Introduction
Head and Neck Squamous Cell Carcinoma (HNSCC) is one
of the most common cancers and accounts for 4% of total malignant
tumours worldwide (1). HNSCC includes
tumours of the various sites of upper aerodigestive tract including
the pharynx, larynx, sinuses, nasal cavity and oral cavity. HNSCC
is an aggressive disease and associated with high mortality and
morbidity rates that are mainly attributed to the late detection.
Lack of reliable biomarkers and simple and accurate diagnostic
tools for the screening of early stage cancers are the major
obstacles to hurdles in reducing the mortality rates (2).
Saliva is known to be capable of mirroring the
status of both oral and systemic health (3). It contains locally expressed proteins
and end-products of different metabolic pathways (i.e.,
metabolites) that are known to alter greatly in their
concentrations in various diseases (4). Therefore, these substances, called as
salivary biomarkers, are good indicators of an individual's health
status. Over the last years, considerable efforts have been made to
clarify the potential of salivary metabolomics as an alternative
diagnostic tool (3–6). Salivary metabolites are powerful in
elucidating the pathways underlying different diseases and thereby
they can be considered as ideal for the early diagnostics of
various diseases, such as HNSCC (7–9). The use
of salivary biomarkers is especially attractive in oral cancer,
since the tumours communicate with saliva. Furthermore,
tumour-derived extracellular vesicles might lead to the development
of tumour-specific salivary biomarkers (10–13).
Nuclear magnetic resonance (NMR) spectroscopy is a
quantitative technique based on the magnetic properties of atomic
nuclei. When the sample is placed in an external magnetic field,
NMR active nuclei (e.g., 1H and 13C) absorb
electromagnetic radiation and move from a low-energy spin stage to
a high-energy spin stage (14). When
exposed with radiofrequency pulses, the nuclei emit electromagnetic
radiation and move back to a low-energy state. The nuclei are said
to be in resonance with external magnetic field. As the resonance
frequencies and chemical shifts are unique or highly characteristic
to individual compounds, NMR spectroscopy is powerful method for
identification of small molecules in biological fluids such as in
saliva (9). Further, as the area
under a signal peak is proportional to the concentration of certain
molecule, NMR spectroscopy allows quantitative analysis of salivary
metabolites (14).
Identification of new salivary biomarkers would help
us to diagnose HNSCC in its early stages, which is highly
advantageous and can help in selecting the most appropriate
treatment modalities. Here, we have used NMR spectroscopy to assess
possible salivary metabolic changes associated with HNSCC. The aim
was to compare the salivary metabolic profile between HNSCC
patients and healthy controls.
Patients and methods
Patients and collection of saliva
samples
A total of 45 consecutive patients with HNSCC were
recruited to the longitudinal case control clinical study. The
investigation was conducted in accordance with the ethical
standards and according to the Declaration of Helsinki. The present
study was approved by the Ethics Committee for Human Studies,
Piracicaba Dental School, State University of Campinas, Sao Paulo
Brazil (protocol no. 142/2010) and written informed consent was
obtained from every participant. Patients' demographic and
clinicopathologic data has been previously described by
González-Arriagada et al (15). For this study, the collection of
saliva samples from all patients was performed after dental
treatment prior to radiotherapy. Unstimulated whole-mouth saliva
sample was collected from all patients and from 30 healthy,
non-smoking subjects (control group) in the morning, between 9 and
11 a.m., using standardized techniques (16). Each subject was asked to let the
naturally produced saliva drain into a sterile glass cup for a
period of 5 min. The collected samples were then centrifuged
(14,000 rpm for 6 min). The supernatants were stored at −20°C for
subsequent NMR analysis.
Sample preparation
To each 450 µl of saliva sample, 50 µl of NMR-buffer
(1.5 M KH2PO4, 2 mM NaN3, 5.8 mM
sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4,
D2O, pH 7.4) was added and then the mixture was
centrifuged at 10,000 g for 5 min at 4°C to remove any solid
debris. The obtained supernatant was then transferred to NMR tubes
(O.D. 5 mm).
