Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancer types globally, and the fifth highest cause of
cancer-associated mortality (1). The
etiology of HCC differs between regions; hepatitis C infection and
alcohol abuse are the most common causes of HCC in western
countries, while in China the majority of patients with HCC harbor
a hepatitis B (HBV)infection (2,3). A variety
of treatments are recommended for HCC, with radical treatments
including radical resection, liver transplantation and
radiofrequency ablation being recommended for the treatment of
early stage HCC. Patients diagnosed with advanced stage HCC are
candidates for palliative treatments, including sorafenib and other
targeted treatments (4,5). In total, ~30% of all patients with HCC
are diagnosed at an advanced stage in China (6), and novel treatments are required in
orderto improve the prognosis of these patients.
Zinc finger proteins (ZNF) are a family of DNA
binding proteins, encoded by 2% of all human genes (7). Generally, ZNFs are divided into eight
categories: C2H2 like (Cys2His2), Gag knuckle, treble clef, zinc
ribbon, Zn2/Cys6, TAZ2 domain like, Zinc binding loops and
metallothionein (8). C2H2 type zinc
finger protein is the largest of these groups. Previous studies
have revealed an association between ZNF expression and HCC
prognosis; Wang et al (9)
demonstrated that increased zinc finger and BTB domain containing
20 (ZBTB20) expression is associated with poor HCC prognosis, and
Kan et al (10) recently
reported that ZBTB20 is an independent prognostic biomarker for
HCC. Yang et al (11) analyzed
92 HCC tumor tissue samples and identified that zinc finger E-box
binding homeo box 2 expression was associated with tumor
metastasis. Recently, Wu et al (12) determined that ZNF191 inhibited HCC
metastasis by inactivating discs large 1-mediated yes-associated
protein.
ZNF689, a C2H2 type ZNF, is a putative transcription
regulating factor. A previous study demonstrated that the knockdown
of ZNF689 results in tumor cell growth inhibition (8). It has also been identified that ZNF689
inhibited tumor cell apoptosis via downregulation of the
pro-apoptotic factors B cell lymphoma (Bcl)2 antagonist/killer 1,
Bcl2 associated X protein (Bax) and BH3 interacting domain death
agonist (Bid) (13). In the
aforementioned study, immunohistochemical analysis was also
performed on HCC tumor samples and it was revealed that 4/12 cases
presented with nuclear expression of ZNF689. In order to further
explore whether ZNF689 is a novel treatment target and its
significance in determining HCC prognosis, the present study used
an increased number of specimens to analyze the association between
ZNF689 expression and the baseline characteristics of patients with
HCC.
Materials and methods
Patients and follow-up
A total of 102 paired HCC tissues and adjacent
non-cancerous tissues were collected from patients who underwent
radical liver resection between January 2011 and December 2012 in
West China Hospital of Sichuan University (Chengdu, Sichuan
Province, China), including 93 males and 9 females. Patients who
were diagnosed by histological detection and with a preserved liver
function (Child A,B), without prior systematic therapy or local
treatments prior to liver resection were included; additionally
patients who withdrew from follow-up or without sufficient clinical
data were excluded. Written informed consent was obtained from all
enrolled patients, and the study was ethically approved by the
Biomedical Ethics Committee of West China Hospital. The median
patient age was 55 years (range, 21–76), with 42 patients ≥60 years
(41.2%). A total of 51 patients (50%) presented with an
α-fetoprotein level >400 ng/ml, and 96 patients (94.1%)
presented with positive expression of the HBV surface antigen.
Among all patients, tumor size ranged from 1.5 to 13.8 cm (median
size, 6.0 cm). Tumor differentiation was classified as high
differentiation, moderate differentiation or low differentiation
(14). According to the Barcelona
Clinical Liver Cancer system (BCLC), HCC was classified as early
stage (BCLC A), intermediate stage (BCLC B), or advanced stage
(BCLC C) (4). A total of 15 patients
(14.7%) were staged as BCLC A, 48 (47.1%) as BCLC B, and 39 (38.2%)
as BCLC C. Liver function was measured by Child-Pugh grade system,
which is the most common evaluating system for liver function. It
comprises five variables total bilirubin, prothrombin time,
albumin, ascites, and hepatic encephalopathy. Child A is defined as
score 5,6, Child B is defined as score 7–9 (15). No patients received radiofrequency
ablation or trans-arterial chemoembolization prior to liver
resection. Liver tissues were collected <15 min after liver
resection and stored at −80°C. Paired non-cancerous tissue was
defined as liver tissue >3 cm from the tumor resection edge. All
frozen liver samples tested in the present study were stored in the
Clinical Sample Bank of West China Hospital. Follow-up was
performed at one to three-month intervals via an outpatient visit
or telephone call. Recurrence was defined as new lesions within the
residual liver or distant organ metastasis detected by ultrasound,
computed tomography or magnetic resonance imaging scans. Follow-up
was completed in May 2016.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from each specimen using
TRIzol® reagent (Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. RNA concentration was determined with a ScanDrop Nuclear
Acid Analyzer (AnalytikJenaAG, Jena, Germany). To determine the
integrity of RNA, 3 µg of each RNA sample was separated on a 1%
denatured agarose gel and then detected via a Chemical Mpimaging
system (Bio-Rad, Laboratories, Inc, Hercules, CA, USA). If the peak
area of the 28S ribosome RNA (rRNA) was approximately twice that of
18S rRNA, the integrity of the total RNA was considered as
acceptable and used for continued investigation. ZNF689 mRNA was
quantified using RT-qPCR, with primers designed via Primer 5.0
software (PremierBiosoft, International, Palo Alto, CA, USA), and
synthesized by Sangon Biotech (Shanghai, China) (Table I). The reference gene used for qPCR is
GAPDH, the detail sequence was listed in Table I. cDNA was synthesized using a
RevertAid First-Strand cDNA Synthesis kit (ThermoFisher Scientific,
Inc.), and qPCR was performed in triplicate for each sample using
Maxima SYBR Green qPCR Master mix (Thermo Fisher Scientific, Inc.)
on the CFX connect Real-Time system (Bio-Rad, Laboratories, Inc.).
Amplification proceeded for 3 min at 95°C for denaturing, followed
by 40 cycles for ZNF689 and GAPDH, at 95°C for 15 sec, 60°C for 30
sec in the CFX connect Real-Time system. Relative expression levels
of each gene were calculated using the 2−ΔΔCq method
(16).
 | Table I.Primers used in RT-qPCR. |
Table I.
Primers used in RT-qPCR.
| Primer | Sequence (5′-3′) | No. of bases |
|---|
| ZNF689-forward |
TGGAACGAAACACCGATGACT | 21 |
| ZNF689-reverse |
CCATTCTTCTTTCTGGTTCTGCT | 23 |
| GAPDH-forward |
ACTCCTCCACCTTTGACGC | 19 |
| GAPDH-reverse |
GCTGTAGCCAAATTCGTTGTC | 21 |
Cell culture and transfection
The normal hepatic cell line LO2 and HCC cell lines
(Huh7, MHCC97H, and Hep3B) were purchased from the Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). HCC cell lines were preserved in Dulbecco's
modified Eagle's medium (DMEM) (Hyclone, GE Healthcare Life
Sciences, Logan, UT, USA). LO2 cells were maintained in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS) (Hyclone; GE Healthcare Life
Sciences), 100 units/ml penicillin, and 100 mg/ml streptomycin.
Cells were placed in a humidified atmosphere containing 5%
CO2 at 37°C. Cells were transfected with lentivirus or
plasmids using Turbo transfection reagent. Lentivirus were
purchased from Shanghai Genechem Co Ltd (Shanghai, China) shRNA
were loaded by GV208, EGFP, and AMP vectors. Trans-KD™ Easy was
used as transfection reagent, and purchased from Shanghai Genechem
Co Ltd. Transfection incubation was set at 37°C for 48 h.
HCC cell migration and wound-healing
assays
To perform a transwell migration assay,
1×105 cells obtained from MHCC-97 shRNA transfection
(M-Si) cell line were seeded into the upper chamber of transwell
plates (BD Biosciences, Franklin Lakes, NJ, USA) with serum-free
DMEM. The lower chamber was filled with DMEM containing 10% FBS.
After incubation for 48 h at 37°C, cells in the upper surface of
the filters were softly removed by a cotton swap, then cells
migrating to the lower surface of the filters were fixed inethanol
for 20 min at room temperature. Cells were randomly counted in
eight fields, from threedifferent membranes. Considering
Wound-healing assay, Cells in logarithmic growth phase were
cultured in a 6-well plate until 90% confluence was reached. Next,
a single wound was created in the monolayer cells by gently
scratching the attached cells with a sterile 10 µl micropipette tip
(time 0 h). After scratching, the cells were incubated with
serum-free DMEM medium for 24 h. Cell migration was photographed
using fluorescence microscopy (IX70; Olympus, Japan) and ax10
objective at 48 h following injury. Remodeling was measured as the
diminishing distance across the induced injury, normalized to the 0
h control, and expressed as relative migration. Each experiment was
performed at least three times independently.
