Introduction
Cervical cancer (CXCA) is the fourth leading cause
of cancer-associated mortality among females worldwide (1). The majority of CXCA cases are associated
with high risk human papillomavirus (HR-HPV) infection. Among
HR-HPV, HPV16 and HPV18 account for 70% of all CXCA cases worldwide
(1), including in Asian populations
(2). Epidemiological data revealed an
increase in multiple HPV infection from 10% of CXCA cases in 2005
(3) to 65% in 2016 (3–5). However,
these figures may reflect a higher sensitivity of detection,
resulting in the co-infection of HPV16 and HPV18 being reported as
the most common HR-HPV in Africa (6)
and Asia (7), and specifically in
Thailand (8), as 31.9, 20.8 and
27.8%, respectively. An increased odds ratio of HPV16 and 18
co-infection compared with single infection was demonstrated by
Chaturvedi et al (9) and
Trottier et al (10), but the
opposite results were reported by Salazar et al (11). These controversial findings implied
that other viral parameters, not only HPV genotype, may serve
important roles in disease progression.
The physical state of HPV infection occurs as one of
two forms: An episomal or an integrated form. The episomal state
involves the complete life cycle of viral replication in the
infected host cells, whereas the integrated form involves
integration of HPV DNA into the host DNA, a major genetic event
leading to cervical carcinogenesis (12). Viral integration and viral load have
previously been reported to be biomarkers for cancer with
high-grade cervical lesions (13).
Several methods have been used for the detection of integrated HPV,
including polymerase chain reaction (PCR) (14), in situ hybridization (15,16) and
amplification of papillomavirus oncogene transcripts (APOT)
(17). These are all qualitative
measurements. Recently, we established a quantitative PCR of E2 and
E6 genes to measure the viral load and physical status of HPV16 DNA
in one tube (18). A ratio of E2/E6
gene of 1.0 is used to define the episomal form, while a decreased
ratio (less than 1.0) indicates the integrated form, due to
deletion of the E2 gene during viral integration. The present study
demonstrated the benefit of using viral numbers and physical status
as surrogate markers of cancer progression.
To the best of our knowledge, the present study was
the first to report the simultaneous measurement of 4 genes, E2 and
E6 genes of HPV16 and HPV18, in a single tube. The development of
multiplex qPCR in the present study provides a total coverage of
70% of HR-HPV-associated CXCA, including single and co-infection of
HPV16 and HPV18. The analytical performance of the multiplex qPCR
was evaluated in clinical samples, compared with simplex qPCR.
Materials and methods
Samples
A total of 20 cervical tissues harboring single or
co-infection of HPV16 and HPV18 were collected from 5 pre-cancerous
lesions (mean, 42.2±6.6 years) and 15 cancerous lesions (mean,
49.5±13.7 years). Samples were collected between 2002 and 2004
under written informed consents approved by the Ethical Committee
of Khon Kaen University, Khon Kaen, Thailand (approval no.
HE562296) and between 2013 and 2014 approved by the the
Ubonratchathani Cancer Hospital, Ubon Ratchathani, Thailand
(approval no. CE012/2013). DNA samples were extracted using a
QIAamp Viral DNA kit (Qiagen GmbH, Hilden, Germany), according to
the manufacturer's protocol. Extracted DNA was used for HPV16 and
HPV18 screening by Nested Multiplex PCR, as previously described
(19).
Cell culture
The human papillomavirus Caski and HeLa cell lines
containing the integrated form of HPV16 (600 DNA copies per cell)
and HPV18 (20–50 DNA copies per cell) were used as internal
standard for determination of physical status of HPV16 and 18,
respectively. HeLa cell line containing HPV18 and CaSki cell line
containing HPV16 were used as HPV positive controls. Cells were
cultured in 25 cm2 flasks at 37°C with 5% CO2
in Dulbecco's Modified Eagle Medium high glucose (DMEM-HG) media
supplemented with 10% fetal bovine serum and 1%
penicillin-streptomycin (10,000 U/ml penicillin and 10 mg/ml
streptomycin; all Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). At 70% confluency, cells were trypsinized with 1
ml of 1X trypsin-EDTA at 37°C for 5 min. The cell pellets were
collected and centrifuged at 2,000 rpm for 3 min and subsequently
used for DNA extraction.
HPV16 and HPV18 integration status
assay
Plasmids containing the whole genomes of HPV16,
HPV18, HPV45 and HPV58 (PBR322 for HPV16 and HPV58, PGM4 for HPV18
and HPV45) were provided by Professor Pientong from the Department
of Microbiology, Faculty of Medicine, Khon Kaen University.