Data acquisition
NMR spectra were acquired using a 600.20 MHz Bruker
AVANCE III HD spectrometer, equipped with a highly sensitive
inverse triple resonance cryoprobe (Bruker CryoProbe Prodigy;
Bruker BioSpin GmbH, Rheinstetten, Germany). The spectrometer was
controlled via TopSpin 3.2 (Bruker BioSpin GmbH) software. An
automated shimming method (Topshim; Bruker BioSpin GmbH) was used
for all saliva samples which were preheated to 25°C about 30 min
before the measurement. NMR data were acquired by employing a
T2-relaxation-filtered pulse sequence that suppresses signals from
macromolecule signals. In order to suppress the water peak, a
Bruker cpmg1d pulse sequence with T2-filter time of 80
msec and irradiation field of 50 Hz was used. For each sample, the
90° pulse was automatically calibrated. A receiver gain setting was
kept as constant for all the samples.
Data processing
Samples were analysed blinded, in a random order.
The raw NMR spectra were manually corrected for phase using TopSpin
3.0 software (Bruker BioSpin GmbH). A line-broadening factor of 1
Hz was applied to measured free induction decays prior to Fourier
transformations. In total of 24 metabolites were identified by
referring to the published literature (17,18). The
total-line-shape fitting tool in PERCH NMR software (PERCH
Solutions Ltd, Kuopio, Finland) was used in the quantification of
the metabolites. This method allows accurate quantification of
identified metabolites even if the baseline is not linear or
signals overlap (19). All the
spectra were referenced to reference compound
[(trimethylsilylpropanoic acid, (TSP)], used as an internal
standard. The final metabolite concentrations are reported as
µmol/l in saliva.
Statistical analysis
Data are reported as the median and interquartile
range (IQR). The distribution of metabolite concentration values
was tested for normality using the Shapiro-Wilk test and the values
of kurtosis and skewness. Salivary metabolite concentrations
between HNSCC patients and healthy controls were compared with a
non-parametric Mann-Whitney U-test. Collinearity between salivary
metabolite pairs was assessed by computing the Pearson correlation
matrix. P<0.05 was considered to indicate a statistically
significant difference. Multivariate discrimination function
analysis (DFA) was employed to clarify which metabolites, when
considering together, give maximum discrimination power between the
groups. In the DFA, stepwise method with Wilk's lambda criterion
was used. Two separate discriminant analyses were performed: i)
Initially all salivary metabolites having no more than one missing
value were entered; and ii) only those metabolites with no
significant correlation with the best single predictor were
entered. The limit for significant correlation coefficient was set
to 0.50. Finally, sensitivity and specificity of proposed
discrimination model was determined. SPSS software, version 23.0
(IBM Corp., Armonk, NY, USA) was employed in all statistical
analyses.
Results
Saliva samples collected from eight male patients
with HNSCC, with a mean age of 61.7±9.6 years (range, 52–76 years),
and from 30 controls, with a mean age of 54.4±9.0 years (range
42–74 years) were included in the present study. There was no
significant difference between the groups with respect to age
(P=0.065). The reason for high number of rejected patient samples
(37/45) was their limited sample volume, being too small for
reliable NMR analysis. Out of 8 HNSCC patients, primary tumour was
located in the larynx in five patients and in oral cavity in three
patients (Table I). All of the
patients, except one, were diagnosed with advanced stage (III/IV)
disease (Table I).
 | Table I.Clinical characteristics of the 8
HNSCC patients analysed. |
Table I.
Clinical characteristics of the 8
HNSCC patients analysed.
| Patient no. | Age (years) | Sex | Tumour
localization | Stage | Smoking | Drinking | Hyposalivation |
|---|
| 1 | 57 | Male | Larynx | I | Yes | Yes | No |
| 14 | 56 | Male | Oral cavity | III | Yes | Yes | No |
| 33 | 52 | Male | Larynx | IV | No | No | Mild |
| 38 | 73 | Male | Oral cavity | III | Yes | Yes | Severe |
| 42 | 65 | Male | Larynx | IV | Yes | Yes | Severe |
| 43 | 57 | Male | Larynx | IV | Yes | Yes | No |
| 44 | 76 | Male | Oral cavity | IV | Yes | Yes | Severe |
| 45 | 53 | Male | Larynx | III | Yes | Yes | No |
From each sample, up to 19 metabolites including
organic acids (acetate, butyrate, formate, lactate, propionate,
pyruvate, succinate), carbohydrates (1,2-propanediol, butanol,
fucose, methanol), amino acids (alanine, glycine, phenylalanine,
taurine, tyrosine) and amines (choline, methylamine, proline) were
successfully quantified (Table II).
