Introduction
Human laryngeal squamous cell carcinoma (LSCC)
originates from the laryngeal epithelium (1,2). Advanced
cervical lymph node metastasis frequently occurs in patients with
LSCC, which may result in a poor prognosis (3). The 5-year survival rate (72.5%) is low
due to tumor metastasis being responsible for >90% of
cancer-associated mortalities in the Southeast Asia region, as
found between 2000 and 2006 (4).
Currently the primary treatment of LSCC is surgery supplemented
with radiotherapy, chemotherapy and biological therapy (5,6); however,
to the best of our knowledge, no significant progress in the
molecular biomarkers of the most common cervical lymph node
metastasis of LSCC and targeted therapies has been proposed for
patients with advanced LSCC (7–9).
Therefore, investigating molecular biological markers, in order to
analyze the invasion and migration of LSCC, is vital for improving
the treatment of local invasion and cervical lymph node metastasis
in patients with LSCC (10,11). Notably, microRNA (miRNA), such as
miRNA-1 and miRNA-205, has been reported to be associated with the
invasion and migration of LSCC, which contributed to the
understanding of the molecular mechanism underlying the invasion
and migration of LSCC (12–14).
Numerous studies have demonstrated that miRNA serves
a critical role in the regulation of LSCC metastasis (15–17). A
report has indicated that miR-153 inhibited the proliferation and
invasion of human LSCC by targeting Kruppel-like factor 5 (18). The microRNA-153 (miR-153) in
glioblastoma is significantly lower, compared with non-neoplastic
brain tissues (19). Previous reports
indicated that miR-153 suppresses breast cancer cell proliferation
and induces apoptosis by targeting HECT domain E3 ubiquitin protein
ligase 3 or myeloid cell leukemia 1 (20,21). In
addition, miR-153 serves as a tumor suppressor and the association
between miR-153 and metadherin is a promising therapeutic target
for breast cancer (22). Furthermore,
the downregulation of miR-153 accelerates epithelial-mesenchymal
transition (EMT) via the regulation of the Snail family
transcriptional repressor 1 (SNAI1) expression level (23). Clinical analysis indicated that the
low expression level of miR-153 in patients with gastric cancer was
associated with poor prognosis and tumor metastasis (24). In addition, a previous report
indicated that miR-153 expression was decreased in lung cancer
tissues compared with that in adjacent noncancerous tissues
(24). Furthermore, miR-153 can
decrease the migration and invasion of non-small-cell lung cancer
cells by targeting ADAM metallopeptidase domain 19 (25,26);
however, the association between miR-153 and SNAI1 in human LSCC
has not been investigated previously.
EMT is associated with the occurrence, development,
invasion and metastasis of numerous malignancy types, including
lung and pancreatic cancer (27,28). In
the present study, the expression level of EMT inducible factor
SNAI1 was detected and the expression pattern of SNAI1 was
determined in LSCC tissues and cells. The association between SNAI1
and tumor metastasis was also investigated in LSCC cells.
Materials and methods
Tissue samples and cell lines
LSCC and adjacent non-tumorous tissues (5 samples)
were collected from the Department of Otorhinolaryngology and
Maxillofacial Oncology, Tianjin Medical University Cancer Institute
and Hospital (Tianjin, China). All cases were pathologically
ascertained. All tissue samples were stored at −80°C until gene
expression analysis. Written informed consent for research purposes
was obtained from each patient. The characteristics of the patients
were summarized (Table I). The study
protocol was approved by the Committee of Tianjin Medical
University Cancer Institute and Hospital. SNAI1 expression levels
were analyzed in LSCC and normal tissues from 148 patients from The
Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Associations among
Twist family BHLH transcription factor 1 (TWST1),
metastasis-associated 1 family member 3 (MTA3), glycogen synthase
kinase 3β (GSK3B), lysine demethylase 1A (KDM1A), histone
deacetylase 1 (HDAC1), ubiquitin C (UBC) and cadherin 1 (CDH1),
SMAD3, SMAD4 and body resistance tumor cluster were analyzed based
on TCGA data on 148 patients with LSCC. The analysis of the
patients' survival curve was conducted on the oncolytic website
(https://www.cancer.org/), according to the
statistical data of the 148 patients with LSCC. Human laryngeal
cancer cell line PCI-13 was purchased from the American Type
Culture Collection (Manassas, VA, USA). The cells were cultured in
RPMI-1640 medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 µg/ml penicillin and
streptomycin (Sigma-Aldrich; Merck KGaA) and 10% glutamine.
 | Table I.Characteristics of the patients with
LSCC (n=22). |
Table I.
Characteristics of the patients with
LSCC (n=22).
|
Characteristics | No. of
patients |
|---|
| Sex |
|
|
Male | 15 |
|
Female | 7 |
| Age (years) |
|
|
Mean | 56 |
|
Range | 42–84 |
| Tumor location |
|
|
Supraglottic | 7 |
|
Glottic | 6 |
|
Subglottic | 9 |
| Histological
differentiation (47) |
|
| Well
differentiation LSCC | 8 |
|
Moderately differentiation
LSCC | 6 |
| Poorly
differentiation LSCC | 3 |
|
Undifferentiation. | 3 |
|
Verrucous
differentiation. | 2 |
Metabolic Gene Rapid Visualizer
(MERAV)
The MERAV website (http://merav.wi.mit.edu/) provided additional and more
advanced tools for analyzing gene expression, and was capable of
analyzing multiple genes in parallel. SNAI1 expression in normal
tissues, primary tumors and cancer cell lines were analyzed, and
the analysis was performed using the MERAV website, in order to
compare the multiple SNAI1 expression levels in LSCC and normal
tissues.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from tissues and PCI-13
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to manufacturer's protocols. Total RNA
was reverse transcribed using TransScript First-Strand cDNA
Synthesis Super Mix kit (Beijing Transgen Biotech Co., Ltd.,
Beijing, China) and TaqMan Human MiRNA Assay kit (both from Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to analyze
miRNA expression according to the manufacturer's protocols. The
Homo sapiens genes primers used were: SNAI1; frizzled class
receptor 2 (FZD2); KDM1A; HDAC1; TWST1; CDH1; MTA3; GSK3B; and
β-actin, which were synthetized from Genewiz, Inc. (South
Plainfield, NJ, USA). The primer sequences are detailed in Table II. The PCR amplification for the
quantification of the gene mRNAs was performed using an ABI 7500
Sequence Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and a SYBR® Premix rx Taq™
II (Perfect Real-Time) kit (Takara Bio, Inc., Otsu, Japan), as
previously described (29). Relative
genes expression levels were calculated using the comparative Cq
method (30). The PCR steps were: 2
min at 95°C; followed by 40 cycles of 10 sec at 95°C, annealing for
20 sec at 55°C and a primer extension for 30 sec at 72°C; and then
10 min at 72°C. Results were expressed as a fold-change relative to
β-actin mRNA expression.
 | Table II.Sequences of the primers used in the
present study. |
Table II.