Western blot analysis
Protein was extracted from tissues using
radioimmunoprecipitation assay (RIPA buffer) (BeyotimeInstitute of
Biotechnology, Shanghai, China) buffer containing a 1/10 Complete
Miniprotease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim,
Germany). RIPA lysate was added into the samples, at 0°C for 30 min
following vortex blending, and then a 10 min intermittent
oscillation was performed. The protein concentration was measured
using a bicinchoninic acid protein assay kit (Beyotime, Institute
of Biotechnology). Following denaturing at 95°C for 10 min, 50 µg
of each protein sample was separated using SDS-PAGE (10% gel) and
then transferred to a polyvinylidene difluoride membrane.
Subsequent to blocking with 5% nonfat dry milk in TBS with 0.1%
Tween-20 (TBST) for 1 h at room temperature, the membrane was
incubated with primary antibodies at 4°C overnight. The primary
antibodies used for western blot included ZNF-689 from
Sigma-Aldrich, Merck KGaA, Darmstadt, Germany (1:1,000, Cat no.
SAB1408243), E-cadherin (1:1,000, Cat no. 3195T), β-catenin
(1:1,000, Cat no. 8480T) and Snail 1 (1:1,000, Cat no. 3879T) from
Cell signaling Technology, Inc., Danvers, MA, USA, the GAPDH
employed as normalized control from Zen BioScience (1:2,000, Cat
no. 220068, Chengdu, China). Subsequent to washing three times with
TBST buffer for 10 min per wash, the membrane was further incubated
at room temperature for 1 h with horseradish peroxidase conjugated
rabbit anti-mouse IgG (cat no. 250097) or mouse anti-rabbit (cat
no. 701051) Secondary antibodies were also purchased from Zen
BioScience, (Chengdu, China) and used at a dilution of 1:5,000.
Subsequently, the results were scanned with a Chemical Mp Imaging
System (Bio-Rad Laboratories). Following treatment with immobilon
ECL ultra western HRP substrate of Millipore (Merck KGaA,
Darmstadt, Germany) according to the manufacturer's protocol the
expression levels of each protein was quantified by Image J
software version 1.8.0 (National Institutes of Health, Bethesda,
MD, USA).
Immunohistochemistry (IHC)
staining
IHC was conducted according to the instructions of
the SP-9001 kit (Beyotime Institute of Biotechnology). Images were
observed at magnification, ×400 under a fluorescence microscope.
10% formalin-fixed, paraffin-embedded tissue sections of
representative areas of tumor were cut into 4 µm-thick sections.
IHC was performed with a standard two-step method. First, the
slides were de-paraffinized with xylene and rehydrated a graded
alcohol series (100, 90, 70 and 50% ethyl alcohol) for 10 min at
room temperature, and antigen retrieval was performed by incubating
samples in sodium citrate buffer (pH 6.0) for 30 min at 98°C.
Inactivation of endogenous peroxidase was performed with a 3%
hydrogen peroxide solution for 20 min at room temperature. After 30
min blocking (5% normal goat serum purchased from Beyotime
Institute of Biotechnology) at room temperature), the tissue
sections were incubated with rabbit polyclonal anti-ZNF689 primary
antibodies (SAB2701570; dilution 1:100) (Sigma-Aldrich; Merck KGaA)
overnight at 4°C. Subsequent to washing, the slides were incubated
with horseradish peroxidase-conjugated secondary antibody for 40
min at 37°C (SAB2701462; dilution 1:500) (Sigma-Aldrich; Merck
KGaA).
IHC staining was evaluated by an immunoreactivity
score (IRS), which was calculated by multiplying the staining
intensity and extent as previously described (17). Deng DW who works in Department of
hepato-biliary-pancrease of Affiliated Hospital of North Sichuan
College performed IHC and he was blinded to the mRNA ZNF689
expression results. The staining intensity was classified as:
0(negative), 1(weak), 2(moderate) or 3(strong). Based on the
percentage of positively stained cells throughout the tumor, the
extent of staining was defined as 0(0%), 1(<10%), 2(10%-50%),
3(51%-80%) or 4(>80%). IRS score is the staining intensity score
multiplied by the extent of staining score, ranging from 0 to 12,
with an IRS score of ≥4 defined as positive expression, and an IRS
score of <4 defined as negative expression.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 21.0; IBM Corp, Armonk, NY, USA) and GraphPad
Prism software (version 5.00; GraphPad Software, Inc., La Jolla,
CA, USA). P<0.05 was considered to indicatea statistically
significant difference. Continuous variables are presented as the
mean ± standard deviation or mean ± standard difference with 95%
confidence intervals (CI). An unpaired Student's t-tests or one-way
analysis of variance tests were performed to compare continuous
variables with parametric distributions, the post-hoc test used
following the analysis of variance was Student Newman Keuls test.
Categorical data were analyzed using the chi-squared or Fisher's
exact tests. Survival data were calculated with the Kaplan-Meier
method and compared by the log-rank test. Univariate and
multivariate analyses were performed using Cox's regression
method.
Results
Expression of ZNF689 in HCC, paired
non-cancerous tissue, and normal liver tissue
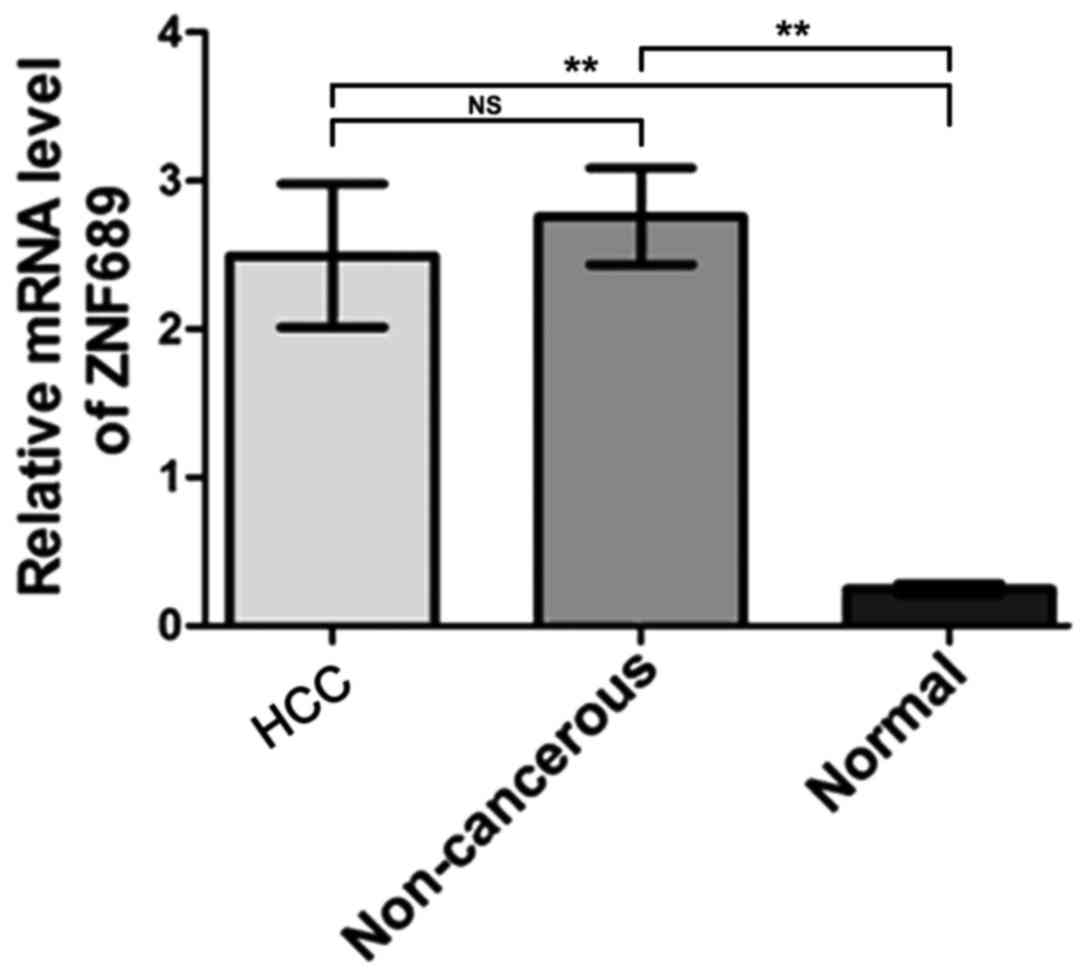
RT-qPCR analysis revealed no significant difference
in the expression of ZNF689 mRNA between HCC and paired
non-cancerous tissues, with a mean difference of −0.26 (95% CI,
−1.32, 0.80; P=0.62). In addition, the expression of ZNF689 mRNA
was detected in 16 normal liver tissues collected from patients
with hemangioma who underwent liver resection in West China
Hospital of Sichuan University from January 2011 to December 2012.
The diagnosis of hemangioma was based on pathological test, and
patients with hemangioma who have not received any systematic or
local therapy were included, those who also complicated with tumor
lesions in other organs were excluded. Of these 16 patients, 10
were female, 6 were male, and the median age was 41.7 years (age
range 27–68 years). All these 16 patients were diagnosed as
hemangioma and not included in the above 102 patients. The results
demonstrated that ZNF689 mRNA expression was increased in HCC
tissues and paired non-cancerous tissues compared with the normal
liver tissues. The differences between means were 2.246±0.4840 (95%
CI, 1.278, 3.214) and 2.510±0.3273 (95% CI, 1.856, 3.165),
respectively (P<0.05; Fig. 1).