Some metabolites, e.g., citrate, were detected only in some cases,
and thus they were omitted in further analyses. In univariate
analysis, the median concentrations of fucose and 1,2-propanediol
were significantly higher (P=0.003, P=0.032, respectively) in the
HNSCC patients compared to the controls. Instead, the proline was
significantly lower (P=0.043) in the HNSCC saliva samples compared
to controls. In respect of other metabolites, no statistically
significant differences were observed.
 | Table II.Comparison of salivary metabolite
concentrations between patients with HNSCC (n=8) and healthy
controls (n=30). |
Table II.
Comparison of salivary metabolite
concentrations between patients with HNSCC (n=8) and healthy
controls (n=30).
| Metabolite | HNSCC patients | Controls | P-value |
|---|
| Butyrate | 74.2
(33.9–266.4) | 58.6
(25.9–128.4) | 0.562 |
| Propionate | 659.0
(319.9–2,157.6) | 527.3
(251.1–1,028.4) | 0.428 |
|
1,2-propanediol | 69.6
(32.7–2,465.4) | 30.1
(21.7–54.1) | 0.032a |
| Fucose | 694.0
(302.0–1,527.2) | 189.1
(100.6–284.7) | 0.003b |
| Lactate | 207.5
(71.5–1,132.9) | 197.4
(140.4–324.6) | 0.986 |
| Alanine | 90.3
(47.1–515.9) | 107.4
(53.0–173.0) | 0.820 |
| Butanol | 59.9
(17.2–190.5) | 36.5
(16.8–84.3) | 0.428 |
| Acetate | 2916.1
(2,559.8–9,344.8) | 3282.4
(1,977.7–5,239.5) | 0.428 |
| Pyruvate | 27.3
(12.6–73.1) | 13.9
(7.2–33.3) | 0.148 |
| Succinate | 50.6
(24.5–214.3) | 58.9
(47.1–71.9) | 0.765 |
| Methylamine | 5.7 (1.7–66.6) | 3.7 (1.9–5.7) | 0.445 |
| Choline | 17.1
(12.2–43.7) | 19.2
(14.2–24.7) | 0.765 |
| Taurine | 133.8
(72.1–195.4) | 170.2
(104.7–205.1) | 0.502 |
| Methanol | 118.0
(36.6–208.1) | 80.4
(51.4–121.5) | 0.515 |
| Proline | 156.9
(104.1–799.9) | 610.1
(318.5–1,244.3) | 0.043a |
| Tyrosine | 113.8
(42.3–173.5) | 96.8
(55.3–165.5) | 0.847 |
| Phenylalanine | 100.9
(41.9–147.6) | 79.7
(59.1–123.6) | 0.880 |
| Formate | 229.7
(191.4–426.2) | 178.3
(77.0–433.3) | 0.428 |
| Glycine | 560.8
(103.2–719.1) | 494.3
(241.1–923.6) | 0.582 |
The first stepwise DFA (in which all salivary
metabolites having no more than one missing value were included)
resulted in four salivary metabolites (fucose, glycine, methanol
and proline) that being considered together results the maximal
discriminating power between the two groups. The second DFA, when
only those metabolites with no significant correlation with the
best single predictor (i.e., fucose) were entered, resulted in just
identical combination of metabolites. 92.1% of the originally
grouped cases were correctly classified. A sensitivity of 87.5%
(i.e., 7/8 cancerous cases were correctly predicted) and
specificity of 93.3% (i.e., 28/30 healthy cases were correctly
predicted) was resulted. When only two metabolites (among the group
of fucose, glycine, methanol and proline) were entered into the
DFA, a pair of fucose-proline resulted in the highest discriminant
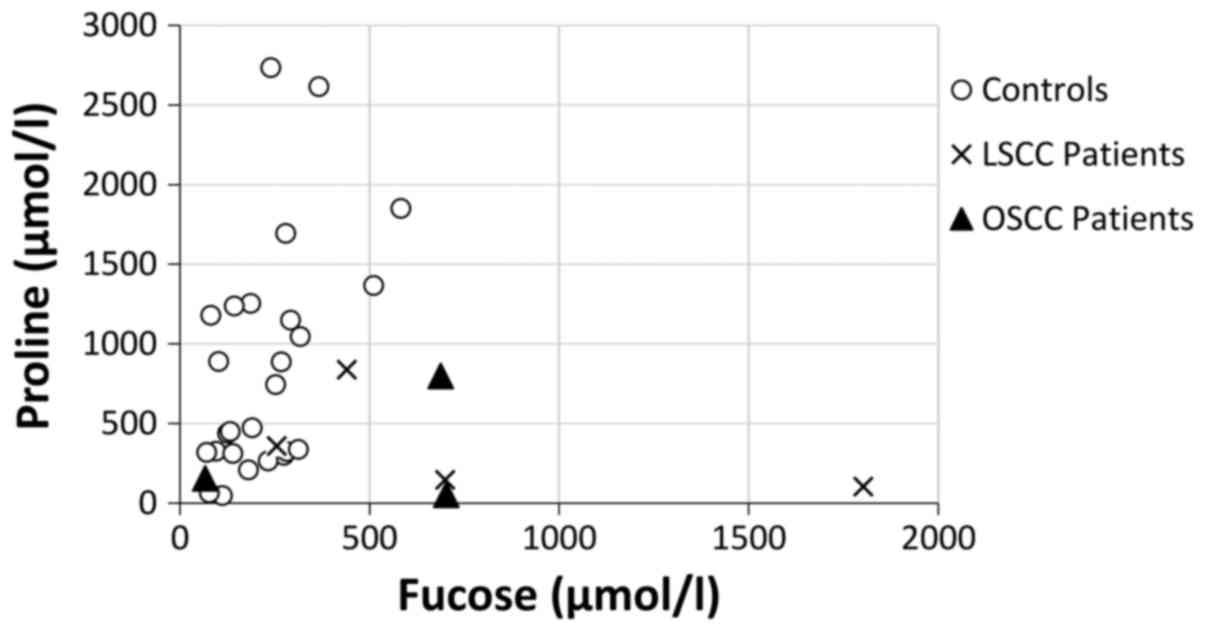
power (90.9%) (Fig. 1).
Discussion
The analysis of salivary metabolomic profile can
offer an early phase approach to assess the changes associated with
a wide range of diseases (8–10). Mass spectrometry (MS) and NMR are the