Sequences of the primers used in the
present study.
|
| Sequence |
|---|
|
|
|
|---|
| Gene name | Reverse | Forward |
|---|
| SNAI1 |
5′-GCGGGATCCATGCCGCGCTC-3′ |
5′-CGCCTCGAGTCATTTGTCATCATCG-3′ |
| KDM1A |
5′-CTGTGGCCATCTGCCTAGT −3′ |
5′-GGGACGCAGCAACTGACATT −3′ |
| HDAC1 |
5′-CGCTTCCACTCCGAGGACTA-3′ |
5′-CTGGGCAGTCATCGCCTAC-3′ |
| TWST1 |
5′-AAGGCATCACTATGGACTTTCTCT-3′ |
5′-GCCAGTTTGATCCCAGTATTTT-3′ |
| CDH1 |
5′-TGATCCCAGGTCTTAGTGAG-3′ |
5′-AGTCTGAACTGACTTCCGCA-3′ |
| MTA3 |
5′-AAGGCGAGAGATTCTTTCCCTG′-3′ |
5′-ACTGGGGACAATTCACTAGAGC-3′ |
| GSK3B |
5′-AAAAGTTTAAAAAGGTCTGGAAAGC-3′ |
5′-TAGTGTTGCTTTTAAGACAGGGTTT-3′ |
| β-actin |
5′-CGGAGTCAACGGATTTGGTC-3′ |
5′-AGCCTTCTCCATGGTCGTGA-3′ |
miR-153, miR-199, small
interfering-SNAI1 (si-SNAI1) and plasmids
miR-153, miR-199 negative control miRNA and si-SNAI1
were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China).
The human SNAI1 3′-untranslated region (3′-UTR) was amplified by
PCR, which was aforementioned, and inserted downstream of the
firefly luciferase gene into the pMIR-REPORT vector (Promega
Corporation, Madison, WI, USA), as described previously (31). The mutated 3′-UTR of SNAI1 in PCI-13
cells were created using overlap PCR, as described previously
(32) and the mutated SNAI13′-UTR was
also inserted into the pMIR-REPORT vector. The constructs of the
pMIR-REPORT-UTR vectors were confirmed via sequencing. Renilla
luciferase-expressing plasmid pRL-TK (used as an internal control;
Promega Corporation) was purchased from Promega Corporation. A
total of three independent experiments were performed for each
assay.
Cell viability assay
PCI-13 cells or miR-153-transfected PCI-13 cells
were cultured in 96-well plates (4,000 cells/well) in RPMI-1640
medium with 10% FBS for 24 h at 37°C. Cell viability was measured
using the Cell-Counting kit-8 (CCK-8) assay (Sigma-Aldrich; Merck
KGaA), according to the manufacturer's protocols. The result was
quantitated spectrophotometrically by measuring the optical density
at 450 nm, with a reference wavelength of 650 nm.
Matrigel invasion assay
Matrigel invasion assays were carried out in 24-well
plates. PCI-13 cells (1×106) in HuMEC Basal serum-free
medium (cat. no. 12753018; Thermo Fisher Scientific, Inc.)
containing Matrigel insert filters at a 1:6 dilution. The lower
chamber contained Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc.) with 10% FBS. After incubation for 48 h at 37°C,
cells were fixed with 5% paraformaldehyde for 10 min at room
temperature and stained with 0.1% crystal violet (Sigma-Aldrich;
Merck KGaA) for 30 min at room temperature. The cells that invaded
through the Matrigel membrane were quantified under a light
microscope at ×40 magnification.
Wound healing assay
The wound healing assay was performed by following
the protocol of a previous study (33). PCI-13 cells were cultured in a 12-well
plate in RPMI-1640 medium with 10% FBS and treated with siRNA-SNAI1
(si-SNAI1) or siRNA-mimic (si-NC; Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. After three-time washing with
DMEM medium to remove cell debris, the cells were allowed to
migrate for 48 h at 37°C, followed by observation under a light
microscope at ×40 magnification.
Luciferase reporter assay
The 3′-UTR sequence of SNAI1 was used to predict to
interact with miR-153 or a mutated sequence within the predicted
target sites, was transfected into the pGL3 control vector (Promega
Corporation). These constructs were designated as wild-type (wt)
SNAI1-3′-UTR or mutant SNAI1-3′-UTR. For the reporter assay, PCI-13
cells (1×104/well) were seeded in 24-well plates and
transfected with the aforementioned constructs, miR-153 expression
vector or negative control using Lipofectamine® 3000
(Thermo Fisher Scientific, Inc.). After 48 h, the cells were
collected and Renilla luciferase activity was measured using the
Dual-Luciferase Reporter Assay system (Promega Corporation),
according to the manufacturer's protocols. Results were obtained
from three independent experiments performed in duplicate.
Western blotting
PCI-13 cells were lysed using
radioimmunoprecipitation assay lysis buffer (Thermo Fisher
Scientific, Inc.). Western blotting was performed according to a
previously described method (34).
Protein concentration was determined using a BCA Protein Assay
Reagent kit (Pierce; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. Proteins (20 µg) were separated using
SDS-PAGE and then transferred to a polyvinylidene fluoride membrane
(EMD Millipore, Billerica, MA, USA). Following blocking with 5%
bull serum albumin (Sigma-Aldrich; Merck KGaA) for 1 h at 37°C, the
primary antibodies used in the immunoblotting assays were: SNAI1
(1:1,000; cat. no. ab117866; Abcam, Cambridge, UK) and β-actin
(1:1,000; cat. no. G8144; United States Biological, Salem, MA,
USA), for 12 h at 4°C. Horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (cat. no. 1662408; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used at a 1:5,000
dilution for 2 h at 37°C and detected using a Western Blotting
Luminol reagent (EMD Millipore). Protein was quantified and
normalized to β-actin using ImageJ software (version 1.38; National
Institutes of Health, Bethesda, MD, USA).