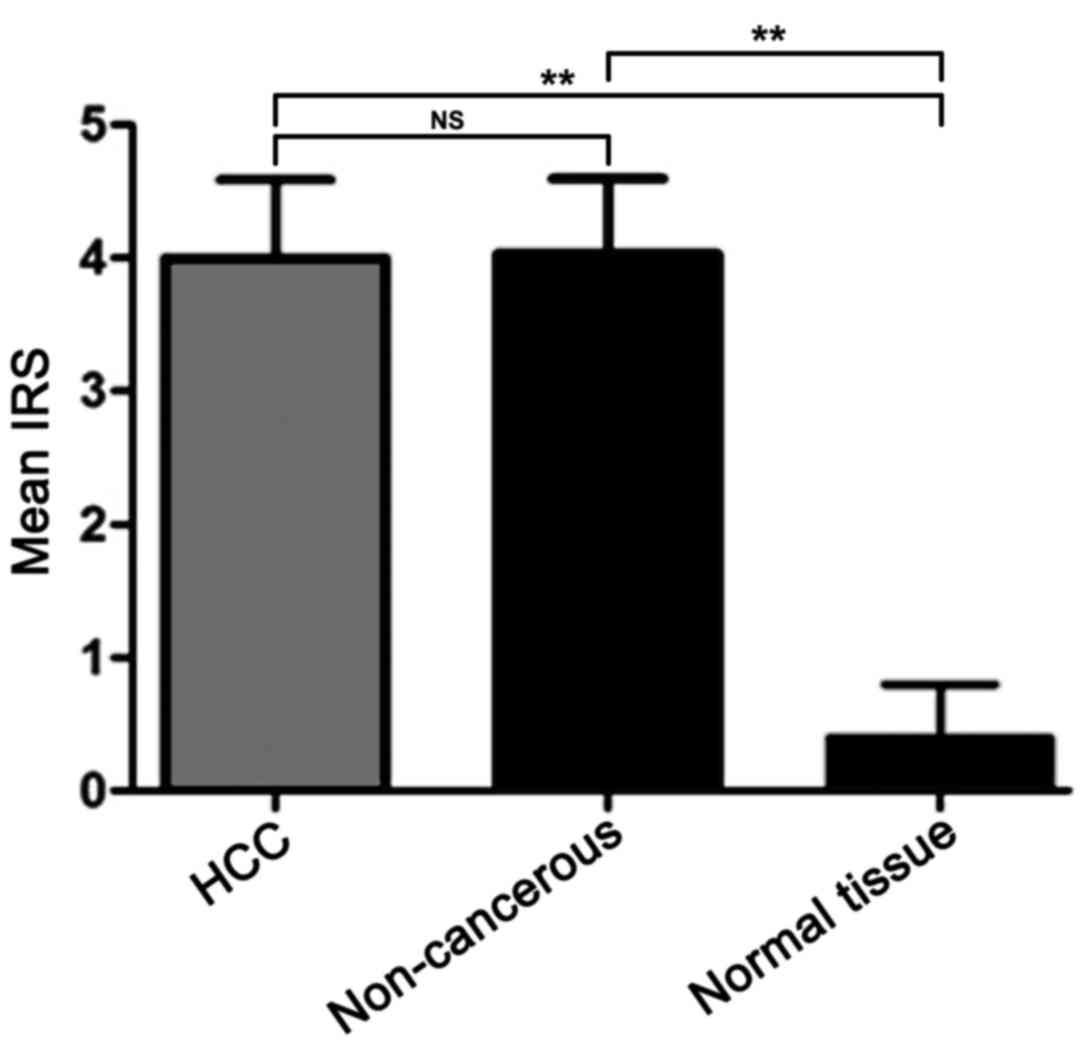
IHC analysis demonstrated that the mean IRS score
was 4, 4.03, and 0.4 in the tumor, paired non-cancerous and normal
liver tissues, respectively (Fig. 2).
IHC revealed positive expression of ZNF689 protein (IRS score ≥4)
in 45 cases (45/102, 44.12%) of HCC and 45 cases (45/102, 44.12%)
of paired non-cancerous tissues. There was no significant
difference in the expression levels of the ZNF689 protein between
HCC and paired non-cancerous tissues, and the mean difference of
the ZNF689 protein in HCC and non-cancerous tissues was −0.02941
(95% CI, −1.587, 1.528, P=0.97). However, the expression levels of
the ZNF689 protein in HCC and non-cancerous tissues were
significantly higher compared with those in normal liver tissues
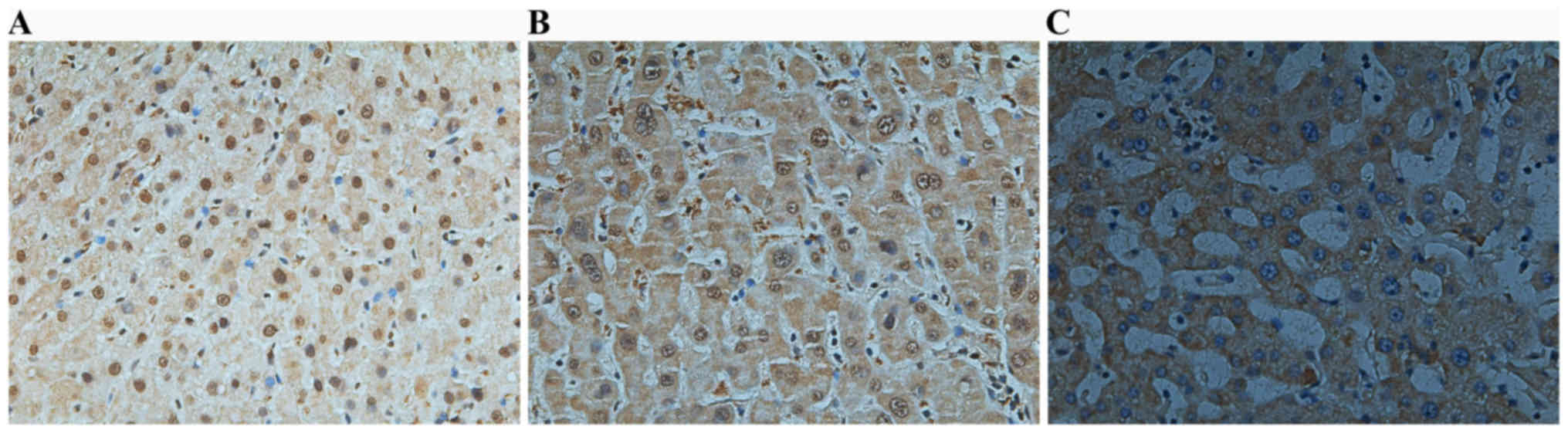
(P<0.05; Fig. 2). Representative
images of the IHC analysis are presented in Fig. 3.
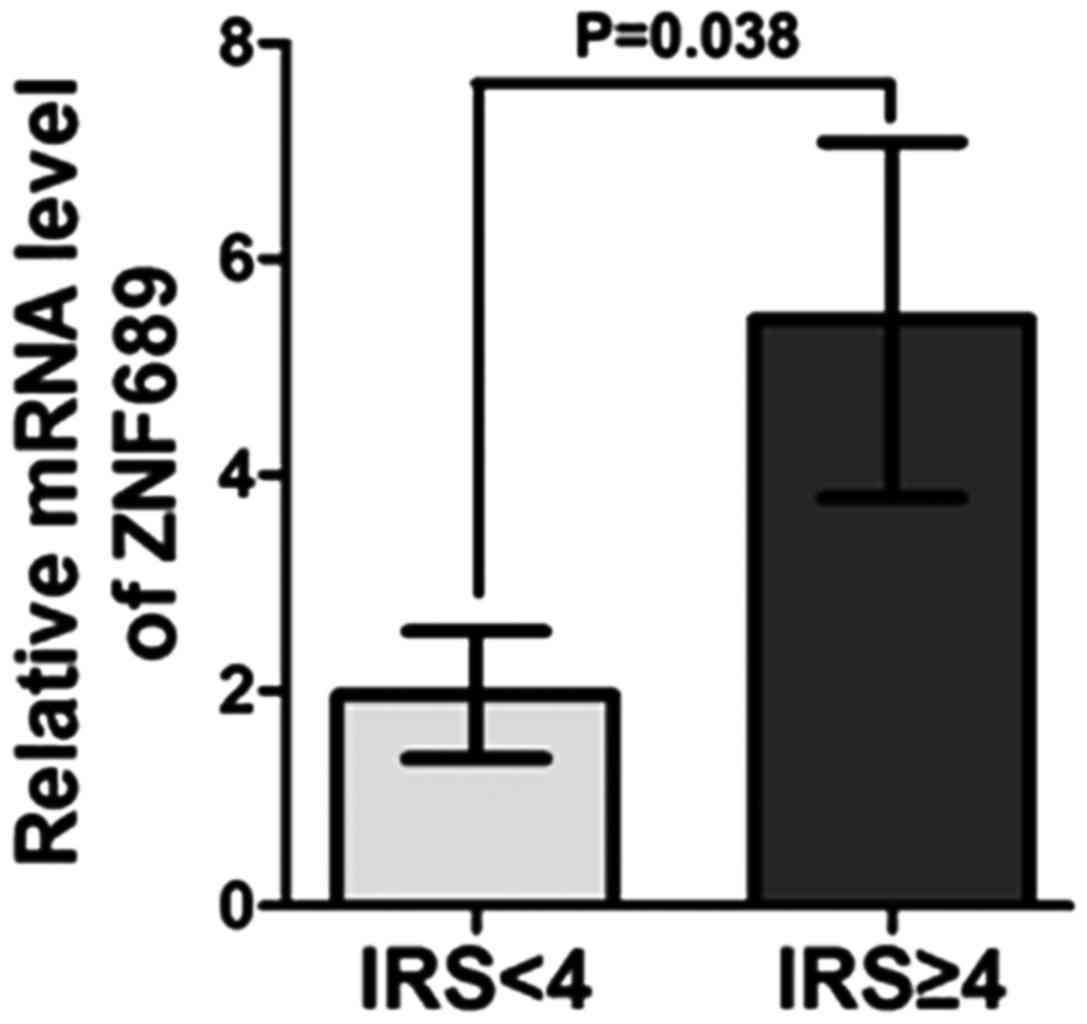
The relative mRNA levels of ZNF689 were analyzed in
the positive and the negative expression group and it was
identified that the mRNA level in the positive expression group was
significantly increased compared with that in the negative
expression group (P=0.038), indicating that ZNF689 expression was
consistent between them RNA and protein level (Fig. 4).