Purified recombinant plasmid copy numbers were estimated by a
spectrophotometer concentration measurement (NanoDrop 2000; Thermo
Fisher Scientific, Inc.). The DNA calculation formula was
6.02×1023 (copies/mol) xA260 (ng/ml)/(DNA
length ×660)=copies/ml. Plasmid DNA was then diluted with sterile
water to obtain between 106 and 102 copies,
and was used to establish calibration curves for measuring E2 and
E6 by multiplex qPCR using a TaqMan® probe assay
(Bioneer Corporation, Daejeon, Korea). The oligonucleotide
sequences of primers and probes were followed as previously
described (20,21), with modifications to the quencher and
reporter fluorescent dyes (Table I).
E2 and E6 DNA were amplified using a qPCR thermocycler (Exicycler™;
Bioneer Corporation). Each 50 µl reaction mixture contained a
premix (AccuPower® DualStar™; Bioneer Corporation) with
0.4 µM probes and primers. The PCR reaction was initiated at 95°C
for 10 min, followed by 40 cycles at 95°C for 15 sec and 63°C for
60 sec. Reactions lacking DNA template were used as negative
controls as previously described. Quantification of E2 and E6 genes
were analyzed using the calibration curve plotted between the
quantification cycle (Cq) on the x-axis and the logarithm of the
standard copy number on the y-axis (102−106
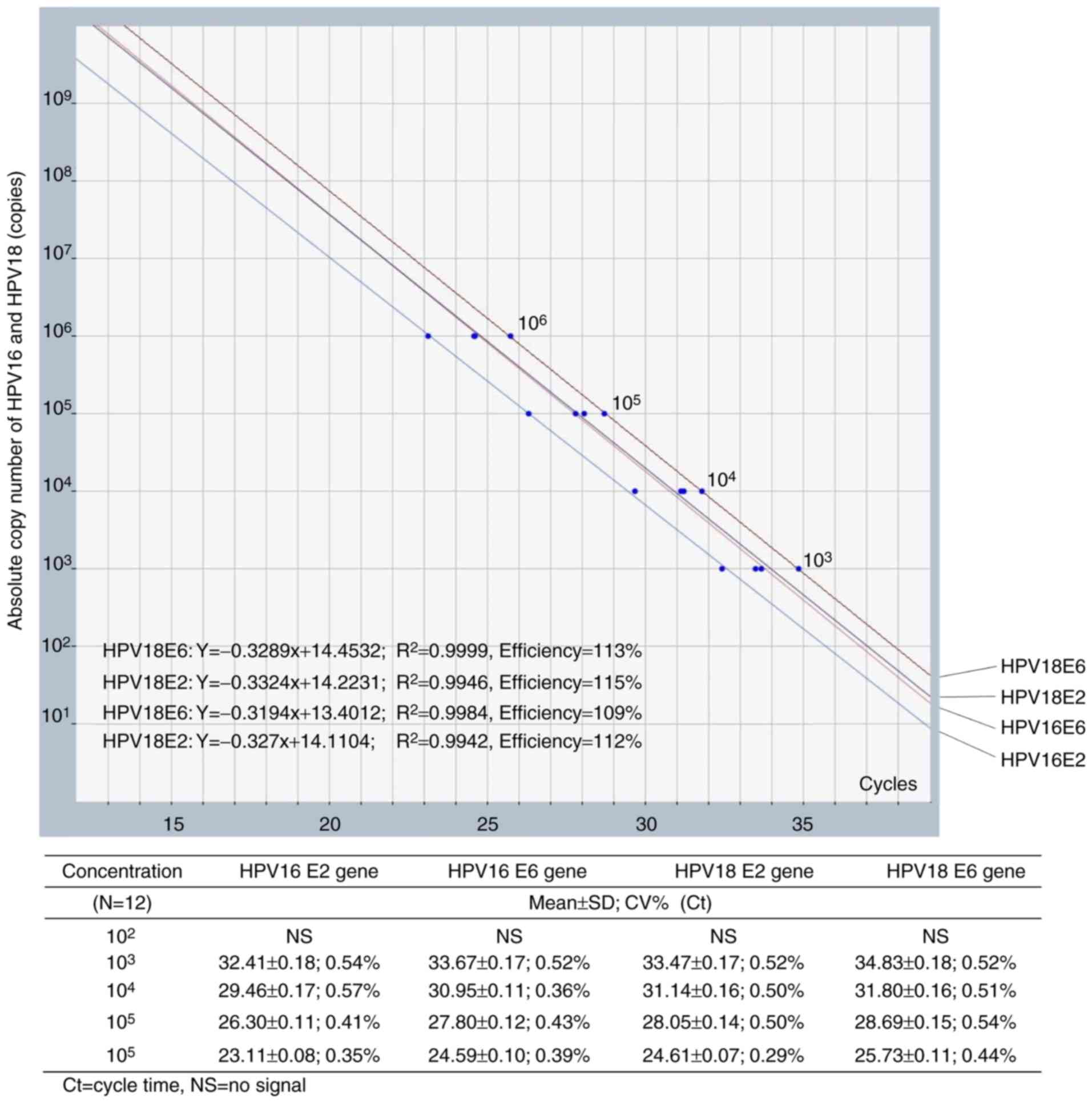
copies). Linear regression equations were estimated as indicated in
Fig. 1. The obtained Cq from samples
were used to calculate E2 and E6 copies from these equations.