Immunofluorescence
PCI-13 cells were fixed with formaldehyde-sucrose
for 4 h at 37°C. Following dehydration in graded ethanol (70, 80,
90, 95 and 100%) and xylene. Cells stained with 1% DAPI at 37°C for
30 min. Cells were washed twice with PBS and then stained with Ki67
for 2 h at 37°C. Immunostained cells were visualized using an
inverted Olympus fluorescence microscope at ×100 magnification (LSM
700; Carl Zeiss AG, Oberkochen, Germany).
Statistical analysis
Results were calculated as the mean ± standard error
of the mean. Significance was established using the SPSS
statistical 18.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism
6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). All data
were analyzed using Student's t-test or one-way analysis of
variance with a Tukey's Honest Significant Difference test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
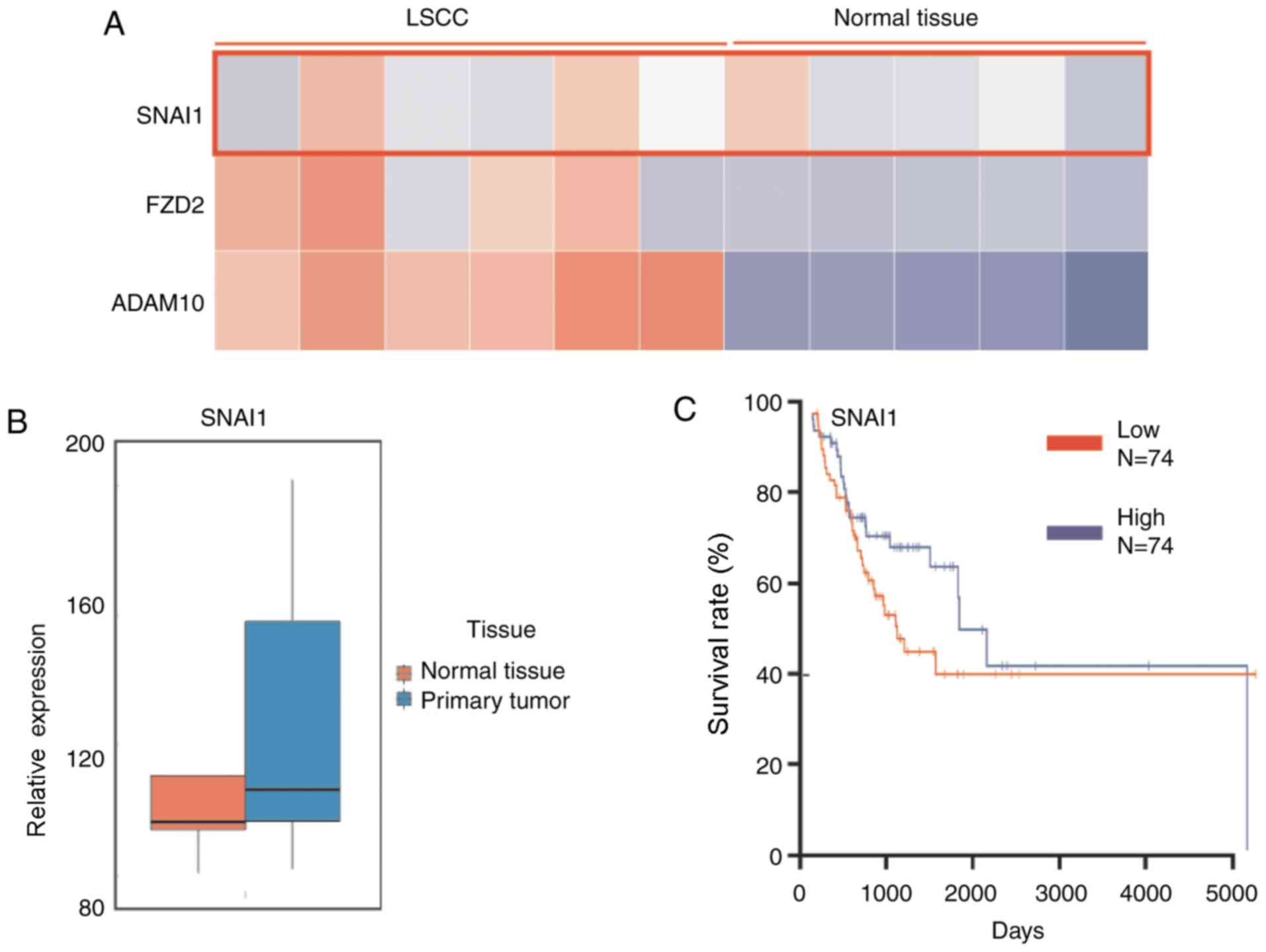
Expression level of SNAI1 in LSCC is
higher, compared with normal tissue
The SNAI1 expression level in LSCC and normal
tissues was analyzed using MERAV image Heat-map database, with
preliminary differential expression analysis described previously
(35). The results demonstrated that
the level of SNAI1 in LSCC tissues was generally higher, compared
with normal tissues (Fig. 1A).
Additionally, two cancer cell invasion-associated genes SNAI1 and
FZD2 were determined to be markedly upregulated in LSCC, which may
explain the high invasiveness of LSCC. Furthermore, the database
was also used for the analysis of mRNA expression levels. The
results demonstrated that LSCC tumor tissues (n=5) exhibited higher
miR-153 level, compared with normal tissues (n=5; Fig. 1B). The analysis of the patients'
survival curve from the oncolytic website demonstrated that a
higher expression level of SNAI1 was negatively associated with a
lower survival rate of patients with LSCC in 148 patients (Fig. 1C). These results indicated that
expression level of SNAI1 is higher in LSCC tissue, compared with
normal tissue, and lower SNAI1 expression presents longer survival,
compared with higher SNAI1 expression.
miR-153 negatively regulates the
expression of SNAI1
As depicted in Fig.