Association between ZNF689 levels in
HCC tissues and clinicopathological parameters
Associations between ZNF689 expression in HCC and
clinicopathological parameters is summarized in Table II. Positive expression of the ZNF689
protein in HCC was significantly associated with a tumor size of
≥10 cm (P=0.028), tumor capsule infiltration (P=0.005) and
microvascular invasion (MVI; P=0.037). No significant association
was observed between expression of the ZNF689 protein and age, sex,
HBV surface antigen, the extent of cirrhosis, tumor number, portal
vein embolus, BCLC stage or differentiation.
 | Table II.Associations between ZNF689 expression
levels and the clinical characteristics of patients with
hepatocellular carcinoma. |
Table II.
Associations between ZNF689 expression
levels and the clinical characteristics of patients with
hepatocellular carcinoma.
| Characteristics | Positive expression
(IRS score ≥4) | Negative expression
(IRS score <4) | P-value |
|---|
| Sex |
|
| 0.495 |
|
Male | 42 | 51 |
|
|
Female | 3 | 6 |
|
| Age (years) |
|
| 0.171 |
|
>60 | 12 | 30 |
|
|
<60 | 33 | 27 |
|
| α-fetoprotein
(ng/ml) |
|
| 0.549 |
|
>400 | 24 | 27 |
|
| <400 | 21 | 30 |
|
| Hepatitis B surface
antigen |
|
| 0.492 |
|
Positive | 45 | 51 |
|
|
Negative | 0 | 6 |
|
| HBV-DNA |
|
| 0.495 |
|
>103/copies | 30 | 30 |
|
|
<103/copies | 15 | 27 |
|
| Child-Pugh
classification |
|
| 0.764 |
| Child
A | 42 | 54 |
|
| Child
B | 3 | 3 |
|
| Liver
cirrhosis |
|
| 0.506 |
|
Yes | 33 | 45 |
|
| No | 12 | 12 |
|
| Tumor number |
|
| 0.613 |
|
Single | 42 | 48 |
|
|
Multiple | 3 | 9 |
|
| Tumor size
(cm) |
|
| 0.028 |
|
>10 | 18 | 3 |
|
|
<10 | 27 | 54 |
|
| Tumor size
(cm) |
|
| 0.104 |
|
>5 | 42 | 39 |
|
|
<5 | 3 | 18 |
|
| Cancer embolus in
portal vein |
|
| 0.718 |
|
Yes | 15 | 15 |
|
| No | 30 | 42 |
|
| BCLC stage |
|
| 0.410 |
| BCLC
A | 3 | 12 |
|
| BCLC
B | 21 | 27 |
|
| BCLC
C | 21 | 18 |
|
| Tumor capsule |
|
| 0.005 |
|
Complete infiltration | 6 | 36 |
|
| No
capsule infiltration | 39 | 21 |
|
| Microvascular
invasion |
|
| 0.037 |
|
Yes | 33 | 18 |
|
| No | 12 | 39 |
|
|
Differentiation |
|
| 0.273 |
|
High | 3 | 3 |
|
|
Moderate | 42 | 45 |
|
|
Low | 0 | 9 |
|
Positive expression of ZNF689 in HCC
is associated with poor prognosis
Follow-up was completed in May 2016. Since the
majority of the patients enrolled in the present study were from a
rural area in China, a lack of compliance with treatment guidelines
and financial difficulties resulted in a relatively high incidence
of loss to follow-up.
Of the 63 patients who completed the follow-up, 47
(74.6%) experienced recurrence and 39 (61.9%) patients had
succumbed to the disease by the follow-updeadline. The median
follow-up time was 33 months (range, 1–65 months). The overall
survival (OS) rate was 38.1% (24/63), with a median OS of 33
months. The median progression-free survival (PFS) was 20
months.
Patients with follow-up data were grouped into two
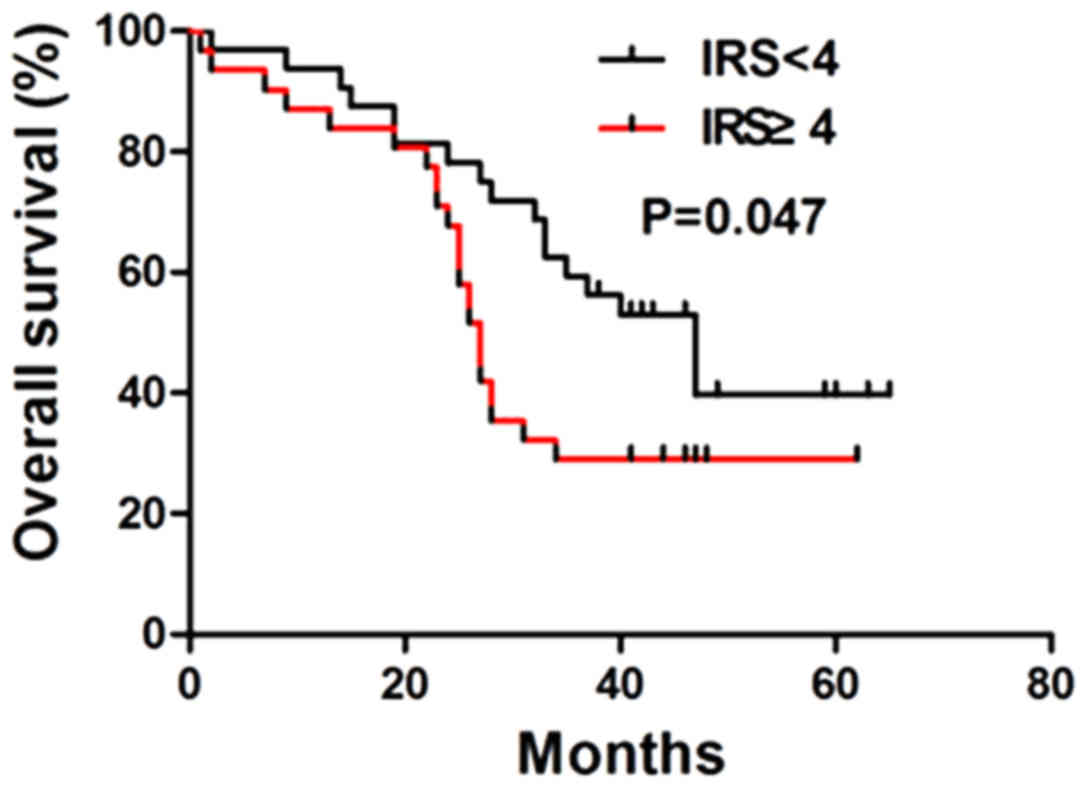
categories according to the IRS in HCC tissues. The median OS in
the positive expression group (27±1.10 months; 95% CI, 24.85–29.15)
was significantly lower compared with that in the negative
expression group (47±6.33 months; 95% CI, 34.59–59.41;
χ2=3.954; P=0.047; Fig.
