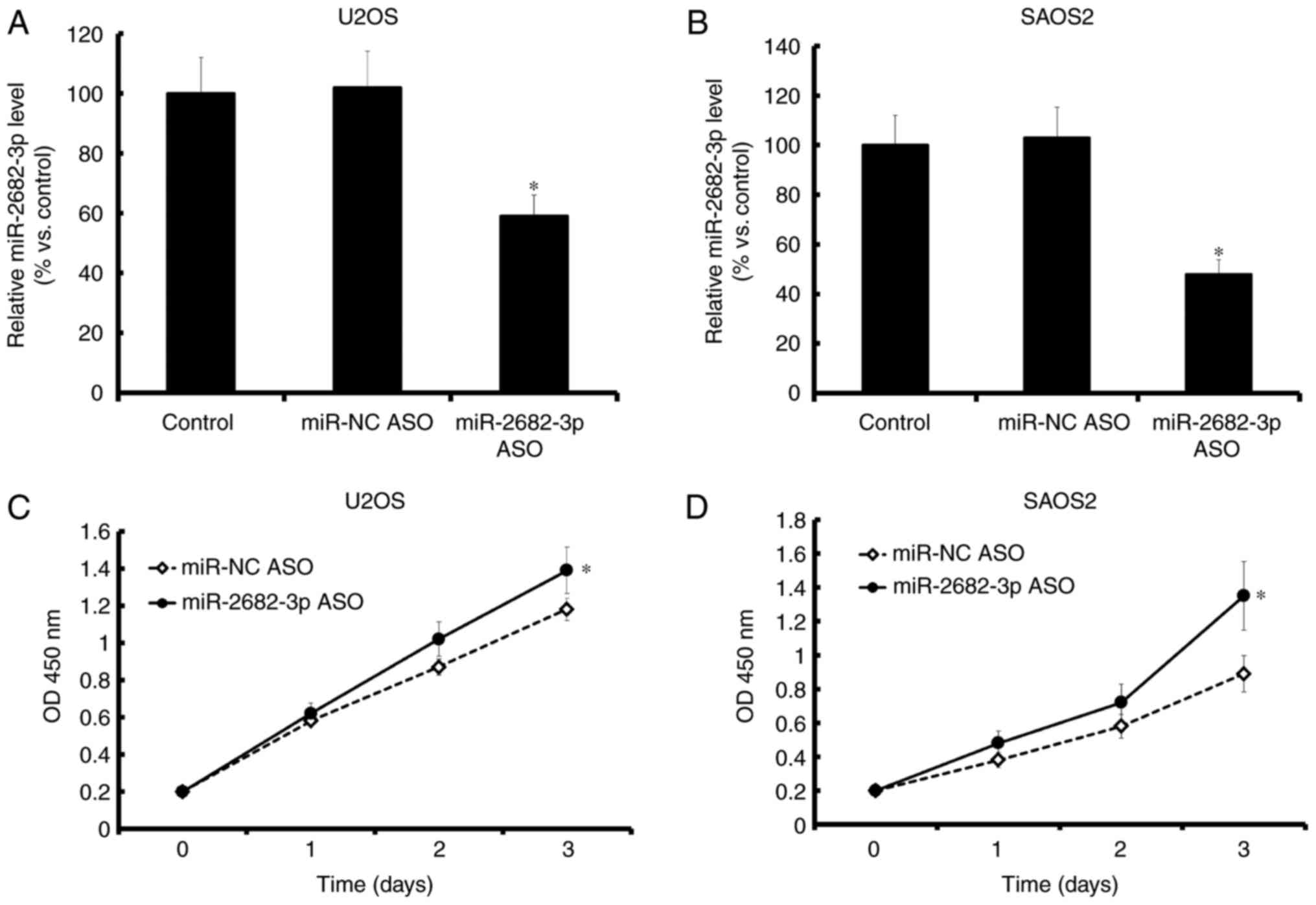
Downregulation of miR-2682-3p promoted
tumor proliferation
Decreased miR-2682-3p expression, which was
confirmed by qRT-PCR (Fig. 3A and B),
promoted the proliferation of tumor cells (MG-63 and U2OS)
(Fig. 3C and D).