Amplification efficiency was determined from the slope of
log-linear calibration curve (10−1/slope−1) (22).
 | Table I.Oligonucleotide sequences and
TaqMan® probes used for amplification of E2 and E6 genes
of HPV16 and HPV18. |
Table I.
Oligonucleotide sequences and
TaqMan® probes used for amplification of E2 and E6 genes
of HPV16 and HPV18.
| HPV type | Name | Sequence (5′-3′) | Amplified product
length (bp) | (Refs.) |
|---|
| HPV16 | HPV16 E2 Forward
primer |
AACGAAGTATCCTCTCCTGAAATTATTAG | 82 | Peitsaro et
al, 2002 (20) |
|
| HPV16 E2 Reverse
primer |
CCAAGGCGACGGCTTTG |
|
|
|
| HPV16 E2 Probe |
(TAMRA)-CACCCCGCCGCGACCCATA-(BHQ) |
|
|
|
| HPV16 E6 Forward
primer |
GAGAACTGCAATGTTTCAGGACG | 81 |
|
|
| HPV16 E6 Reverse
primer |
TGTATAGTTGTTTGCAGCTCTGTGC |
|
|
|
| HPV16 E6 gene
Probe | (Texus
red)-CAGGAGCGACCCAGAAAGTTACCACAGTT-(BHQ) |
|
|
| HPV18 | HPV18 E2 Forward
primer |
AGAAGCAGCATTGTGGACCT | 167 | Damay et al,
2009 (21) |
|
| HPV18 E2 Reverse
primer |
GGTCGCTATGTTTTCGCAAT |
|
|
|
| HPV18 E2 Probe |
(TeT)-TCAACC-CACTTCTCGGTGCAGC-(BHQ) |
|
|
|
| HPV18 E6 Forward
primer |
TCACAACATAGCTGGGCACT | 91 |
|
|
| HPV18 E6 Reverse
primer |
CTTGTGTTTCTCTGCGTCGT |
|
|
|
| HPV18 E6 Probe |
(FAM)-GCCATTCGTGCTGCAACCGA-(BHQ) |
|
|
Optimization of temperature for
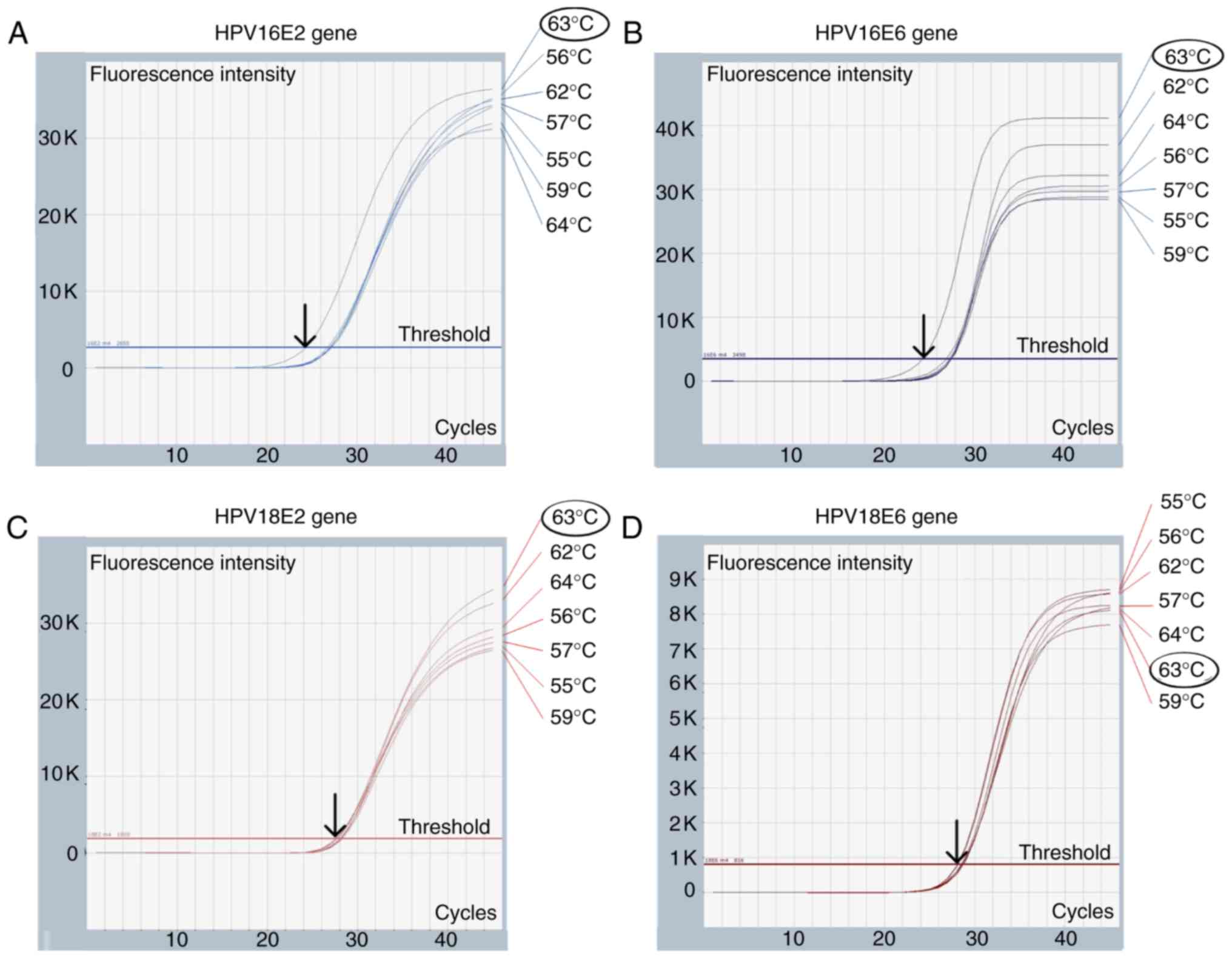
multiplex qPCR
Multiplex qPCR temperature for HPV16 (E2 and E6
genes) and HPV18 (E2 and E6 genes) was optimized. A mixture of
105 copies of plasmids containing the whole genomes of
HPV16 and HPV18 was used to optimize the annealing temperature from
55 to 64°C. The temperature that produced the lowest Cq was
selected as the optimal annealing temperature.
Evaluation of multiplex qPCR
performances
Multiplex qPCR analytical range
Ten-fold dilutions of mixed HPV16 and HPV18 whole
genomic DNA (from 102 to 106 copies) were