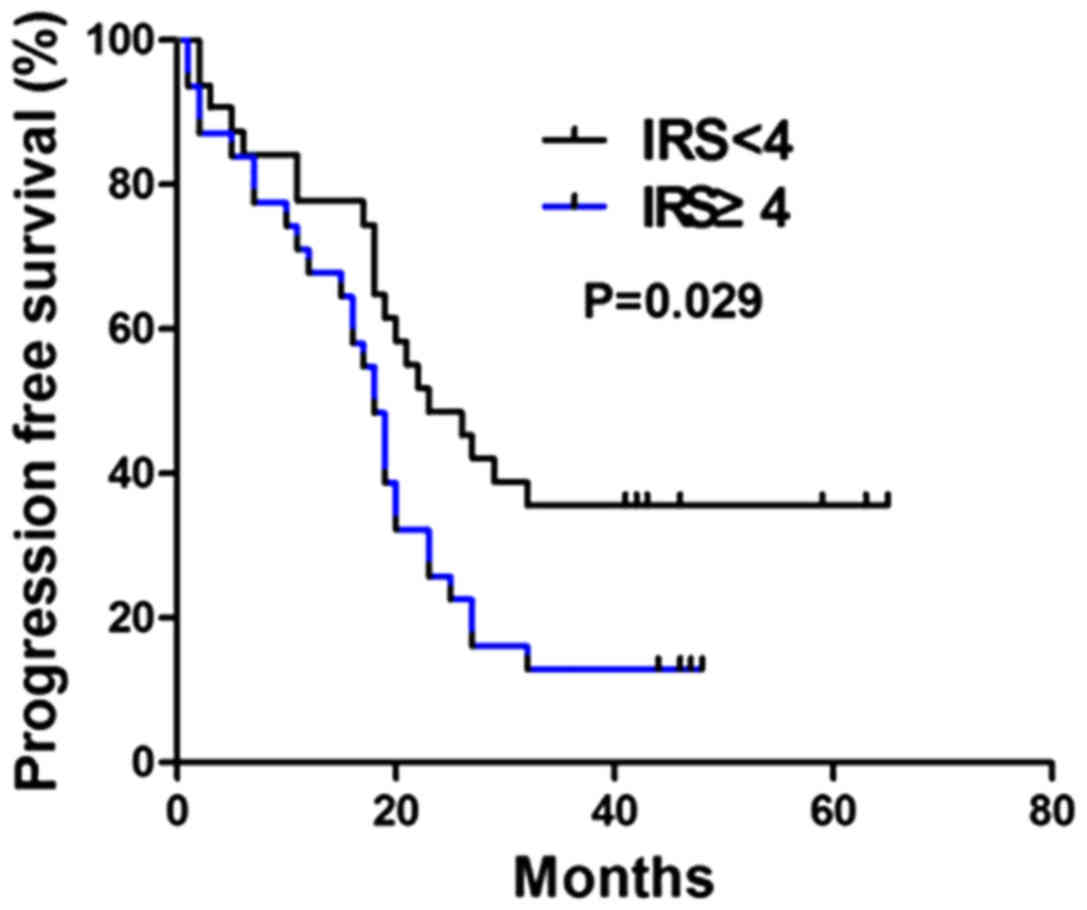
5). The median PFS in the positive expression group (19±1.40
months; 95% CI, 15.28–20.73) was also significantly lower compared
with that in the negative expression group (23±4.17; 95% CI,
14.85–31.16; χ2=4.762; P=0.029; Fig. 6).
Prognostic factors for OS and PFS
Univariate analysis indicated that cancer embolus in
the portal vein [hazard ratio (HR), 3.894; P=0.013], MVI (HR,
3.109; P=0.040) and positive expression of ZNF689 (HR, 2.033;
P=0.041) were prognostic factors for OS. Multiple tumors (HR,
2.399; P=0.019), cancer embolus in the portal vein (HR, 2.388;
P=0.009), tumor capsule infiltration (HR, 2.398; P=0.013), MVI (HR,
2.799; P=0.002) and positive expression of ZNF689 (HR, 1.967;
P=0.036) were prognostic factors for PFS detected by univariate
analysis.
For multivariate analysis, the known prognostic
factors by univariate analysis were selected together with the
known significant clinical variables. Cancer embolus in the portal
vein (HR, 2.298; P=0.038), MVI (HR, 2.178; P=0.047) and positive
expression of ZNF689 (HR, 1.961; P=0.048) were significantly
associated with OS, whereas multiple tumors (HR, 1.398; P=0.021),
cancer embolus in the portal vein (HR, 1.561; P=0.045), MVI (HR,
2.108; P=0.014) and positive expression of ZNF689 (HR, 1.902;
P=0.041) were prognostic factors for PFS (Table III).
 | Table III.Prognostic factors for overall
survival and progression-free survival by univariate and
multivariate analysis. |
Table III.
Prognostic factors for overall
survival and progression-free survival by univariate and
multivariate analysis.
| A, Univariate
analysis |
|---|
|
|---|
|
| Overall
survival | Progression free
survival |
|---|
|
|
|
|
|---|
| Factors | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years, >60
vs. <60) | 0.458 | (0.156,1.348) | 0.156 | 1.246 | (0.700,2.219) | 0.455 |
| α-fetoprotein
(ng/ml, >400 vs. <400) | 0.774 | (0.280,2.138) | 0.621 | 0.956 | (0.518,1.764) | 0.886 |
| Hepatitis B surface
antigen (positive vs. negative) | 0.653 | (0.085,5.015) | 0.682 | 2.402 | (0.329,17.528) | 0.387 |
| HBV-DNA (copies,
>103 vs. <103) | 0.613 | (0.217,1.730) | 0.355 | 0.810 | (0.441,1.488) | 0.497 |
| Child-Pugh
classification (Child B vs. Child A) | 2.466 | (0.317,19.198) | 0.389 | 1.451 | (0.349,6.039) | 0.609 |
| Liver cirrhosis
(Yes vs. no) | 1.989 | (0.440,8.989) | 0.372 | 1.639 | (0.725,3.704) | 0.235 |
| Tumor number
(multiple vs. single) | 1.681 | (0.465,6.081) | 0.428 | 2.399 | (1.153,4.989) | 0.019 |