2A, two miRNAs (miR-153 and miR-199) were directly targeted by
the 3′UTR of SNAI1 (Fig. 2A), using
software programs TargetScan (http://www.targetscan.org), miRDB4.0 (http://www.mirdb.org/) and Miranda (http://www.microrna.org/microrna/home.do). The dual
luciferase reporter assay demonstrated that the relative luciferase
activity containing the wt SNAI1 3′-UTR was markedly decreased via
miR-153 in PIC-13 cells, compared with miRNA-mimic; however, the
relative activity of the mutated SNAI1 3′-UTR reporter was not
altered by miR-153. Additionally, miR-199-transfected cells did not
significantly change the luciferase activity (Fig. 2B). The miR-153 and miR-199 expression
in LSCC tissues was measure and compared with corresponding
non-cancer tissues. It was determined that the miR-53 expression
level was downregulated in cancer tissues, compared with non-cancer
tissue (Fig. 2C). The association
between miR-153 and SNAI1 in clinical specimens was analyzed.
RT-qPCR was performed to measure the expression of SNAI1 in five
LSCC samples. The results demonstrated that the miR-153 expression
was inversely associated with SNAI1 mRNA expression in the five
LSCC samples (Fig. 2D). These results
indicated that miR-153 may negatively regulate the expression of
SNAI1.
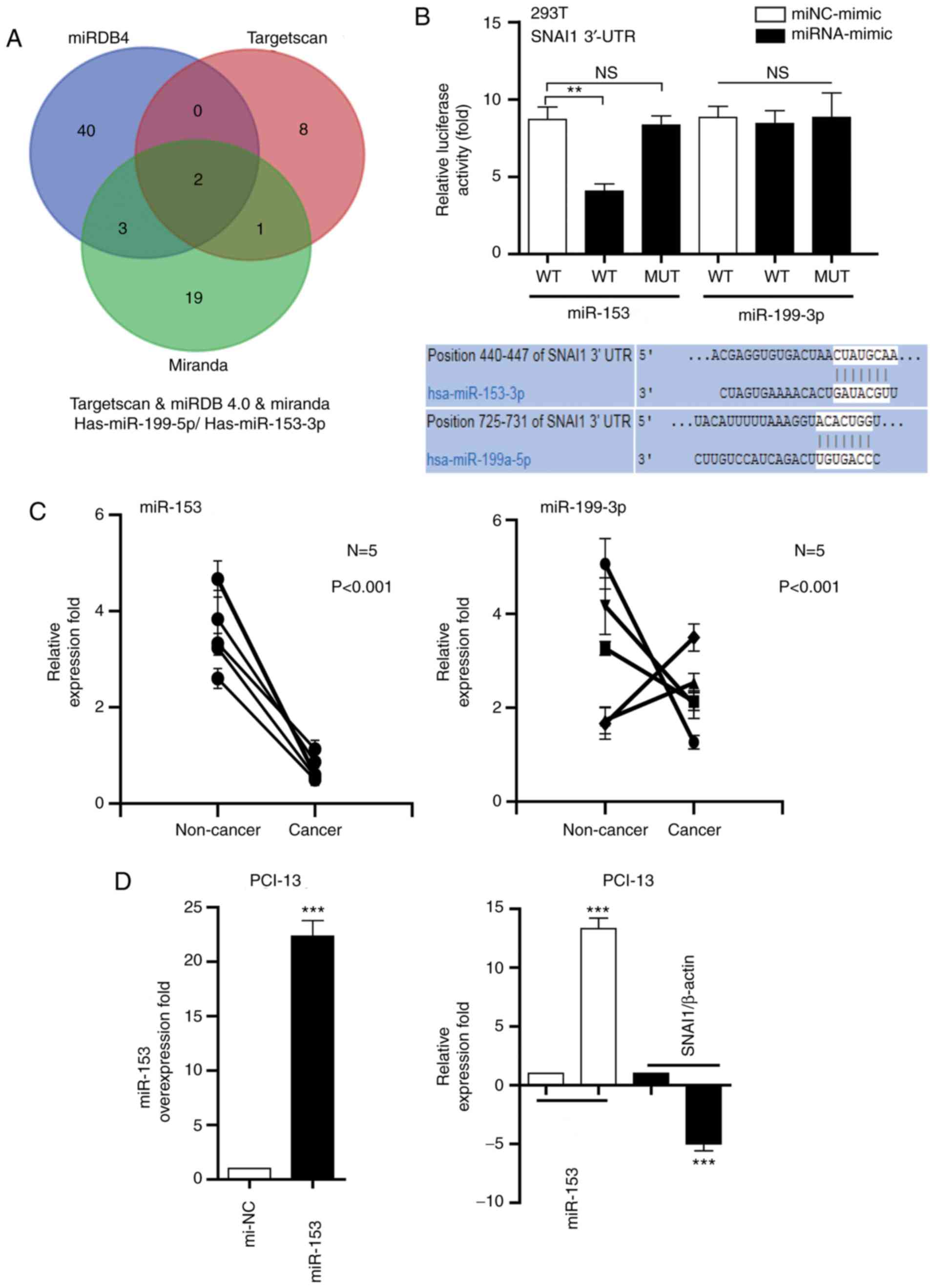 | Figure 2.miR-153 negatively regulates the
expression of SNAI1 in LSCC cells. (A) miR-153 or miR-199 directly
targets the 3′-UTR of SNAI1. (B) Luciferase activity of miR-153 and
miR-199 in HEK293T cells. (C) Analysis of miR-153 and miR-199
expression in cancer tissues, compared with corresponding
non-cancer tissues. (D) Association between miR-153 and SNAI1 in
clinical specimens. **P<0.01, ***P<0.001 vs. mi-NC. miR,
microRNA; NS, no significance; WT, wild-type; mut, mutated; UTR,
untranslated; SNAI1, Snail family transcriptional repressor 1; NC,
negative control; LSCC, laryngeal squamous cell carcinoma. |
miR-153 inhibits PCI-13 cell
proliferation, migration and invasion in vitro
To determine the role of miR-153 in PCI-13 cells,
miR-153 and si-SNAI1 were transfected into PCI-13 cells via adding
the miR-153 or si-SNAI1. As measured by RT-qPCR and western blot
analysis, the expression of miR-153 and si-SNAI1 were overexpressed
by miR-153 and si-SNAI1 mimics in PCI-13 cells (Fig. 3A). CCK-8 assay and Ki67 staining were
used to detect cell proliferation and it was determined that after
72 h, miR-153 and si-SNAI1 markedly inhibited cell proliferation,
compared with si-NC (Fig. 3A and B).