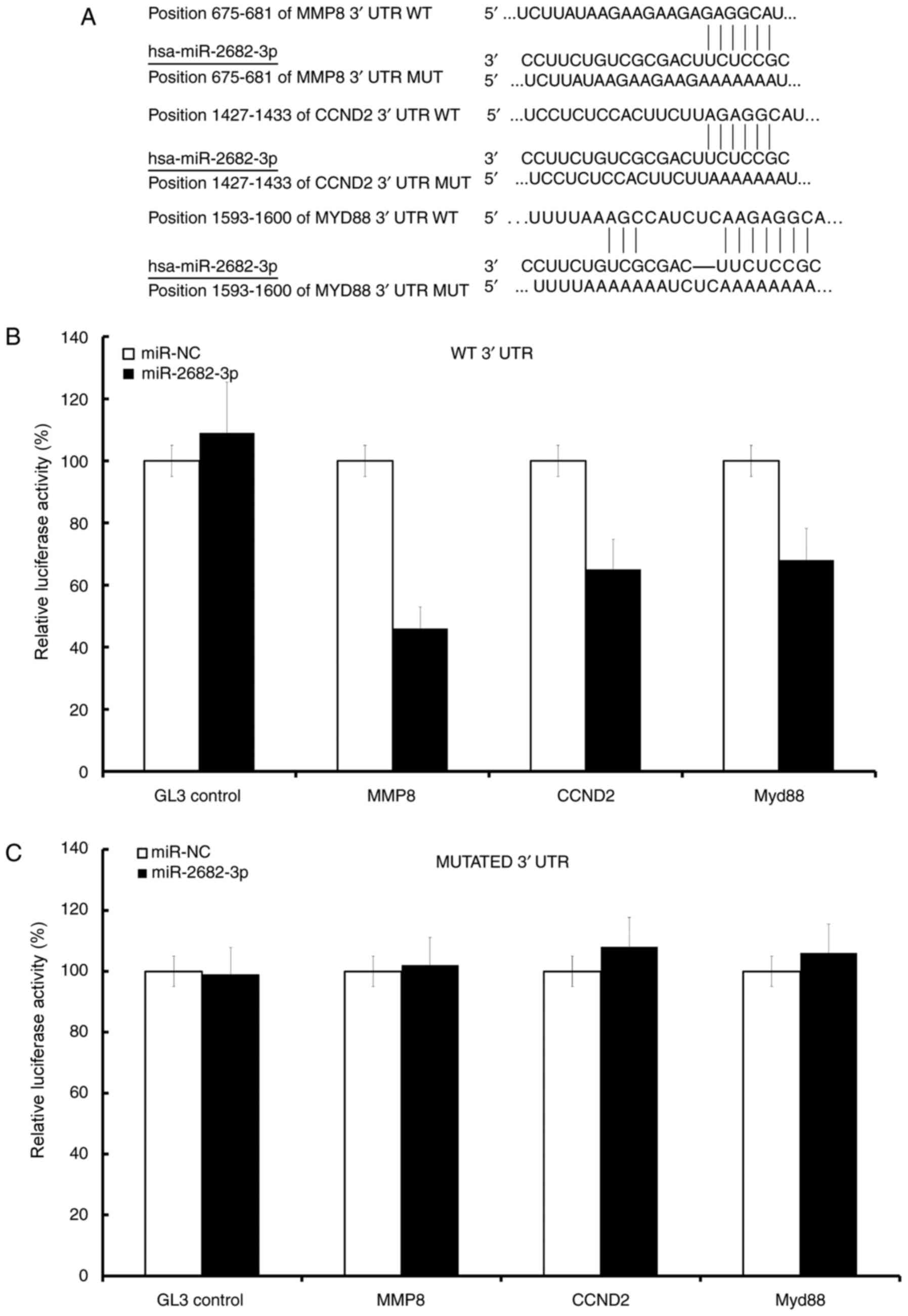
CCND2, MMP8, and Myd88 are the direct
targets of miR-2682-3p in osteosarcoma cells
CCND2, MMP8, and Myd88 were predicted to be
miR-2682-3p targets (Fig. 4A). The
mRNA levels of CCND2, MMP8, and Myd88 were inhibited in the
miR-2682-3p mimic groups but not in the control groups (Fig. 4B), the mutation of 3′UTR conversely
(Fig. 4C).