| Tumor size (cm,
>10 vs. <10) | 1.291 | (0.569,2.930) | 0.541 | 1.405 | (0.655,3.015) | 0.383 |
| Tumor size (cm,
>5 vs. <5) | 0.899 | (0.300,2.696) | 0.850 | 1.432 | (0.715,2.869) | 0.311 |
| Cancer embolus in
portal vein (Yes vs. no) | 3.849 | (1.331,11.126) | 0.013 | 2.388 | (1.238,4.605) | 0.009 |
| Tumor capsule
(infiltration vs. complete) | 1.579 | (0.537,4.643) | 0.406 | 2.398 | (1.201,4.788) | 0.013 |
| Microvascular
invasion (Yes vs. no) | 3.109 | (1.108,9.976) | 0.040 | 2.799 | (1.464,5.351) | 0.002 |
| Differentiation
High+moderate vs. low | 1.313 | (0.455,3.792) | 0.615 | 1.155 | (0.616,2.169) | 0.653 |
| ZNF689 expression
(positive vs. negative) | 2.033 | (1.054,3.920) | 0.041 | 1.967 | (1.092,3.543) | 0.036 |
|
| B, Multivariate
analysis |
|
| Overall
survival | Progression free
survival |
|
| Factors | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Tumor number
(multiple vs. single) | 1.473 | (0.116,1.929) | 0.297 | 1.398 | (1.181,2.872) | 0.021 |
| Cancer embolus in
portal vein (Yes vs. no) | 2.298 | (1.481,4.835) | 0.038 | 1.561 | (1.058,3.922) | 0.045 |
| Tumor capsule
(infiltration vs. complete) | 0.946 | (0.294,3.014) | 0.926 | 1.597 | (0.750,3.401) | 0.225 |
| Microvascular
invasion (Yes vs. no) | 2.178 | (1.118,4.245) | 0.047 | 2.108 | (1.162,3.825) | 0.014 |
| ZNF689 expression
(positive vs. negative) | 1.961 | (1.023,3.758) | 0.048 | 1.902 | (1.061,3.412) | 0.041 |
Underlying mechanism of ZNF689
regulating invasion and migration of HCC
The ZNF689 expression was evaluated in LO2 cells and
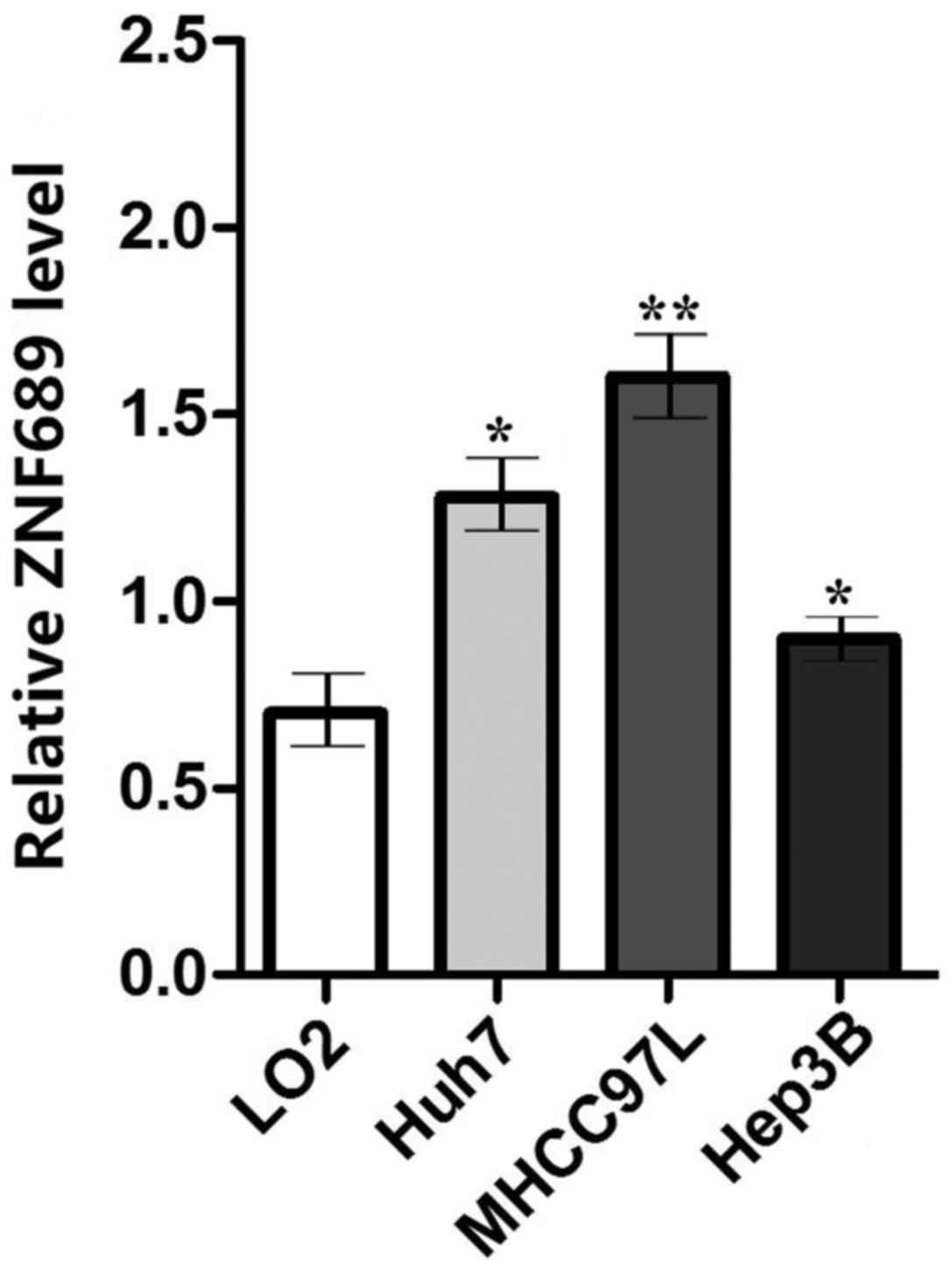
three HCC cell lines (Huh7, MHCC97H and Hep3B). It was identified
that the expression of ZNF689 in HCC cell lines was significantly
higher compared with that in LO2 (P<0.05; Fig. 7). ZNF689 was knocked down and
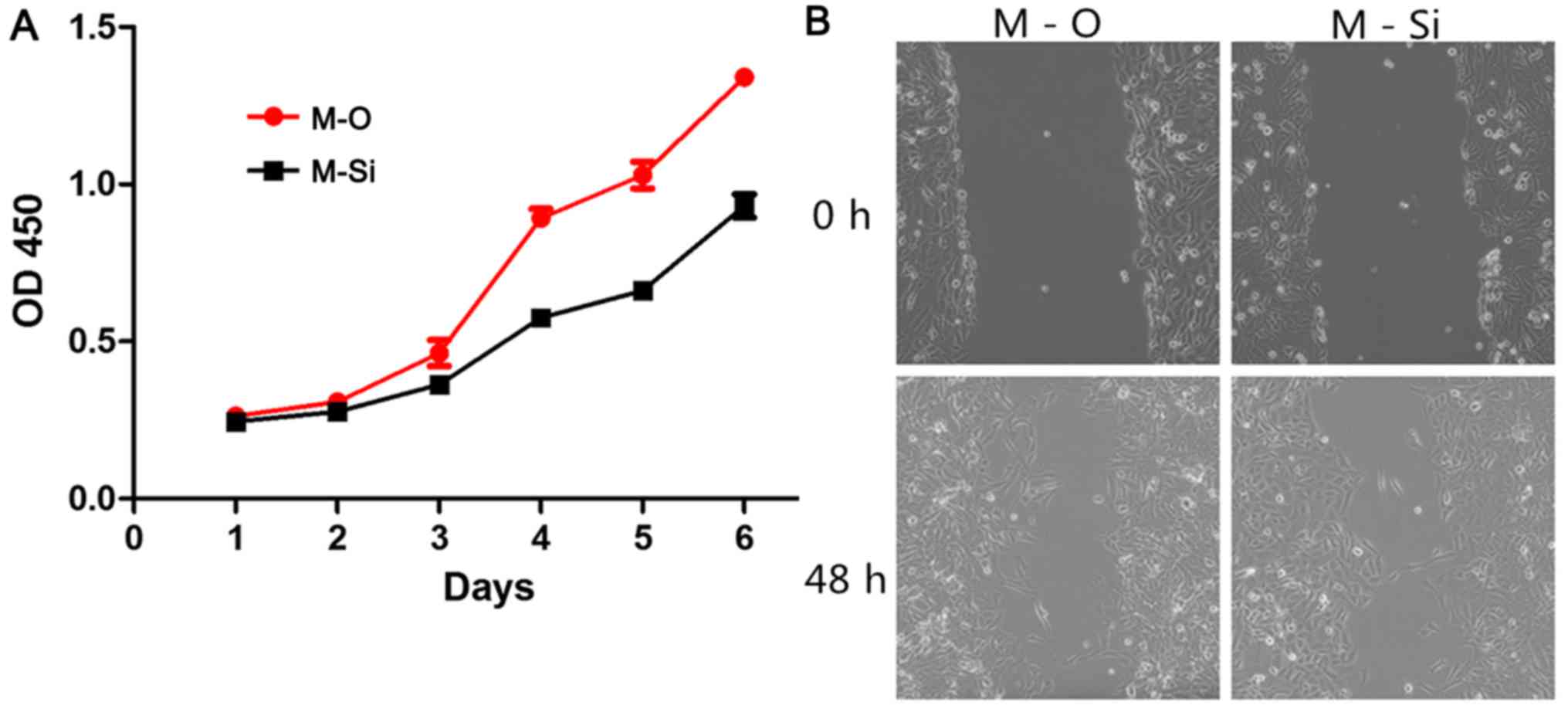
over-expressed in the HCC cell line MHCC97L, and it was revealed
that knockdown of ZNF689 substantially suppressed the proliferation
and migration of HCC cells (Fig.
8).
To investigate the potential underlying molecular
mechanism of ZNF689 regulating the invasion and migration of HCC,
the expression of E-cadherin, which is the most important biomarker
of the epithelial-mesenchymal transition (EMT), was investigated in
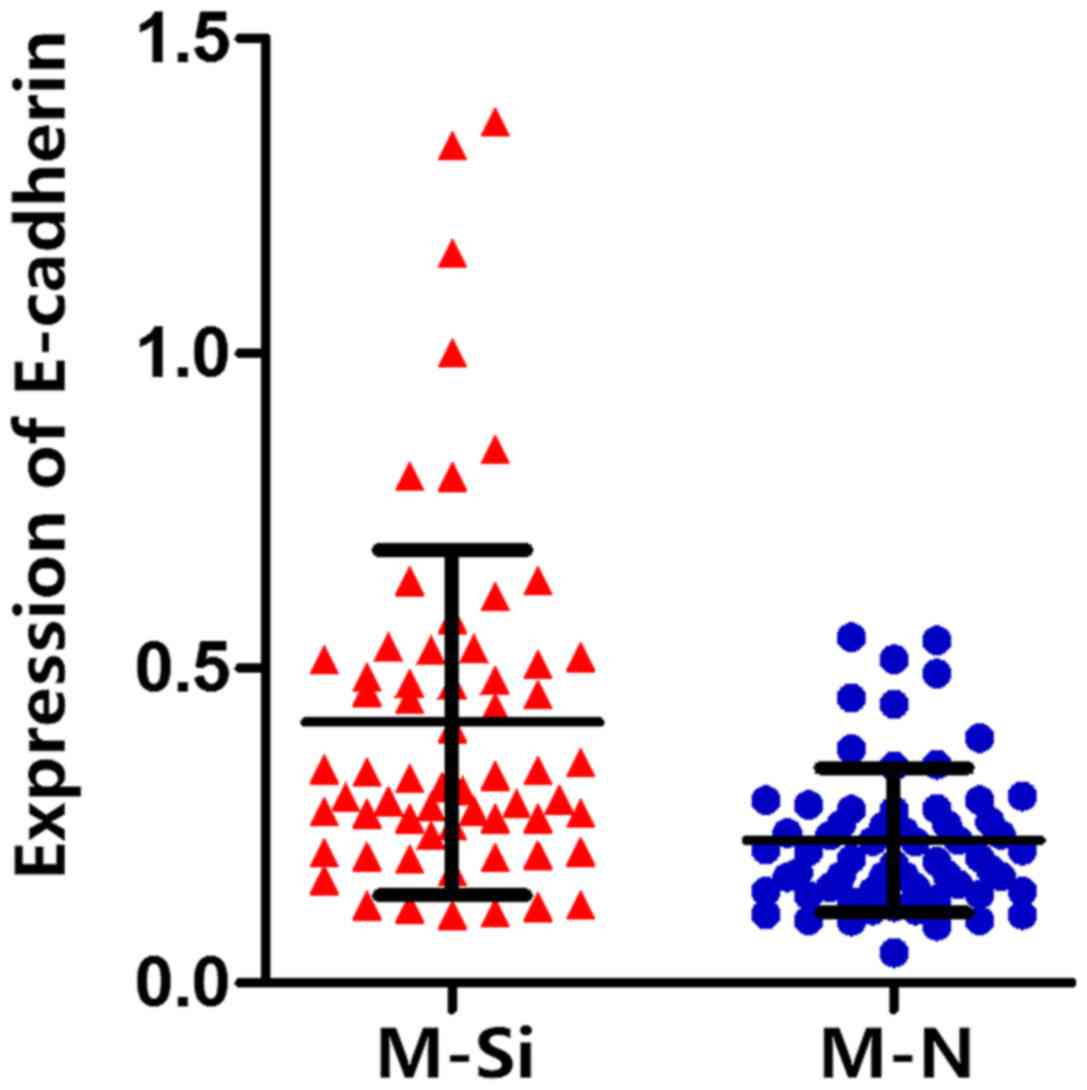
normal and ZNF689 knockdown MHCC97L cells. The expression of
E-cadherin was higher in ZN689 knockdown MHCC97L cells compared
with that in normal MHCC97L cells (Fig.