used as templates for determining the analytical range.
Multiplex qPCR analytical
imprecision
Within-run and between-run precision were each
determined using low (103 copies) and high
(106 copies) concentrations of whole genome HPV16 and
HPV18 DNA mixtures.
Multiplex qPCR analytical
specificity
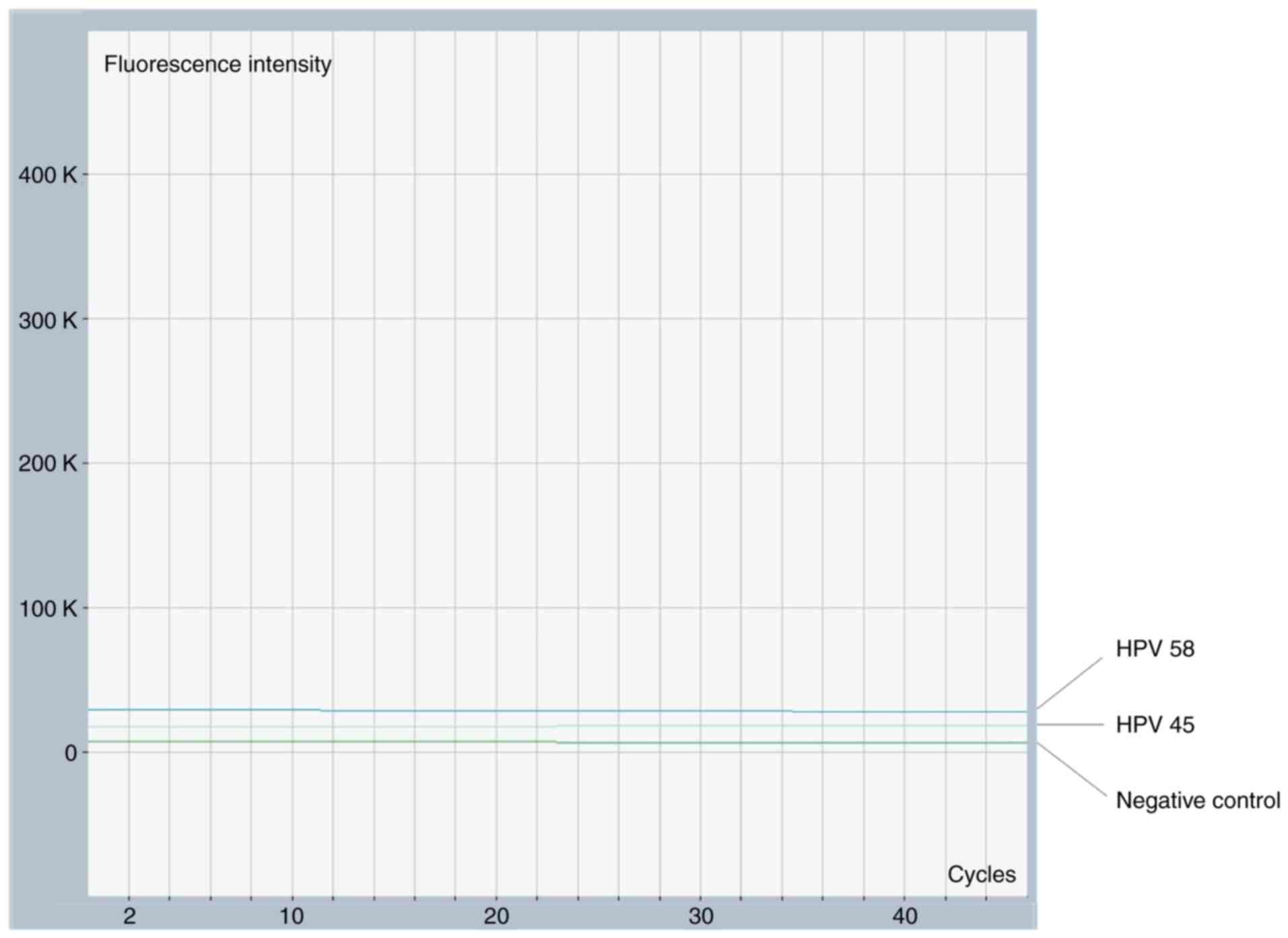
Cross-reactivity with two other HPV genotypes, HPV45
and HPV58, was tested. No fluorescent signal indicated a lack of
cross-reaction, and uninfected HPV DNA was used as a negative
control.
Competitive effect of HPV16 and HPV18
in multiplex qPCR
A mixture of unequal concentrations of HPV16 and
HPV18 DNA (from 103 to 106 copies) was used
to evaluate competitive effects in multiplex qPCR. E2 and E6 copies
obtained from unequal HPV16 and HPV18 templates (test) were
compared to those determined using a single template of HPV16 or
HPV18 (control) by a paired t-test. No significant difference
(P>0.05) indicated no competitive effect.
Evaluation of physical status using
multiplex qPCR in clinical samples
The cut-off value for an episomal form (complete E2
and E6 sequence) was first determined using plasmid DNA containing
whole HPV16 and HPV18 genomes. The E2/E6 ratio was calculated by
the 95% confidence interval (CI) and used to interpret physical
status in clinical samples as previously described (18). To verify the accuracy of the multiplex
qPCR of HPV16 and HPV18, 105 copies of Caski and HeLa
cells containing pure integrated HPV16 and HPV18, respectively,
were prepared according to the DNA calculation formula and used as
internal standards for the integrated form. A total of
105 copies of whole plasmid genome HPV16 and HPV18 were
used as internal standards for the episomal form. The two cell
lines were provided by Professor Pientong. A total of 20 cervical
samples with single and mixed infection were used to compare the