Cell wounding assays were performed to determine the effect of
increased miR-153 and si-SNAI1 levels on tumor cell migration. It
was demonstrated that the upregulation of miR-153 and si-SNAI1
resulted in a significant reduction of migration of PCI-13 cells,
compared with mi-NC (Fig. 3C).
Furthermore, as determined by Matrigel assays, the number of
invaded PCI-13 cells decreased in cells transfected with miR-153
and si-SNAI1 mimics, compared with mi-NC (Fig. 3D). These results indicated that
miR-153 could inhibit PCI-13 cell proliferation, migration and
invasion in vitro.
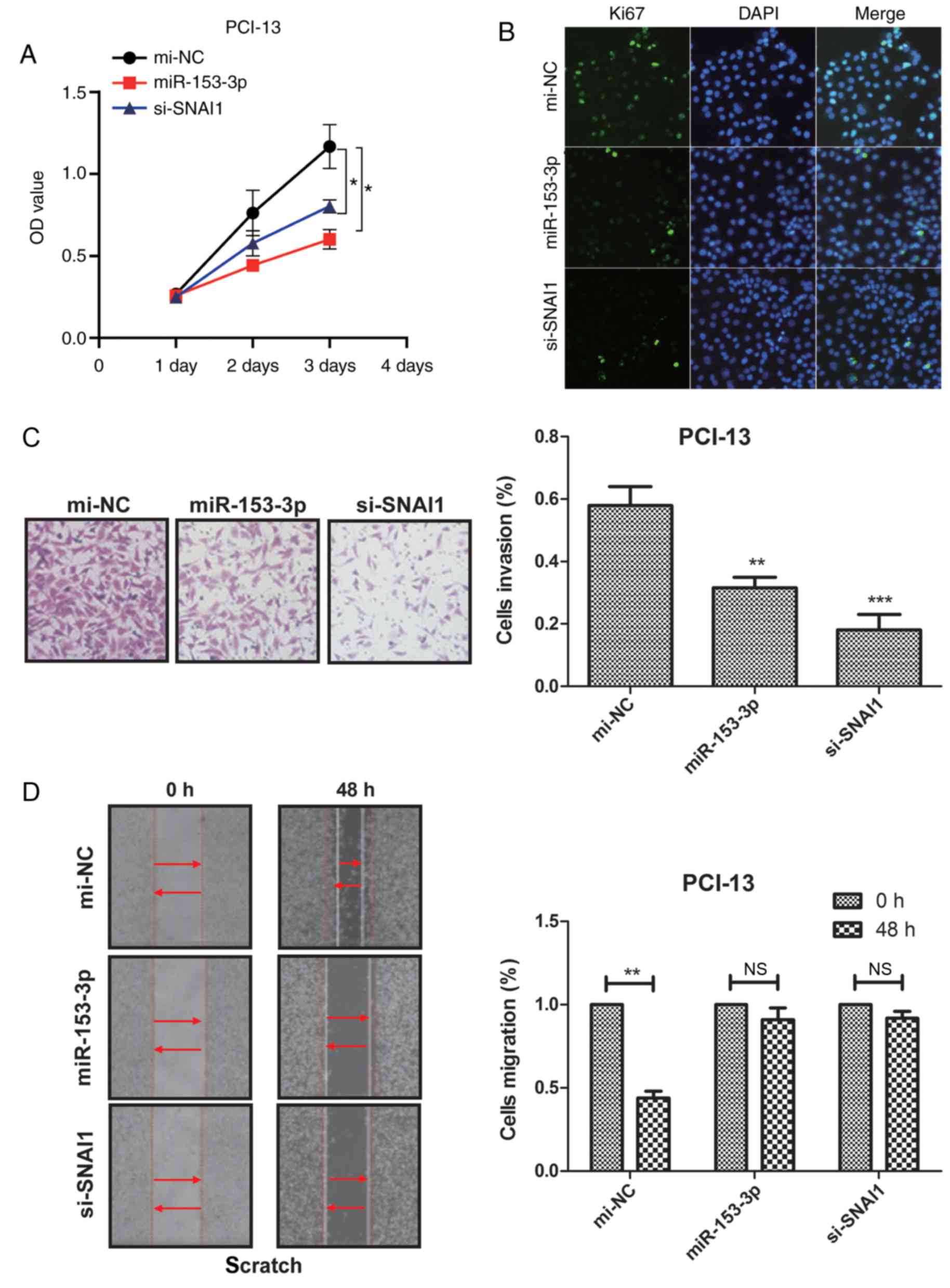 | Figure 3.miR-153 inhibits PCI-13 cell
proliferation, migration and invasion in vitro. (A)
Expression of miR-153 and si-SNAI1 in miR-153 and si-SNAI1 mimics
in PCI-13 cells. (B) miR-153 or si-SNAI1 inhibits PCI-13 cells
proliferation. (C) miR-153 and si-SNAI1 significantly reduces the
cell invasion of PCI-13 cells. (D) miR-153 and si-SNAI1 mimics
decrease the migration of PCI-13 cells. *P<0.05, **P<0.01,
***P<0.001, vs. mi-NC. miR, microRNA; NC, negative control;
SNAI1, Snail family transcriptional repressor 1; OD, optical
density; si, small interfering; NS, no significance. |
Association analysis of the SNAI1
gene
A number of genes were determined to be associated
with the SNAI1 gene using STRING (http://string-db.org/) and a protein-associated online
analysis software (Fig. 4A). The
expression level of genes were verified and it was determined that
the expression levels of KDM1A, HDAC1 and CDH1 were not
significantly altered by si-SNAI1, whilst the expressions levels of
TWST1, MTA3 and GSK3B were markedly downregulated by si-SNAI1 in
PCI-13 cells. These genes were associated with cell invasion,
indicating that SNAI affected cell invasion (Fig. 4B). The results demonstrated that
changes in the expression of metastatic proteins TWST1, MTA3,
GSK3B, KDM1A, HDAC1 and CDH1 were in accordance with gene
expression (Fig. 4C). These results
indicated that the SNAI1 gene is associated with LSCC cell
invasion.