most common analytical techniques used in metabolomics. The two
techniques have distinct advantages and limitations. Compared to
NMR, MS is significantly more sensitive to identify broader variety
of the metabolites (18). However, MS
analyses of complex biofluids requires a metabolite separation
prior to measurement, and thus MS is rarely used alone but usually
coupled to separation techniques like gas chromatography (GC-MS) or
liquid chromatography (LC-MS). This may bias any analysis.
Furthermore, the quantification by MS is challenging. The main
benefits of NMR spectroscopy include its minimal sample handling,
unbiased quantification of low molecular weight compounds and high
reproducibility (17–19). In this study, we assessed salivary
metabolic alterations associated with HNSCC by NMR spectroscopy.
Our study showed that NMR analysis is a robust approach to study
the metabolome of saliva that is sensitive to metabolic changes in
HNSCC. In a univariate analysis, two metabolites, i.e., fucose and
1,2-propanediol were significantly upregulated, whereas proline was
significantly downregulated in HNSCC. However, in a multivariate
analysis, a combination of four salivary metabolites (fucose,
glycine, methanol and proline) together provided maximum
discrimination among HNSCC patients and healthy controls.
6-deoxy-L-galactose (fucose) is a monosaccharide and
an important constituent of glycoproteins. Fucosylation of
glycoproteins, i.e., a process of adding fucose units at the
terminal end of the oligosaccharide chain mediates several specific
biologic functions (20) and known to
occur during the development of cancer. Tumour cells modulate their
surface by increasing fucosylation levels that leads to several
abnormal cellular characteristics, such as decreased adhesion and
uncontrolled tumour growth (21). In
normal tissues, fucosylation levels are relatively low, but rapidly
increases during carcinogenesis. Therefore, several researchers
have speculated that the monitoring of serum fucose levels could be
a potential approach for the early detection, diagnosis, and
prognosis of cancers (22–25). It has been suggested that the
increased presence of fucose is caused more by local synthesis by
tumour cells than destruction of the malignant cells (26,27). Shah
et al (23) analysed blood
samples collected from 130 patients with untreated oral cancer
(OC), from 75 patients with oral precancerous conditions and from
100 healthy controls. They found that serum fucose levels were
significantly elevated in OC patients OC compared with patients
having oral precancerous lesions or healthy controls. Shetty et
al (28) estimated serum L-fucose
glycoprotein levels among 50 HNSCC patients in comparison of 50
age- and sex-matched healthy controls. They reported over 2-fold
increase in serum glycoprotein L-fucose in patients compared to
controls. Findings reported by other researchers are also
consistent (29,30). However, according to our best
knowledge, this study, for the first time, demonstrates that
HNSCC-induced alterations in fucose levels can be also detected in
unstimulated whole-mouth saliva.