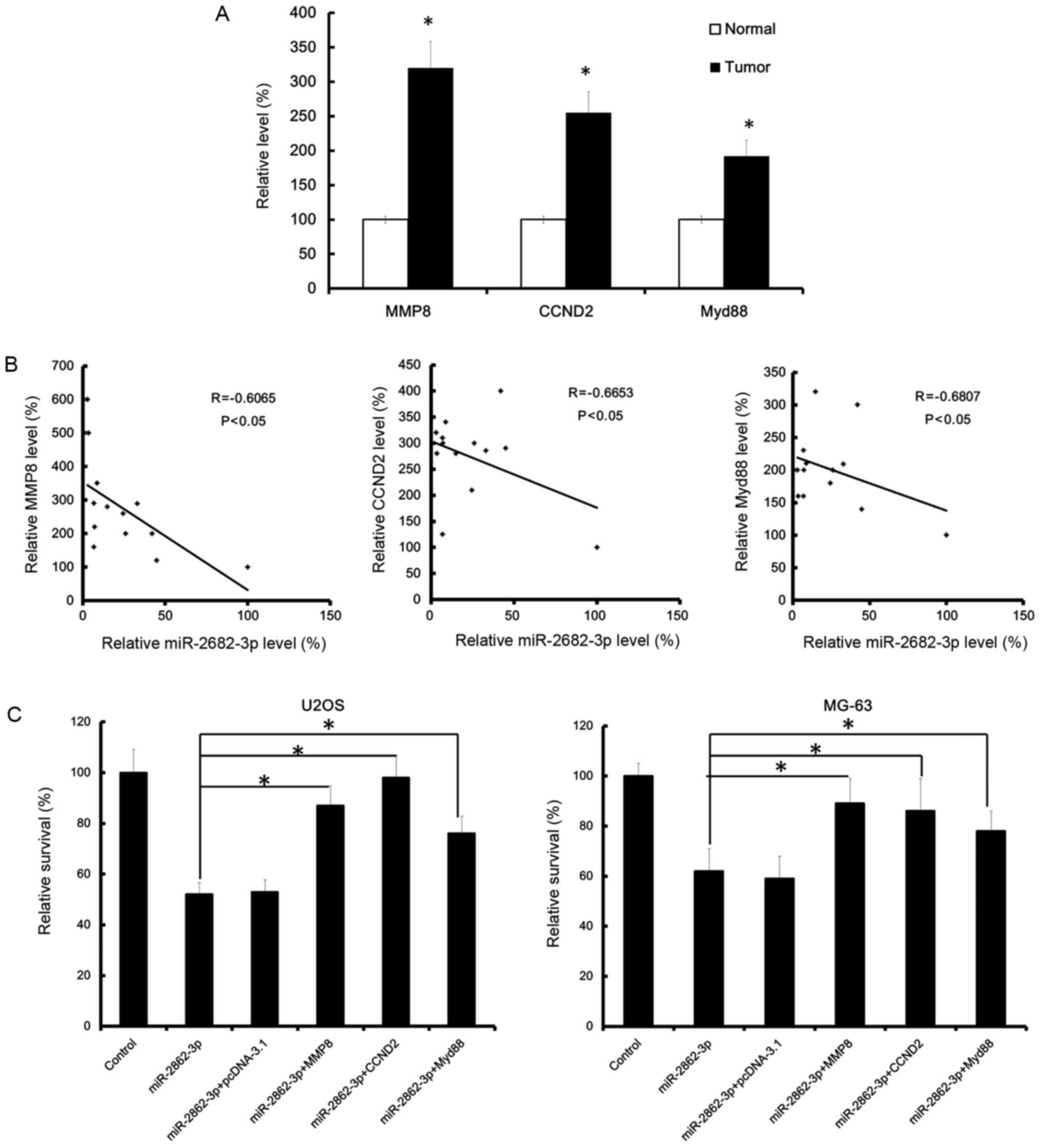
MMP8, CCND2, and Myd88 were
upregulated in osteosarcoma and inversely correlated with
miR-2682-3p expression
MMP8, CCND2, and Myd88 were upregulated in
osteosarcoma tissues. Moreover, the upregulation of MMP8, CCND2,
and Myd88 and miR-2682-3p expression were inversely correlated in
osteosarcoma tissues of patients (Fig. 5A
and B).
miR-2682-3p restrained osteosarcoma
cell proliferation by targeting CCND2, MMP8 and Myd88
CCND2, MMP8, and Myd88 promoted
osteosarcoma cell proliferation. The miR-2682-3p mimic that
was added into tumor cells inhibited osteosarcoma cell
proliferation and invasion. Consistent with our data, the survival
rate was significantly decreased in the miR-2682-3p groups.
However, in the co-transfected group, the survival rate was
significantly increased, indicating that CCND2, MMP8 and
Myd88 partly rescued the effect of miR-2682-3p on
osteosarcoma (Fig. 5C).
Discussion
Accumulating evidence has shown that
miR-2682-3p expression is downregulated in osteosarcoma
tissues and cell lines (17). The
data obtained in this study show that overexpression of
miR-2682-3p inhibits cell proliferation in MG-63 and U2OS
cells. Moreover, CCND2, MMP8, and Myd88 are
considered to be direct targets of miR-2682-3p. When
miR-2682-3p mimic and CCND2, MMP8, and Myd88 were added into
tumor cells, the effect of miR-2682-3p was partly rescued in
osteosarcoma. These results indicate that miR-2682-3p is a
potential tumor suppressor gene of osteosarcoma. However, further
investigations of the role of miR-2682-3p in vivo are
needed.