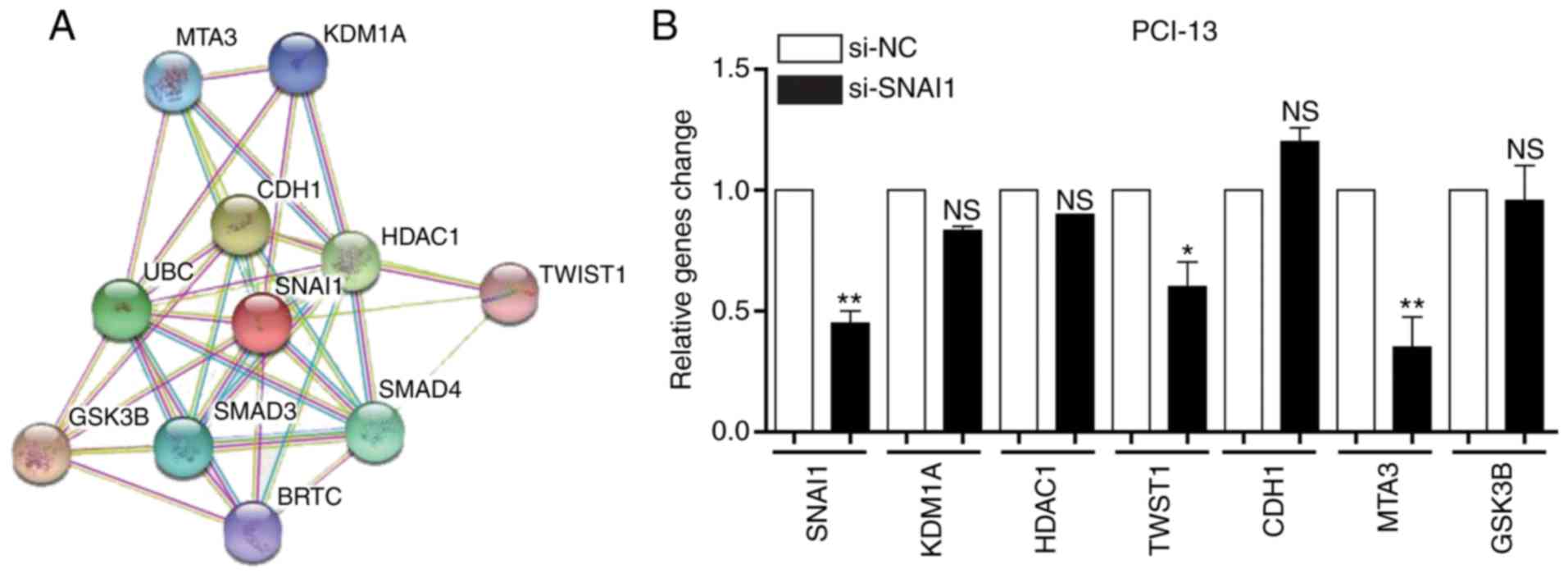 | Figure 4.Analysis of the association between
the SNAI1 gene and SNAI1-associated genes in PCI-13 cells. (A)
Association of SNAI1 and its associated genes in PCI-13 cells. (B)
Effects of knockdown of SNAI1 on its associated genes in PCI-13
cells. (C) Effects of knockdown of SNAI1 on its associated proteins
in PCI-13 cells. *P<0.05, **P<0.01 vs. NC. SNAI1, Snail
family transcriptional repressor 1; si, small interfering; NC,
negative control; NS, no significance; MTA3, metastasis-associated
1 family member 3; KDM1A, lysine demethylase 1A; CDH1, cadherin 1;
HDAC1, histone deacetylase 1; UBC, ubiquitin C; TWST1, Twist family
BHLH transcription factor 1; GSK3B, glycogen synthase kinase 3β;
BRTC, body resistance tumor cluster. |
Discussion
Local invasion and cervical lymph node metastasis of
LSCC are the primary risk factors for the recurrence and prognosis
of patients with LSCC (36). The
molecular mechanisms underlying tumor development have become a
popular research topic, and research regarding the molecular
mechanisms underlying the invasion and metastasis of LSCC also
focused on the regulatory role of miRNA (12,13,37).
Although the tumor suppressor effects of miR-153 have been studied
in human breast cancer, colorectal cancer and epithelial cancer
cells, the miR-153-mediated SNAI1 signal pathway has not been
reported in LSCC cells (22,38,39). In
the present study, miR-153 expression was demonstrated to be
inversely associated with SNAI1 mRNA expression in the five LSCC
samples. Notably, it was determined that miR-153 could inhibit the
proliferation, migration and invasion of PCI-13 cells via
inhibition of SNAI1 expression.
Currently, an increasing number of reports have
demonstrated the association between miRNA and tumor metastasis,
which is regulated by miRNA mediating a number of signal pathways
(40,41). Liu et al (42) indicated that miR-153 could enhance the
therapeutic effects of gemcitabine, by targeting SNAI1 in
pancreatic cancer and gastric carcinoma. In the present study, the
expression differences of SNAI1 in cancer and normal tissues were
examined. SNAI1 serves a key role in understanding the
intercellular matrix signal pathway through regulation of
E-cadherin and Slug, which induces the EMT processes in LSCC. A
previous study demonstrated that oxidase-like 2 can be used as a
poor prognosis indicator for human squamous cell carcinomas, which
could promote malignant transformation by SNAI1-dependent and
SNAI1-independent pathways (43). The
present study indicated that the SNAI1 gene may be the target of
miR-153. The association between the SNAI1 gene and miR-153 was
verified by a dual-luciferase experiment. This method can be
extended to the interaction of other associated genes. Previous
studies indicated that EMT markers are associated with human tumor
metastasis (44–46). In the present study, only the
association between miR-153 and SNAI1 in LSCC cells was analyzed.
Future studies should analyze the association between miR-153 and
other EMT markers in LSCC cells.