Previous saliva-based studies aimed to monitor
aberrant glucosylation in cancer diagnostics have focused on sialic
acid (N-acetyl neuraminic acid) that is a negatively charged
nine-carbon monosaccharide. Sialic acids are also important
terminal sugars in cell membrane glycoproteins and glycolipids
(31). Previous studies have showed
elevated levels of serum and salivary sialic acid in various
carcinomas, including oral pre-cancer and OC (32–37).
Unfortunately, sialic acid was not among these 19 metabolites that
we managed to identify and quantify in the present study. Recently,
Dame et al (18) identified
and quantified by 1H-NMR a total of 76 different
metabolites from saliva samples including short chain organic
acids, amino acids, alcohols, amines, sugars and pharmaceutical
adjuvants. Among these metabolites, 41 were unique, i.e., they were
not detected by other metabolomics methods. In contrast, some
compounds were detected by GC-MS or LC-MS but not by NMR. These
compounds either do not consist of NMR-detectable protons or their
concentrations were below the detection limit (18).
Altered serum proline levels are known to be related
to cancer metabolism. In previous studies, significantly descended
levels of proline have been observed in serum samples taken from
patients suffering from renal cell carcinoma (38), oral cancer (39) and esophageal cancer (40). The lowered proline level is expected
to be an indicator of overutilization of amino acids in the tumor
tissue (40). We consistently
observed significantly lowered salivary proline levels in patients
with HNSCC in comparison to healthy individuals. Interestingly,
salivary proline concentration against fucose concentration seems
to provide a promising linear discrimination power for HNSCC, even
though present study is limited due to the low number of patient
samples. We had to reject major part (82%) of patient samples due
to the limited sample volume that was not adequate for reliable NMR
analysis. NMR analysis requires a relatively large saliva sample
(~0.5 ml) that may be challenging when collecting unstimulated
saliva, especially patients with dry mouth. NMR experiments were
not originally planned to this patient population and thus
sufficient sample volume collection was not systematically ensured.
Furthermore, this study consists of saliva samples taken from
patients whose primary tumour was located either in the larynx or
in oral cavity. We acknowledge that the etiology of these diseases
differ in their etiopathogenesis. However, whole saliva is a
complex biofluid deriving from the secretion of salivary glands,
gingival folds and oral mucosal transudate. In addition, it
includes exudates from mucous of the nasal cavity and pharynx,
blood cells, bacterial metabolites, food remainders, desquamated
epithelial cells, traces of medications or chemical products
(41). Therefore, it is rational that
also laryngeal squamous cell carcinoma origin alterations can be
found in saliva.
As a diagnostic media, saliva fulfills essential
criteria such as an ease and non-invasive collection and low-cost
handling and storage of samples. Saliva consists of a numerous
compounds such as proteins, peptides, nucleic acids, electrolytes,
and hormones originating from multiple local and systemic sources
that can be used as disease specific biomarkers in diagnosis and
disease monitoring (41). Unlike
blood, saliva does not clot and saliva analytes are stable and
cost-efficient to store. Problems of low concentration of relevant
biomarker compounds in saliva have been largely surpassed through
several instrumental and analytical advancements in the field of
omics technologies (42).
Furthermore, pain, anxiety and infection risk closely related to
traditional methods, i.e., blood collection or tissue biopsy can be
avoided in saliva-based diagnostics. Feasibility of multiple
repetition sampling is also a significant bonus for disease
screening, diagnostics and follow-up of treatment and
rehabilitation outcomes. All in all, the collection, processing and
analysis of saliva can be considered as easier than corresponding
procedures for blood or any other biological fluid (43).
Besides oral diseases, salivary analysis is highly
potential also in diagnostics of various systemic diseases like
Sjögren's syndrome (44,45) as well as distant malignancies, such as
breast cancer (46), lung cancer
(47,48) and pancreatic cancer (49,50).
Although saliva is able to reflect well the overall health, its use
as a diagnostic media is still rare. The current evidence about the
diagnostic potential of reported salivary biomarkers in various
pathologies is still weak and needs to be strengthen in further
validation studies with larger number of samples. Further studies
with various state-of-the-art ‘omics’ methods can help in
developing a prospective disease-specific biomarker pattern based
on these molecules (51). It is
evident that metabolic map of cancer contains more than one
biomarker molecule and therefore, salivary metabolomics based on
NMR or MS is a useful quantitative technique to screen wide variety
of salivary components (18).