Increasing evidence indicates that miR-2682-3p is
involved in the progression of some cancers, such as osteosarcoma
(9). The same study recently
suggested that CCND2, MMP8, and MyD88 can promote cancer metastasis
and that the proliferation of many cancer cells can be induced by
regulating the composition of extracellular matrix (9). However, the underlying mechanisms by
which CCND2, MMP8, and Myd88 stimulate MG-63 cell growth still need
to be clarified. The present results show that one of the tumor
suppressor miRNAs is miR-2682-3p in osteosarcoma.
In conclusion, miR-2682-3p expression was decreased
in osteosarcoma tissues and cell lines. Overexpression of
miR-2682-3p leads to the inhibition cell proliferation of
osteosarcoma. CCND2, MMP8, and Myd88 are potential targets of
miR-2682-3p. Thus, miR-2682-3p has potential value as a new target
for the treatment of osteosarcoma in future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
FZ analyzed and interpreted the patient data
regarding the osteosarcoma. YZ performed the cytology examination
of the osteosarcoma. GF was a major contributor in writing the
manuscript and helped with the experiment and statistical analysis.
SH as the leader of this study, designed the experiment and
reviewed the article. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients participated in the study provided
written informed consent, and the study was approved by the Ethics
Committee of Shanghai Tenth People's Hospital.
Consent for publication
All the patients and researchers consented for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gorlick R and Khanna C: Osteosarcoma. J
Bone Miner Res. 25:683–691. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 Suppl 7:Vii320–Vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kager L, Zoubek A, Dominkus M, Lang S,
Bodmer N, Jundt G, Klingebiel T, Jürgens H, Gadner H and Bielack S:
COSS Study Group: Osteosarcoma in very young children: Experience
of the cooperative osteosarcoma study group. Cancer. 116:5316–5324.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Botter SM, Neri D and Fuchs B: Recent
advances in osteosarcoma. Curr Opin Pharmacol. 16:15–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moore DD and Luu HH: Osteosarcoma. Cancer
Treat Res. 162:65–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gianferante DM, Mirabello L and Savage SA:
Germline and somatic genetics of osteosarcoma-connecting aetiology,
biology and therapy. Nat Rev Endocrinol. 13:480–191. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Szewczyk M, Lechowski R and Zabielska K:
What do we know about canine osteosarcoma treatment? review. Vet
Res Commun. 39:61–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niu G, Li B, Sun L and An C: MicroRNA-153
inhibits osteosarcoma cells proliferation and invasion by targeting
TGF-β2. PLoS One. 10:e01192252015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng K, Wang H, Guo X and Xia J: The cross
talk between long, non-coding RNAs and microRNAs in gastric cancer.
Acta Biochim Biophys Sin (Shanghai). 48:111–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jothy SL, Chen Y, Vijayarathna S, Kanwar
JR and Sasidharan S: MicroRNAs: Association with radioresistant and
potential uses of natural remedies as green gene therapeutic
approaches. Curr Gene Ther. 15:15–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou P, Ding J and Fu S: Elevated
expression of microRNA-19a predicts a poor prognosis in patients
with osteosarcoma. Pathol Res Pract. 213:194–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lian D, Wang ZZ and Liu NS: MicroRNA-1908
is a biomarker for poor prognosis in human osteosarcoma. Eur Rev
Med Pharmacol Sci. 20:1258–1262. 2016.PubMed/NCBI
|
|
15
|
Chen J, Yan D, Wu W, Zhu J, Ye W and Shu
Q: MicroRNA-130a promotes the metastasis and epithelial-mesenchymal
transition of osteosarcoma by targeting PTEN. Oncol Rep.
35:3285–3292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang S, Zhao Y and Wang L: MicroRNA-198
inhibited tumorous behaviors of human osteosarcoma through directly
targeting ROCK1. Biochem Biophys Res Commun. 472:557–565. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Waresijiang N, Sun J, Abuduaini R, Jiang
T, Zhou W and Yuan H: The downregulation of miR-125a-5p functions
as a tumor suppressor by directly targeting MMP-11 in osteosarcoma.
Mol Med Rep. 13:4859–4864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Hou W, Chai M, Zhao H, Jia J, Sun
X, Zhao B and Wang R: MicroRNA-127-3p inhibits proliferation and
invasion by targeting SETD8 in human osteosarcoma cells. Biochem
Biophys Res Commun. 469:1006–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Duan J, Shi J, Fiorentino A, Leites C,
Chen X, Moy W, Chen J, Alexandrov BS, Usheva A, He D, et al: A rare
functional noncoding variant at the GWAS-implicated MIR137/MIR2682
locus might confer risk to schizophrenia and bipolar disorder. Am J
Hum Genet. 95:744–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu J, Lv G, Zhou S, Zhou Y, Nie B, Duan H,
Zhang Y and Yuan X: The Downregulation of MiR-182 is associated
with the growth and invasion of osteosarcoma cells through the
regulation of TIAM1 expression. PLoS One. 10:e01211752015.
View Article : Google Scholar : PubMed/NCBI
|