9), which revealed the potential role of ZNF689 in regulating
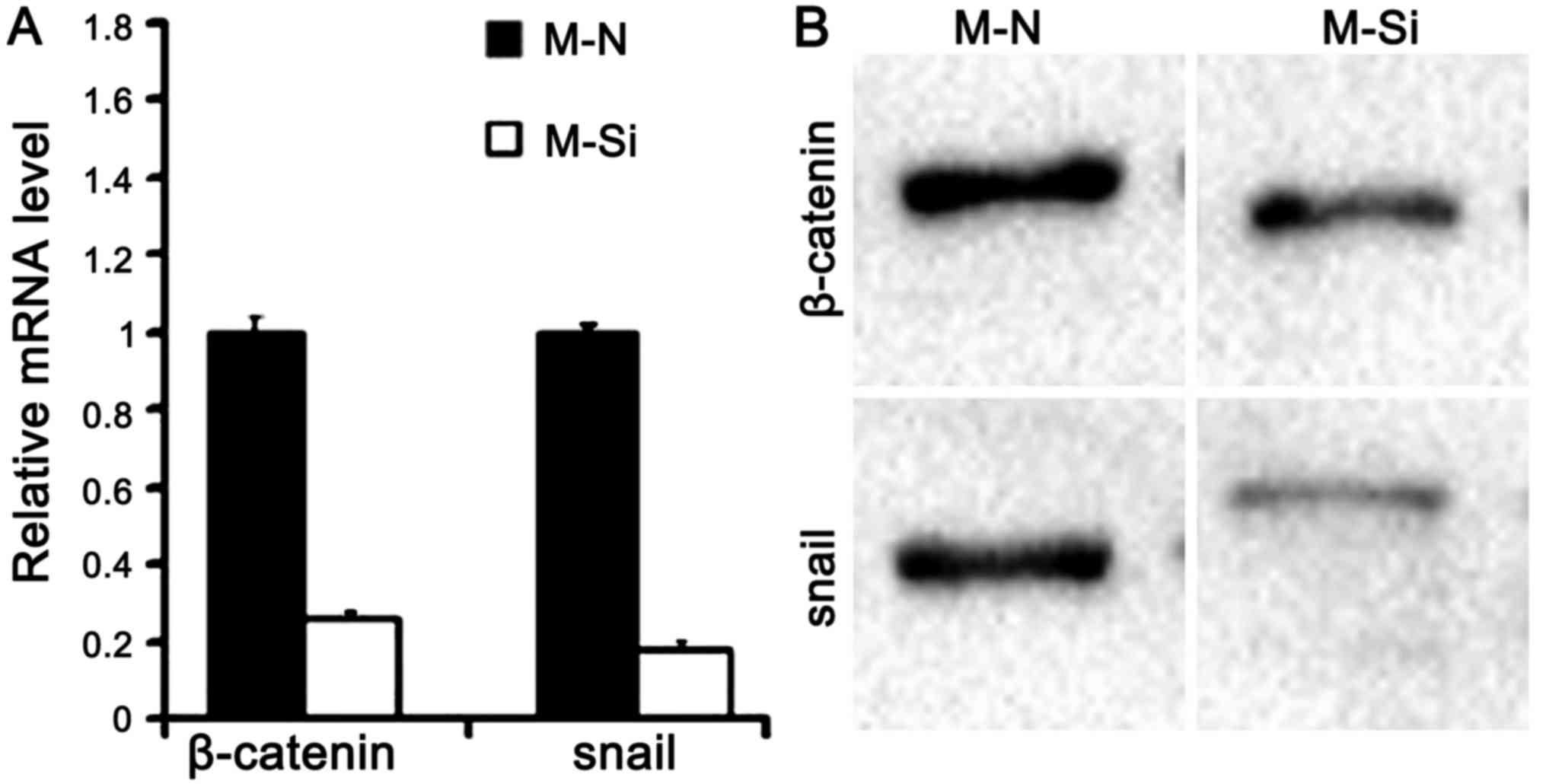
the EMT process. Further supporting the function of ZNF689 in
regulating EMT, the expression of β-catenin and the target gene
SNAIL1 were substantially decreased following knockdown of ZNF689
in MHCC97L cells, SNAIL1 is the abbreviation of snail family
transcriptional repressor 1, which is a member of transcription
factors, is critical for inducing and sustaining cancer EMT
(18) (Fig. 10). It is hypothesized that ZNF689 may
regulate the EMT of HCC via the Wnt-β-catenin-snail signaling
pathway.
Discussion
The aim of the present study was to investigate the
expression of ZNF689 in HCC at the mRNA and protein expression
levels. Its expression did not differ significantly between HCC
tissues and paired non-cancerous tissues. However, the expression
of ZNF689 in HCC tissues and paired non-cancerous tissues was
significantly higher compared with that in normal liver tissues.
Positive expression of ZNF689 was significantly associated with a
tumor size >10 cm, MVI and tumor capsule infiltration.
Additionally, the positive expression of ZNF689 in HCC was
associated with the poor prognosis of HCC.
ZNF689 has previously been implicated in the
development of HCC. Silva et al (8) analyzed the gene-expression profiles of
20 HCC tissue samples, and identified a novel gene
(transcription-involved protein upregulated in HCC) that encoded a
500-amino-acid protein containing 12 zinc-finger domains and a
Kruppel-associated box domain, which was later termed ZNF689.
ZNF689 was identified to be involved in suppressing the apoptosis
of HCC cells via downregulation of the expression of pro-apoptotic
factors.
ZNF689 protein expression was detected by IHC in 102
cases of HCC tissues and paired non-cancerous tissues in the
present study and 45 of the HCC cases were positive for expression
of ZNF689, which was consistent with previous results (13), including a study by Shigematsu et
al (13), reporting that ZNF689
knockdown induced expression of the pro-apoptotic factors of the
Bcl-2 family, Bax, Bcl-2 antagonist/killer 1 and Bid, resulting in
tumor cell apoptosis. Suppression of tumor cell apoptosis by ZNF689
is reflected by increased tumor burden, as indicated by tumor size.
Hu et al (19) demonstrated
that Bcl-2 associated death promoter (Bad) serves a key function in
HCC development, and that low expression of Bad is associated with
larger tumor size and poor prognosis. It was also identified that
HCC tumor cell growth was inhibited by microRNA-204 via
downregulation of Bcl-2, and low expression of microRNA-204 is
markedly associated with increased tumor size (20). These data are consistent with the
results of the present study, indicating an association between HCC
cell apoptosis and solid tumor size.
MVI is an important indicator of poor prognosis for
patients receiving radical liver resection; patients without MVI
have improved short- and long-term survival outcomes and a lack of
MVI is associated with short- and long-term recurrence rate
(21). Previous studies have revealed
that tumor size is a predictor of MVI (22–24). For
example, a cohort of patients from Australia was retrospectively
analyzed, and investigators identified that a tumor size >5 cm
was independently associated with MVI (25). Since positive expression of ZNF689 was
associated with larger tumor size, it may also be directly
associated with the presence of MVI. Although a variety of studies
have investigated the association between tumor angiogenesis and
MVI, the mechanism of MVI has not yet been completely elucidated.
The question of whether ZNF689 serves a key function in the
angiogenesis of HCC requires further investigation.
The tumor capsule is a unique characteristic of HCC,
functioning as a barrier preventing cancer cell invasion of
adjacent tissues or distant organs (26). Investigators have reached a consensus
that tumor capsule infiltration is an important prognostic factor
of HCC following liver resection. Iguchi et al (26) suggested that extracapsular penetration
is a prognostic factor of overall and disease-free survival, and
demonstrated that the percentage of tumor capsule infiltration in
larger tumor types is increased compared with that in smaller tumor
types (27). Consequently, tumor
capsule infiltration may be considered as a result of tumor cell
growth (28). Consistent with these
results, ZNF689 was identified to participate in suppressing the
apoptosis of HCC tumor cells, and inhibition of cell apoptosis may
increase the diameter of solid tumor types, which may also lead to
tumor capsule infiltration.
Considering the mechanism of ZNF689 in regulating
the invasive ability of HCC, a series of in vitro
experiments were performed. ZNF689 knockdown and overexpressed HCC
cell lines were established and the expression of biomarkers and
important components of EMT were compared. The present study
demonstrated that the knockdown of ZNF689 may inhibit proliferation
and invasion of HCC cells by the Wnt-β-catenin-snail pathway. It is
known that EMT is associated with the progressive ability of
numerous malignant tumors (29).