Identification of reliable, disease-specific biomarkers can allow
the development of novel point-of-care (POC) platforms enabling
simple and cost-effective quantification of target biomarker
molecules. Incorporating these POC approaches into the part of
primary health screening programs, the burden on health care sector
in terms of costly equipment and invasive testing procedures can be
significantly reduced in the future (3,6,43). Although this study provides promising
preliminary results, controlled longitudinal trials with higher
number of patients are needed to ensure the true diagnostic
accuracy and feasibility to build up a real diagnostic saliva test.
Moreover, these biomarkers need to be further examined in other
aspects of HNSCC such as monitoring of therapy response and
classification of disease severity.
Acknowledgements
The authors acknowledge the contribution of NMR
Metabolomics Laboratory at the University of Eastern Finland
(Kuopio, Finland).
Funding
The present study was supported by the Finnish
Funding Agency for Technology and Innovation (Tekes) project ‘Novel
spectroscopic methods for early detection and screening of oral
cancer’ (grant no. 52/31/2014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TS, WAGA, MAL, AMK and SM designed the study. JJWM,
RA, WAGA, MAL, AMK and SM performed the experiments and acquired
the data. JJWM, SPS, RA, RL, AMK and SM analyzed and interpreted
the data. JJWM, SPS and SM were the major contributors in writing
the manuscript. All authors critically reviewed and approved the
final manuscript.
Ethics approval and consent to
participate
The investigation was conducted in accordance with
ethical standards and according to the Declaration of Helsinki. The
present study was approved by the Ethics Committee for Human
Studies, Piracicaba Dental School, State University of Campinas,
Brazil (protocol no. 142/2010) and written informed consent was
obtained from every participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abbey LM, Kaugars GE, Gunsolley JC, Burns
JC, Page DG, Svirsky JA, Eisenberg E, Krutchkoff DJ and Cushing M:
Intraexaminer and interexaminer reliability in the diagnosis of
oral epithelial dysplasia. Oral Surg Oral Med Oral Pathol Oral
Radiol Endod. 80:188–191. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu J and Duan Y: Saliva: A potential
media for disease diagnostics and monitoring. Oral Oncol.
48:569–577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang CZ, Cheng XQ, Li JY, Zhang P, Yi P,
Xu X and Zhou: Saliva in the diagnosis of diseases. Int J Oral Sci.
8:133–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoshizawa JM, Schafer CA, Schafer JJ,
Farrell JJ, Paster BJ and Wong DT: Salivary biomarkers: Toward
future clinical and diagnostic utilities. Clin Microbiol Rev.
26:781–791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cuevas-Córdoba B and Santiago-García J:
Saliva: A fluid of study for OMICS. OMICS. 18:87–97. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guerra EN, Acevedo AC, Leite AF, Gozal D,
Chardin H and De Luca Canto G: Diagnostic capability of salivary
biomarkers in the assessment of head and neck cancer: A systematic
review and meta-analysis. Oral Oncol. 51:805–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel S and Ahmed S: Emerging field of
metabolomics: Big promise for cancer biomarker identification and
drug discovery. J Pharm Biomed Anal. 107:63–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mikkonen JJW, Herrala M, Soininen P,
Lappalainen R, Tjäderhane L, Seitsalo H, Niemelä R, Salo T, Kullaa
A and Myllymaa S: Metabolic profiling of saliva in patients with
primary Sjögren's syndrome. Metabolomics. 3:1282013.
|
|
10
|
Spielmann N and Wong DT: Saliva:
Diagnostics and therapeutic perspectives. Oral Dis. 17:345–354.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonne NJ and Wong DT: Salivary biomarker
development using genomic, proteomic and metabolomic approaches.
Genome Med. 4:822012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Winck FV, Ribeiro Prado AC, Domingues
Ramos R, Ling LY, Riaño-Pachón DM, Rivera C, Brandão TB, Gouvea AF,
Santos-Silva AR, Coletta RD and Paes Leme AF: Insights into immune
responses in oral cancer through proteomic analysis of saliva and
salivary extracellular vesicles. Sci Rep. 5:163052015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zlotogorski-Hurvitz A, Dayan D, Chaushu G,
Salo T and Vered M: Morphological and molecular features of oral
fluid-derived exosomes: Oral cancer patients versus healthy
individuals. J Cancer Res Clin Oncol. 142:101–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Günther H: NMR spectroscopy: Basic
principles, concepts, and applications in chemistry. 2 edition.