copy number and physical status between the simplex and multiplex
qPCR using paired t-tests and χ2 tests.
Statistical analysis
The linear regression equation was estimated from
standard curves between viral copy number (y-axis) and cycle time
(x-axis). The percentage of efficiency between 80–120% and
correlation (R2) >0.98 were used to determine the
standard curve. The comparison of viral copy number between
multiplex and simplex was performed using paired t-tests, while
physical status was compared by χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was performed using SPSS 19 software (IBM
Corp., Armonk, NY, USA) under a Khon Kaen University license.
Results
Multiplex qPCR performance for the
detection of HPV16 and HPV18 (E2 and E6 genes)
The optimization of Cq was performed under annealing
temperatures from 55 to 64°C, as demonstrated in Fig. 2. The optimal Cq for the E2 and E6
genes of HPV16 was 63°C, whereas the optimal Cq for HPV18 was
between 55 and 64°C. Therefore, 63°C was used to construct a
standard curve for HPV16 and HPV18 multiplex qPCR, as demonstrated
in Fig. 1. According to the
guidelines for validation of quantitative PCR methods, linear
regression and correlation (R2) analyses for each gene
revealed an acceptable efficiency of 109–115% (23). The analytical range was verified at
1,000-1,000,000 copies for HPV16 and HPV18, with average
imprecision from 0.42 to 0.50% CV as demonstrated in Fig. 1. The imprecision of measured HPV
copies is presented in Table II. The
average CV of within-run and between-run was 10.2 and 12.1,
respectively. No cross reactivity was observed with HPV58 and HPV45
(Fig. 3).
 | Table II.Analytical imprecision of the HPV16
(E2 and E6 genes) and HPV18 (E2 and E6 genes) measurement using
multiplex quantitative polymerase chain reaction. |
Table II.