Currently, three signaling pathways have been demonstrated to
regulate the EMT process (30,31). The
wnt-β-catenin signaling pathway has been demonstrated to serve key
functions in regulating HCC invasion and migration by previous
studies. Snail is a common target gene in the wnt-β-catenin
pathway, which serves critical functions at the
post-transcriptional level (32). The
present study identified the association between ZNF689 alteration
and the wnt-β-catenin pathway, however, in vivo experiments
should be performed for further confirmation.
The present study had several limitations. First, a
comparatively high rate of loss to follow-up reduced the
reliability of the survival analysis. Furthermore, the majority of
the patients in the present study suffered from a HBV infection,
with underlying liver cirrhosis. Whether a difference in ZNF689
expression between HCC tissue and normal liver tissue also exists
in non-cirrhotic HCC requires further investigation.
In conclusion, the present study demonstrated that
ZNF689 was upregulated in HCC tissues compared with normal liver
tissues, and the expression level of ZNF689 was associated with
tumor size, MVI and tumor capsule infiltration. Positive expression
of ZNF689 was identified to be a poor prognostic factor for OS and
progression-free survival. These data suggest that ZNF689 may be a
novel factor in the design of treatment strategies for HCC and in
predicting the prognosis of patients with HCC following liver
resection. Inhibitors of ZNF689 may be designed to knockdown
expression of ZN689 and serve as a complementary strategy to HCC
treatment.
Acknowledgements
The authors would like to acknowledge the support of
the Department of Pathology and Clinical Sample Bank of West China
Hospital.
Funding
The present study was funded by grants from the
National Natural Science Foundation of China (grant. no. 71673193),
and the Key Technology Research and Development Program of the
Sichuan Province (grant. no. 2015SZ0131).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PY wrote the paper and undertook the majority of
experiments, BW and GZ were responsible for data collection, DD
analyzed the available data, JL proposed the idea and approved the
submission of the present study.
Ethics approval and consent to
participate
Written informed consent was obtained from all
enrolled patients, and the study was ethically approved by the
Biomedical Ethics Committee of West China Hospital.
Patient consent for publication
Written informed consent was obtained from all study
participants for publication associated data and images in this
study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mazzanti R, Arena U and Tassi R:
Hepatocellular carcinoma: Where are we? World J Exp Med. 6:21–36.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruix J, Reig M and Sherman M:
Evidence-based diagnosis, staging, and treatment of patients with
hepatocellular carcinoma. Gastroenterology. 150:835–853. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yi PS, Zhang M and Xu MQ: Management of
the middle hepatic vein in right lobe living donor liver
transplantation: A meta-analysis. J Huazhong Univ Sci Technolog Med
Sci. 35:600–605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Bru C and Bruix J: Prognosis of
hepatocellular carcinoma: The BCLC staging classification. Semin
Liver Dis. 19:329–338. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu MD, Jia LH, Liu HB, Zhang KH and Guo
GH: Sorafenib in combination with transarterial chemoembolization
for hepatocellular carcinoma: A meta-analysis. Eur Rev Med
Pharmacol Sci. 20:64–74. 2016.PubMed/NCBI
|
|
7
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Silva FP, Hamamoto R, Furukawa Y and
Nakamura Y: TIPUH1 encodes a novel KRAB zinc-finger protein highly
expressed in human hepatocellular carcinomas. Oncogene.
25:5063–5070. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Q, Tan YX, Ren YB, Dong LW, Xie ZF,
Tang L, Cao D, Zhang WP, Hu HP and Wang HY: Zinc finger protein
ZBTB20 expression is increased in hepatocellular carcinoma and
associated with poor prognosis. BMC Cancer. 11:2712011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kan H, Huang Y, Li X, Liu D, Chen J and
Shu M: Zinc finger protein ZBTB20 is an independent prognostic
marker and promotes tumor growth of human hepatocellular carcinoma
by repressing FoxO1. Oncotarget. 7:14336–14349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Sun B, Li Y, Zhao X, Zhao X, Gu Q,
An J, Dong X, Liu F and Wang Y: ZEB2 promotes vasculogenic mimicry
by TGF-β1 induced epithelial-to-mesenchymal transition in
hepatocellular carcinoma. Exp Mol Pathol. 98:352–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu D, Liu G, Liu Y, Saiyin H, Wang C, Wei
Z, Zen W, Liu D, Chen Q, Zhao Z, et al: Zinc finger protein 191
inhibits hepatocellular carcinoma metastasis through discs large
1-mediated yes-associated protein inactivation. Hepatology.
64:1148–1162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shigematsu S, Fukuda S, Nakayama H, Inoue
H, Hiasa Y, Onji M and Higashiyama S: ZNF689 suppresses apoptosis
of hepatocellular carcinoma cells through the down-regulation of
Bcl-2 family members. Exp Cell Res. 317:1851–1859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang BG, Wan ZH, Huang J, Li LM, Liu H,
Fu SY, Yang Y, Zhang J, Yuan SX, Wang RY, et al: Elevated ZC3H15
increases HCC growth and predicts poor survival after surgical
resection. Oncotarget. 7:37238–37249. 2016.PubMed/NCBI
|
|
15
|
Papatheodoridis GV, Cholongitas E,
Dimitriadou E, Touloumi G, Sevastianos V and Archimandritis AJ:
MELD vs Child-Pugh and creatinine-modified Child-Pugh score for
predicting survival in patients with decompensated cirrhosis. World
J Gastroenterol. 11:3099–3104. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Specht E, Kaemmerer D, Sanger J, Wirtz RM,
Schulz S and Lupp A: Comparison of immunoreactive score, HER2/neu
score and H score for the immunohistochemical evaluation of
somatostatin receptors in bronchopulmonary neuroendocrine
neoplasms. Histopathology. 67:368–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu W, Liu H, Liu ZG, Wang HS, Zhang F,
Wang H, Zhang J, Chen JJ, Huang HJ, Tan Y, et al: Histone
deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation
and stabilization of Snail to promote metastasis of hepatoma cells.
Cancer Lett. 420:1–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu W, Fu J, Lu SX, Liu LL, Luo RZ, Yun JP
and Zhang CZ: Decrease of Bcl-xL/Bcl-2-associated death promoter in
hepatocellular carcinoma indicates poor prognosis. Am J Cancer Res.
5:1805–1813. 2015.PubMed/NCBI
|
|
20
|
Li K, Xyu Q, Liu X, Liu Q and Wang M:
Growth inhibition of human hepatocellular carcinoma by miRNA-204
via down-regulation of Bcl-2 and Sirt1 expression. Xi Bao Yu Fen Zi
Mian Yi Xue Za Zhi. 31:168–172. 2015.(In Chinese). PubMed/NCBI
|
|
21
|
Yu YQ, Wang L, Jin Y, Zhou JL, Geng YH,
Jin X, Zhang XX, Yang JJ, Qian CM, Zhou DE, et al: Identification
of serologic biomarkers for predicting microvascular invasion in
hepatocellular carcinoma. Oncotarget. 7:16362–16371.
2016.PubMed/NCBI
|
|
22
|
Shen J, Wen J, Li C, Wen T, Yan L, Li B,
Yang J and Lu C: The prognostic value of microvascular invasion in
early-intermediate stage hepatocelluar carcinoma: A propensity
score matching analysis. BMC Cancer. 18:2782018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Giannini EG, Bucci L, Garuti F, Brunacci
M, Lenzi B, Valente M, Caturelli E, Cabibbo G, Piscaglia F, Virdone
R, et al: Patients with advanced hepatocellular carcinoma need a
personalized management: A lesson from clinical practice.
Hepatology. 67:1784–1796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Imai K, Yamashita YI, Yusa T, Nakao Y,
Itoyama R, Nakagawa S, Okabe H, Chikamoto A, Ishiko T and Baba H:
Microvascular invasion in small-sized hepatocellular carcinoma:
Significance for outcomes following hepatectomy and radiofrequency
ablation. Anticancer Res. 38:1053–1060. 2018.PubMed/NCBI
|
|
25
|
Schlichtemeier SM, Pang TC, Williams NE,
Gill AJ, Smith RC, Samra JS, Lam VW, Hollands M, Richardson AJ,
Pleass HC, et al: A pre-operative clinical model to predict
microvascular invasion and long-term outcome after resection of
hepatocellular cancer: The Australian experience. Eur J Surg Oncol.
42:1576–1583. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iguchi T, Aishima S, Taketomi A, Nishihara
Y, Fujita N, Sanefuji K, Maehara Y and Tsuneyoshi M: Extracapsular
penetration is a new prognostic factor in human hepatocellular
carcinoma. Am J Surg Pathol. 32:1675–1682. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iguchi T, Aishima S, Sanefuji K, Fujita N,
Sugimachi K, Gion T, Taketomi A, Shirabe K, Maehara Y and
Tsuneyoshi M: Both fibrous capsule formation and extracapsular
penetration are powerful predictors of poor survival in human
hepatocellular carcinoma: A histological assessment of 365 patients
in Japan. Ann Surg Oncol. 16:2539–2546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015.PubMed/NCBI
|
|
29
|
Brabletz T: Metastasis: EMT, microRNAs and
cancer stem cells. Clin Experimental Metastasis. 32:187. 2015.
|
|
30
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang JD, Nakamura I and Roberts LR: The
tumor microenvironment in hepatocellular carcinoma: Current status
and therapeutic targets. Semin Cancer Biol. 21:35–43. 2011.
View Article : Google Scholar : PubMed/NCBI
|