John Wiley and Sons Ltd.; Chichester: 1995
|
|
15
|
González-Arriagada WA, Ramos LM, Silva AA,
Vargas PA, Coletta RD, Bingle L and Lopes MA: Salivary BPIFA1
(SPLUNC1) and BPIFA2 (SPLUNC2 A) are modified by head and neck
cancer radiotherapy. Oral Surg Oral Med Oral Pathol Oral Radiol.
119:48–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Navazesh M: Methods for collecting saliva.
Ann N Y Acad Sci. 694:72–77. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Silwood CJ, Lynch E, Claxson AW and
Grootveld MC: 1H and (13)C NMR spectroscopic analysis of human
saliva. J Dent Res. 81:422–427. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dame ZT, Aziat F, Mandal RS, Krishnamurthy
R, Bouatra S, Borzouie S, Guo AC, Sajed T, Deng L, Lin H, et al:
The human saliva metabolome. Metabolomics. 11:1864–1883. 2015.
View Article : Google Scholar
|
|
19
|
Soininen P, Haarala J, Vepsäläinen J,
Niemitz M and Laatikainen R: Strategies for organic impurity
quantification by 1H NMR spectroscopy: Constrained total-line-shape
fitting. Anal Chim Acta. 542:178–185. 2005. View Article : Google Scholar
|
|
20
|
Ma B, Simala-Grant JL and Taylor DE:
Fucosylation in prokaryotes and eukaryotes. Glycobiology.
16:158R–184R. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Miyoshi E, Moriwaki K and Nakagawa T:
Biological function of fucosylation in cancer biology. J Biochem.
143:725–729. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fernández-Rodríguez J, de la Cadena Páez
M, Martínez-Zorzano VS and Rodríguez-Berrocal FJ: Fucose levels in
sera and in tumours of colorectal adenocarcinoma patients. Cancer
Lett. 121:147–153. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shah M, Telang S, Raval G, Shah P and
Patel PS: Serum fucosylation changes in oral cancer and oral
precancerous conditions: Alpha-L-fucosidase as a marker. Cancer.
113:336–346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Manjula S, Monteiro F, Aroor Rao A, Rao S,
Annaswamy R and Rao A: Assessment of serum L-fucose in brain tumor
cases. Ann Indian Acad Neurol. 13:33–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Listinsky JJ, Siegal GP and Listinsky CM:
The emerging importance of α-L-fucose in human breast cancer: A
review. Am J Transl Res. 3:292–322. 2011.PubMed/NCBI
|
|
26
|
Shetlar MR, Foster JV, Kelly KH, Shetlar
CL, Bryan RS and Everett MR: The serum polysaccharide level in
malignancy and in other pathological conditions. Cancer Res.
9:515–519. 1949.PubMed/NCBI
|
|
27
|
Sawke NG and Sawke GK: Serum fucose level
in malignant diseases. Indian J Cancer. 47:452–457. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shetty RK, Bhandary SK and Kali A:
Significance of serum L-fucose glycoprotein as cancer biomarker in
head and neck malignancies without distant metastasis. J Clin Diagn
Res. 7:2818–2820. 2013.PubMed/NCBI
|
|
29
|
Parwani RN and Parwani SR: Quantitative
evaluation of serum fucose in oral squamous cell carcinoma
patients. J Can Res Ther. 7:143–147. 2011. View Article : Google Scholar
|
|
30
|
Rai NP, Anekar J, Shankara Shivaraja YM,
Divakar DD, Al Kheraif AA, Ramakrishnaiah R, Sebastian R, Raj AC,
Al-Hazmi A and Mustafa SM: Comparison of serum fucose levels in
leukoplakia and oral cancer patients. Asian Pac J Cancer Prev.
16:7497–7500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chugh S, Gnanapragassam VS, Jain M,
Rachagani S, Ponnusamy MP and Batra SK: Pathobiological
implications of mucin glycans in cancer: Sweet poison and novel
targets. Biochim Biophys Acta. 1856:211–225. 2015.PubMed/NCBI
|
|
32
|
Rajpura KB, Patel PS, Chawda JG and Shah
RM: Clinical significance of total and lipid bound sialic acid
levels in oral pre-cancerous conditions and oral cancer. J Oral
Pathol Med. 34:263–267. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sanjay PR, Hallikeri K and Shivashankara
AR: Evaluation of salivary sialic acid, total protein, and total
sugar in oral cancer: A preliminary report. Indian J Dent Res.