Analytical imprecision of the HPV16
(E2 and E6 genes) and HPV18 (E2 and E6 genes) measurement using
multiplex quantitative polymerase chain reaction.
|
| HPV16 E2 gene | HPV16 E6 gene |
|---|
|
|
|
|
|---|
| Precision | Mean ± SD (CV%)
Cq | Mean ± SD (CV%)
Copy number | Mean ± SD (CV%)
Cq | Mean ± SD (CV%)
Copy number |
|---|
| Within-run
(n=12)a |
|
|
|
|
| Low
levelc | 32.41±0.18
(0.54) | 961±127.1
(13.23) | 33.67±0.17
(0.52) | 1,084±149.1
(13.74) |
| High
leveld | 23.11±0.08
(0.35) | 952,490±59,040.35
(6.19) | 24.59±0.10
(0.39) | 1,097,119±81,784.1
(7.45) |
| Between-run
(n=15)b |
|
|
|
|
| Low
levelc | 32.61±0.20
(0.60) | 973±141.36
(14.53) | 34.26±0.19
(0.54) | 929±137.45
(14.79) |
| High
leveld | 23.37±0.14
(0.59) | 911,140±91,626.87
(10.06) | 25.04±0.14
(0.55) |
1,040,597±112,028.74 (10.77) |
|
|
| HPV18 E2
gene | HPV18 E6
gene |
|
|
|
|
|
Precision | Mean ± SD (CV)
Cq | Mean ± SD (CV)
Copy number | Mean ± SD (CV)
Cq | Mean ± SD (CV)
Copy number |
|
| Within-run
(n=12)a |
|
|
|
|
| Low
levelc | 33.47±0.17
(0.52) | 1,159±160.1
(13.81) | 34.83±0.18
(0.52) | 1,067±144.35
(13.52) |
| High
leveld | 24.61±0.07
(0.29) | 1,123,790±60,126.32
(5.4) | 25.73±0.11
(0.44) | 1,023,242±86,472.54
(8.45) |
| Between-run
(n=15)b |
|
|
|
|
| Low
levelc | 34.03±0.19
(0.57) | 996±147.1
(14.76) | 34.77±0.19
(0.53) | 958±134.18
(14.00) |
| High
leveld | 25.00±0.11
(0.43) | 992,011±81,890.14
(8.25) | 25.42±0.13
(0.51) |
1,054,498±101,053.01 (9.58) |
Evaluation of competitive effect of
unequal HPV16 and HPV18 template concentrations in multiplex
qPCR
To mimic the presence of HPV16 and HPV18
co-infection up to 1,000-fold difference in the same sample,
measurement of mixed HPV16 and HPV18 DNA was compared in parallel
with that of single HPV16 and HPV18. Different concentrations of
HPV16 and HPV18 exhibited no effect on the quantification of E2
(P=0.319 and P=0.526, respectively) and E6 genes (P=0.347 and
P=0.146, respectively), as demonstrated in Tables III and IV. Therefore, our established multiplex
qPCR platform provided an accurate measurement for the presence of
HPV16 and HPV18 co-infection.
 | Table III.Competitive effect of unequal DNA
copies between HPV16 and HPV18 on the measurement of HPV16 E2 and
E6 genes using multiplex quantitative polymerase chain
reaction. |
Table III.
Competitive effect of unequal DNA
copies between HPV16 and HPV18 on the measurement of HPV16 E2 and
E6 genes using multiplex quantitative polymerase chain
reaction.