To conclude, it was determined that miR-153 may
regulate the expression of SNAI1 in the LSCC cells, which may be
considered as a tumor suppressor agent (34). Data indicated that miR-153 is
downregulated in LSCC, whilst overexpression of miR-153 can
significantly inhibit the proliferation and migration of PCI-13
cells. Data also provided a direction for investigating the
molecular mechanism underlying LSCC invasion; however, further LSCC
samples are require to be analyzed, to determine the association
between miR-153 and SNAI1, in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
BZ and TF performed the experiments and analyzed
experimental data. LZ designed this experiments.
Ethics approval and consent to
participate
This study was approved by Ethics Committee of
Tianjin Medical University Cancer Institute and Hospital. Written
informed consent for research purposes was obtained from each
patient.
Patient consent for publication
All patients have provided consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rees L, Birchall M, Bailey M and Thomas S:
A systematic review of case-control studies of human papillomavirus
infection in laryngeal squamous cell carcinoma. Clin Otolaryngol
Allied Sci. 29:301–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miura K, Kum Y, Han G and Tsutsui Y:
Radiation-induced laryngeal angiosarcoma after cervical
tuberculosis and squamous cell carcinoma: Case report and review of
the literature. Pathol Int. 53:710–715. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Joos B, Joos N, Bumpous J, Burns C, French
CA and Farghaly H: Laryngeal squamous cell carcinoma in a 13
year-old child associated with human papillomaviruses 16 and 18: A
case report and review of the literature. Head Neck Pathol.
3:37–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Larbcharoensub N, Cheewaruangroj W and
Nitiyanant P: Laryngeal sarcocystosis accompanying laryngeal
squamous cell carcinoma: Case report and literature review.
Southeast Asian J Trop Med Public Health. 42:1072–1076.
2011.PubMed/NCBI
|
|
5
|
Si-Mohamed A, Badoual C, Hans S, Péré H,
Tartour E and Brasnu D: An unusual human papillomavirus type 82
detection in laryngeal squamous cell carcinoma: Case report and
review of literature. J Clin Virol. 54:190–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sipaul F, Birchall M and Corfield A: What
role do mucins have in the development of laryngeal squamous cell
carcinoma? A systematic review. Eur Arch Otorhinolaryngol.
268:1109–1117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Furusaka T, Matsuda A, Tanaka A, Matsuda H
and Ikeda M: Superselective intra-arterial chemoradiation therapy
for functional laryngeal preservation in advanced squamous cell
carcinoma of the glottic larynx. Acta Otolaryngol. 133:633–640.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xian JM, Zhou GY, Liang CY and Liu SX:
Study on interference therapy induced by epidermal growth factor
receptor-antisense cDNA in signal transduction of laryngeal
squamous cell carcinoma. Hua Xi Kou Qiang Yi Xue Za Zhi.
25:540–543. 5472007.(In Chinese). PubMed/NCBI
|
|
9
|
Louw L and Claassen J: Rationale for
adjuvant fatty acid therapy to prevent radiotherapy failure and
tumor recurrence during early laryngeal squamous cell carcinoma.
Prostaglandins Leukot Essent Fatty Acids. 78:21–26. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Butler A, Rigby MH, Scott J, Trites J,
Hart R and Taylor SM: A retrospective review in the management of
T3 laryngeal squamous cell carcinoma: An expanding indication for
transoral laser microsurgery. J Otolaryngol Head Neck Surg.
45:342016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gross BC, Olsen SM, Lewis JE, Kasperbauer
JL, Moore EJ, Olsen KD and Price DL: Level IIB lymph node
metastasis in laryngeal and hypopharyngeal squamous cell carcinoma:
Single-institution case series and review of the literature.
Laryngoscope. 123:3032–3036. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Chen Y, Yu J, Liu G and Huang Z:
Integrated transcriptome analysis reveals miRNA-mRNA crosstalk in
laryngeal squamous cell carcinoma. Genomics. 104:249–256. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang F, Song G, Liu M, Li X and Tang H:
miRNA-1 targets fibronectin1 and suppresses the migration and
invasion of the HEp2 laryngeal squamous carcinoma cell line. FEBS
Lett. 585:3263–3269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang B, Lv K, Chen W, Zhao J, Luo J, Wu J,
Li Z, Qin H, Wong TS, Yang W, et al: miR-375 and miR-205 Regulate
the Invasion and Migration of Laryngeal Squamous Cell Carcinoma
Synergistically via AKT-Mediated EMT. Biomed Res Int.
2016:96527892016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS,
Yang W, Fu QL and Lei W: miR-375 suppresses IGF1R expression and
contributes to inhibition of cell progression in laryngeal squamous
cell carcinoma. Biomed Res Int. 2014:3745982014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li JZ, Gao W, Lei WB, Zhao J, Chan JY, Wei
WI, Ho WK and Wong TS: MicroRNA 744-3p promotes MMP-9-mediated
metastasis by simultaneously suppressing PDCD4 and PTEN in
laryngeal squamous cell carcinoma. Oncotarget. 7:58218–58233.
2016.PubMed/NCBI
|
|
17
|
Janiszewska J, Szaumkessel M,
Kostrzewska-Poczekaj M, Bednarek K, Paczkowska J, Jackowska J,
Grenman R, Szyfter K, Wierzbicka M, Giefing M and Jarmuz-Szymczak
M: Global miRNA expression profiling identifies miR-1290 as novel
potential oncomiR in laryngeal carcinoma. PloS One.
10:e01449242015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu JY, Lu JB and Xu Y: MicroRNA-153
inhibits the proliferation and invasion of human laryngeal squamous
cell carcinoma by targeting KLF5. Exp Ther Med. 11:2503–2508. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghasemi A, Fallah S and Ansari M: MiR-153
as a tumor suppressor in glioblastoma multiforme is downregulated
by DNA methylation. Clin Lab. 62:573–580. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu X, Li L, Li Y and Liu Z: MiR-153
promotes breast cancer cell apoptosis by targeting HECTD3. Am J
Cancer Res. 6:1563–1571. 2016.PubMed/NCBI
|
|
21
|
Zou Y, Liu W, Zhang J and Xiang D: miR-153
regulates apoptosis and autophagy of cardiomyocytes by targeting
Mcl-1. Mol Med Rep. 14:1033–1039. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li W, Zhai L, Zhao C and Lv S: MiR-153
inhibits epithelial-mesenchymal transition by targeting metadherin
in human breast cancer. Breast Cancer Res Treat. 150:501–509. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xia W, Ma X, Li X, Dong H, Yi J, Zeng W
and Yang Z: miR-153 inhibits epithelial-to-mesenchymal transition
in hepatocellular carcinoma by targeting Snail. Oncol Rep.