19:288–291. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sawhney H and Kumar CA: Correlation of
serum biomarkers (TSA & LSA) and epithelial dysplasia in early
diagnosis of oral precancer and oral cancer. Cancer Biomark.
10:43–49. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taqi SA: Clinical evaluation of total and
lipid bound sialic acid levels in oral precancer and oral cancer.
Indian J Med Paediatr Oncol. 33:36–41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dhakar N, Astekar M, Jain M, Saawarn S and
Saawarn N: Total sialic acid, total protein and total sugar levels
in serum and saliva of oral squamous cell carcinoma patients: A
case control study. Dent Res J (Isfahan). 10:343–347.
2013.PubMed/NCBI
|
|
37
|
Dadhich M, Prabhu V, Pai VR, D'Souza J,
Harish S and Jose M: Serum and salivary sialic acid as a biomarker
in oral potentially malignant disorders and oral cancer. Indian J
Cancer. 51:214–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mustafa A, Gupta S, Hudes GR, Egleston BL,
Uzzo RG and Kruger WD: Serum amino acid levels as a biomarker for
renal cell carcinoma. J Urol. 186:1206–1212. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tiziani S, Lopes V and Günther UL: Early
stage diagnosis of oral cancer using 1H NMR-based metabolomics.
Neoplasia. 11(269–276): 4p2009.
|
|
40
|
Liang S, Sanchez-Espiridion B, Xie H, Ma
J, Wu X and Liang D: Determination of proline in human serum by a
robust LC-MS/MS method: Application to identification of human
metabolites as candidate biomarkers for esophageal cancer early
detection and risk stratification. Biomed Chromatogr. 29:570–577.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Humphrey SP and Williamson RT: A review of
saliva: Normal composition, flow, and function. J Prosthet Dent.
85:162–169. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tiwari M: Science behind human saliva. J
Nat Sci Biol Med. 2:53–58. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koneru S and Tanikonda R: Salivaomics-A
promising future in early diagnosis of dental diseases. Dent Res J
(Isfahan). 11:11–15. 2014.PubMed/NCBI
|
|
44
|
Hu S, Gao K, Pollard R, Arellano-Garcia M,
Zhou H, Zhang L, Elashoff D, Kallenberg CG, Vissink A and Wong DT:
Preclinical validation of salivary biomarkers for primary Sjögren's
syndrome. Arthritis Care Res (Hoboken). 62:1633–1638. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mikkonen JJ, Singh SP, Herrala M,
Lappalainen R, Myllymaa S and Kullaa AM: Salivary metabolomics in
the diagnosis of oral cancer and periodontal diseases. J
Periodontal Res. 51:431–437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bigler LR, Streckfus CF, Copeland L, Burns
R, Dai X, Kuhn M, Martin P and Bigler SA: The potential use of
saliva to detect recurrence of disease in women with breast
carcinoma. J Oral Pathol Med. 31:421–431. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang L, Xiao H, Zhou H, Santiago S, Lee
JM, Garon EB, Yang J, Brinkmann O, Yan X, Akin D, et al:
Development of transcriptomic biomarker signature in human saliva
to detect lung cancer. Cell Mol Life Sci. 69:3341–3350. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xiao H, Zhang L, Zhou H, Lee JM, Garon EB
and Wong DT: Proteomic analysis of human saliva from lung cancer
patients using two-dimensional difference gel electrophoresis and
mass spectrometry. Mol Cell Proteomics. 11(M111):
0121122012.PubMed/NCBI
|
|
49
|
Sugimoto M, Wong DT, Hirayama A, Soga T
and Tomita M: Capillary electrophoresis mass spectrometry-based
saliva metabolomics identified oral, breast and pancreatic
cancer-specific profiles. Metabolomics. 6:78–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lau C, Kim Y, Chia D, Spielmann N, Eibl G,
Elashoff D, Wei F, Lin YL, Moro A, Grogan T, et al: Role of
pancreatic cancer-derived exosomes in salivary biomarker
development. J Biol Chem. 288:26888–26897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Grant MM: What do'omic technologies have
to offer periodontal clinical practice in the future? J Periodontal
Res. 47:2–14. 2012. View Article : Google Scholar : PubMed/NCBI
|