| HPVE2 gene | HPVE6 gene |
|---|
|
|
|---|
| Unequal mixture
HPV16 and HPV18 | Single HPV16 | Unequal mixture
HPV16 and HPV18 | Single HPV16 |
|---|
| Copy Ratio | Measured | Copy of | Measured
copies | Copy Ratio | Measured | Copy | Measured
copies |
| (HPV16:HPV18) | copies
(Test)a | HPV16 |
(Control)a | (HPV16:HPV18) | copies
(Test)a | of HPV16 |
(Control)a |
|
106:103 | 945,367 | 106 | 1,026,287 |
106:103 | 979,497 | 106 | 1,039,326 |
|
106:104 | 901,450 | 106 | 915,772 |
106:104 | 900,328 | 106 | 913,776 |
|
106:105 | 921,335 | 106 | 897,221 |
106:105 | 914,673 | 106 | 911,439 |
|
106:106 | 979,241 | 106 | 980,160 |
106:106 | 1,039,326 | 106 | 964,189 |
|
105:103 | 89,554 | 105 | 90,112 |
105:103 | 87,556 | 105 | 89,341 |
|
105:104 | 95,749 | 105 | 103,356 |
105:104 | 91,586 | 105 | 98,371 |
|
105:105 | 96,968 | 105 | 105,065 |
105:105 | 97,092 | 105 | 103,174 |
|
105:106 | 94,159 | 105 | 92,332 |
105:106 | 112,708 | 105 | 100,329 |
|
104:103 | 8,832 | 104 | 9,149 |
104:103 | 11,219 | 104 | 10,439 |
|
104:104 | 8,836 | 104 | 10,626 |
104:104 | 9,175 | 104 | 9,303 |
|
104:105 | 8,913 | 104 | 9,319 |
104:105 | 8,641 | 104 | 10,322 |
|
104:106 | 11,347 | 104 | 10,861 |
104:106 | 10,989 | 104 | 9,018 |
|
103:103 | 1,143 | 103 | 1,200 |
103:103 | 1,039 | 103 | 1,153 |
|
103:104 | 867 | 103 | 912 |
103:104 | 907 | 103 | 865 |
|
103:105 | 881 | 103 | 1,122 |
103:105 | 848 | 103 | 973 |
|
103:106 | 848 | 103 | 901 |
103:106 | 846 | 103 | 908 |
 | Table IV.Competitive effect of unequal copies
between HPV16 and HPV18 on the measurement of HPV18 E2 and E6 genes
using multiplex quantitative polymerase chain reaction. |
Table IV.
Competitive effect of unequal copies
between HPV16 and HPV18 on the measurement of HPV18 E2 and E6 genes
using multiplex quantitative polymerase chain reaction.
| HPVE2 gene | HPVE6 gene |
|---|
|
|
|---|
| Unequal mixture
HPV18 and HPV16 | Single HPV18 | Unequal mixture
HPV18 and HPV16 | Single HPV18 |
|---|
| Copy ratio | Measured
copies | Copy of | Measured copies
of | Copy ratio | Measured
copies | Copy of | Measured copies
of |
| (HPV18:HPV16) | of HPV18E2
(Test)a | HPV18 | HPV18E2
(Control)a | (HPV18:HPV16) | of HPV18E6
(Test)a | HPV18 | HPV18E6
(Control)a |
|
106:103 | 1,135,661 | 106 | 1,075,620 |
106:103 | 1,015,464 | 106 | 1,107,241 |
|
106:104 | 1,097,976 | 106 | 1,181,450 |
106:104 | 1,107,170 | 106 | 1,125,579 |
|
106:105 | 877,385 | 106 | 1,135,265 |
106:105 | 911,742 | 106 | 893,768 |
|
106:106 | 1,092,086 | 106 | 993,922 |
106:106 | 965,678 | 106 | 1,040,114 |
|
105:103 | 104,611 | 105 | 96,998 |
105:103 | 90,546 | 105 | 94,778 |
|
105:104 | 101,164 | 105 | 113,785 |
105:104 | 97,952 | 105 | 113,933 |
|
105:105 | 105,464 | 105 | 101,469 |
105:105 | 110,175 | 105 | 87,996 |
|
105:106 | 102,760 | 105 | 113,715 |
105:106 | 87,492 | 105 | 107,587 |
|
104:103 | 9,129 | 104 | 8,789 |
104:103 | 10,201 | 104 | 9,746 |
|
104:104 | 9,550 | 104 | 9,443 |
104:104 | 9,732 | 104 | 10,395 |
|
104:105 | 8,639 | 104 | 9,988 |
104:105 | 10,403 | 104 | 8,665 |
|
104:106 | 10,700 | 104 | 9,812 |
104:106 | 8,910 | 104 | 10,550 |
|
103:103 | 912 | 103 | 1,120 |
103:103 | 937 | 103 | 1,056 |
|
103:104 | 936 | 103 | 1,078 |
103:104 | 910 | 103 | 938 |
|
103:105 | 849 | 103 | 831 |
103:105 | 901 | 103 | 833 |
|
103:106 | 896 | 103 | 1,168 |
103:106 | 869 | 103 | 1,063 |
Evaluation of physical status in
clinical samples
Cut-off values for viral status were calculated as
previously described by Wanram et al (18) and are shown in Table V. The E2/E6 ratio was identified as
0.78–1.10 and 0.85–1.18 for HPV16 and HPV18, respectively. An E2/E6
ratio of 0 was defined as the absolute integrated form, whereas
E2/E6 >0 and less than the cut-off value was interpreted as the
mixed form of episomal and integrated HPV (18). Comparisons of HPV16 and HPV18 copy
numbers between the simplex and multiplex qPCR in CXCA samples are
summarized in Table VI. No
significant difference between simplex and multiplex qPCR was
observed for HPV16 E2 and E6 (P=0.307 and P=0.288; paired t-test)
and HPV18 E2 and E6 genes (P=0.396 and P=0252; paired t-test). The
physical status obtained from multiplex qPCR was also compared with
that from the simplex qPCR. The cut-off value for the episomal form
of HPV16 (0.79–1.10) and HPV18 (0.85–1.18) was calculated as
previously described for multiplex qPCR. Interpretation of physical
status was similar in 95% (19/20) of cases between multiplex and
simplex qPCR assays, and differed in one case, CX-1 (P=0.372;
χ2 test; Table VI).