34:655–662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z and Liu C: MiR-153 regulates
metastases of gastric cancer through Snail. Tumour Biol. Aug
2–2015.(Epub ahead of print).
|
|
25
|
Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang
Y, Wang H, Ju J, Zhao L, Wang Z, et al: Suppression of AKT
expression by miR-153 produced anti-tumor activity in lung cancer.
Int J Cancer. 136:1333–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shan N, Shen L, Wang J, He D and Duan C:
MiR-153 inhibits migration and invasion of human non-small-cell
lung cancer by targeting ADAM19. Biochem Biophys Res Commun.
456:385–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam
D, Lee J, Lee SG, Yang WM, Um JY, et al: Ginkgolic acid inhibits
invasion and migration and TGF-β-induced EMT of lung cancer cells
through PI3K/Akt/mTOR inactivation. J Cell Physiol. 232:346–354.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rumman M, Jung KH, Fang Z, Yan HH, Son MK,
Kim SJ, Kim J, Park JH, Lim JH, Hong S and Hong SS: HS-173, a novel
PI3K inhibitor suppresses EMT and metastasis in pancreatic cancer.
Oncotarget. 7:78029–78047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee HH, Lee YJ, Chan P and Lin CY: Use of
PCR-based amplification analysis as a substitute for the southern
blot method for CYP21 deletion detection in congenital adrenal
hyperplasia. Clin Chem. 50:1074–1076. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stanewsky R: Analysis of rhythmic gene
expression in adult Drosophila using the firefly luciferase
reporter gene. Methods Mol Biol. 362:131–142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao YH, Yin MH, Hou L, Luo M and Pei Y:
Asymmetric overlap extension PCR method bypassing intermediate
purification and the amplification of wild-type template in
site-directed mutagenesis. Biotechnol Lett. 29:925–930. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sticker D, Lechner S, Jungreuthmayer C,
Zanghellini J and Ertl P: Microfluidic migration and wound healing
assay based on mechanically induced injuries of defined and highly
reproducible areas. Anal Chem. 89:2326–2333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang G: Erratum: Differential expression
of miR-21 and miR-75 in esophageal carcinoma patients and its
clinical implication. Am J Transl Res. 9:19612017.PubMed/NCBI
|
|
35
|
Shaul YD, Yuan B, Thiru P, Nutter-Upham A,
McCallum S, Lanzkron C, Bell GW and Sabatini DM: MERAV: A tool for
comparing gene expression across human tissues and cell types.
Nucleic Acids Res. 44:D560–D566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McGarey PO Jr, O'Rourke AK, Owen SR,
Shonka DC Jr, Reibel JF, Levine PA and Jameson MJ: Rigid
esophagoscopy for head and neck cancer staging and the incidence of
synchronous esophageal malignant neoplasms. JAMA Otolaryngol Head
Neck Surg. 142:40–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cybula M, Wieteska L,
Józefowicz-Korczyńska M, Karbownik MS, Grzelczyk WL and Szemraj J:
New miRNA expression abnormalities in laryngeal squamous cell
carcinoma. Cancer Biomark. 16:559–568. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang L, Pickard K, Jenei V, Bullock MD,
Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J,
et al: miR-153 supports colorectal cancer progression via
pleiotropic effects that enhance invasion and chemotherapeutic
resistance. Cancer Res. 73:6435–6447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Q, Sun Q, Zhang J, Yu J, Chen W and
Zhang Z: Downregulation of miR-153 contributes to
epithelial-mesenchymal transition and tumor metastasis in human
epithelial cancer. Carcinogenesis. 34:539–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mühlberg L, Kühnemuth B, Costello E, Shaw
V, Sipos B, Huber M, Griesmann H, Krug S, Schober M, Gress TM and
Michl P: miRNA dynamics in tumor-infiltrating myeloid cells
modulating tumor progression in pancreatic cancer. Oncoimmunology.
5:e11601812016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chakraborty C, Chin KY and Das S:
miRNA-regulated cancer stem cells: Understanding the property and
the role of miRNA in carcinogenesis. Tumour Biol. 37:13039–13048.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu F, Liu B, Qian J, Wu G, Li J and Ma Z:
miR-153 enhances the therapeutic effect of gemcitabine by targeting
Snail in pancreatic cancer. Acta Biochim Biophys Sin (Shanghai).
49:520–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Peinado H, Moreno-Bueno G, Hardisson D,
Pérez-Gómez E, Santos V, Mendiola M, de Diego JI, Nistal M,
Quintanilla M, Portillo F and Cano A: Lysyl oxidase-like 2 as a new
poor prognosis marker of squamous cell carcinomas. Cancer Res.
68:4541–4550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L,
Zhang L and Ji J: MicroRNA-130a-3p suppresses cell migration and
invasion by inhibition of TBL1XR1-mediated EMT in human gastric
carcinoma. Mol Carcinog. 57:383–392. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang L, Wang T, Zhang J and Wang X: BTBD7
silencing inhibited epithelial- mesenchymal transition (EMT) via
regulating Slug expression in human salivary adenoid cystic
carcinoma. Cancer Biomark. 20:461–468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Murakami K, Wu Y, Imaizumi T, Aoki Y, Liu
Q, Yan X, Seino H, Yoshizawa T, Morohashi S, Kato Y and Kijima H:
DEC1 promotes hypoxia-induced epithelial-mesenchymal transition
(EMT) in human hepatocellular carcinoma cells. Biomed Res.
38:221–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun C, Han C, Wang P, Jin Y, Sun Y and Qu
L: HOXB9 expression correlates with histological grade and
prognosis in LSCC. Biomed Res Int. 2017:36803052017. View Article : Google Scholar : PubMed/NCBI
|