 | Table V.Estimation of cut-off values (E2 and
E6 ratios) for the interpretation of physical status using various
concentrations of whole genome plasmid DNA from 104 to
106 copies. |
Table V.
Estimation of cut-off values (E2 and
E6 ratios) for the interpretation of physical status using various
concentrations of whole genome plasmid DNA from 104 to
106 copies.
|
|
| HPV16 | HPV18 |
|---|
|
|
|
|
|
|---|
| No. | Mixture of equal
concentration of HPV16 and HPV18 | E2 gene (copy) | E6 gene (copy) | E2/E6 | E2 gene (copy) | E6 gene (copy) | E2/E6 |
|---|
| 1 | 103 | 1,115 | 1,103 | 1.01 | 1,153 | 1,233 | 0.94 |
| 2 | 103 | 1,115 | 1,185 | 0.94 | 1,019 | 1,210 | 0.84 |
| 3 | 103 | 908 | 1,044 | 0.87 | 1,193 | 1,211 | 0.99 |
| 4 | 103 | 1,168 | 1,169 | 1 | 1,155 | 1,134 | 1.02 |
| 5 | 103 | 1,038 | 1,047 | 0.99 | 1,146 | 1,179 | 0.97 |
| 6 | 104 | 8,882 | 10,814 | 0.82 | 10,760 | 9,858 | 1.09 |
| 7 | 104 | 8,789 | 10,438 | 0.84 | 9,187 | 9,408 | 0.98 |
| 8 | 104 | 9,019 | 10,379 | 0.87 | 8,703 | 9,062 | 0.96 |
| 9 | 104 | 9,233 | 9,268 | 1 | 8,513 | 8,220 | 1.04 |
| 10 | 104 | 9,117 | 9,337 | 0.98 | 9,687 | 11,622 | 0.83 |
| 11 | 105 | 94,603 | 105,819 | 0.89 | 110,462 | 107,970 | 1.02 |
| 12 | 105 | 108,055 | 108,928 | 0.99 | 107,699 | 102,882 | 1.05 |
| 13 | 105 | 88,246 | 100,449 | 0.88 | 106,389 | 98,269 | 1.08 |
| 14 | 105 | 87,938 | 105,370 | 0.83 | 114,166 | 112,276 | 1.02 |
| 15 | 105 | 106,246 | 100,978 | 1.05 | 109,971 | 113,442 | 0.97 |
| 16 | 106 | 1,098,531 | 1,059,641 | 1.04 | 1,086,195 | 991,320 | 1.1 |
| 17 | 106 | 926,410 | 982,153 | 0.94 | 1,097,976 | 996,524 | 1.1 |
| 18 | 106 | 912,783 | 1,076,185 | 0.85 | 1,085,161 | 927,645 | 1.17 |
| 19 | 106 | 1,020,080 | 931,143 | 1.1 | 1,086,195 | 1,005,826 | 1.08 |
| 20 | 106 | 940,240 | 997,234 | 0.94 | 1,146,435 | 1,142,588 | 1 |
|
|
|
| Mean | 0.94 |
| Mean | 1.01 |
|
|
|
| SD | 0.08 |
| SD | 0.08 |
|
|
|
| Mean ± 2SD | 0.78–1.10 |
| Mean ± 2SD | 0.85–1.18 |
 | Table VI.Comparison of viral copy number and
physical status determination between multiplex qPCR (test) and
simplex qPCR (control) in 20 cervical tissues samples harboring
single and co-infection of HPV16 and HPV18. |
Table VI.
Comparison of viral copy number and
physical status determination between multiplex qPCR (test) and
simplex qPCR (control) in 20 cervical tissues samples harboring
single and co-infection of HPV16 and HPV18.
|
| Simplex qPCR
(Control) | Multiplex qPCR
(Test) |
|---|
|
|
|
|
|---|
|
| Copies/ng DNA | Physical
status | Copies/ng DNA | Physical
status | Copies/ng DNA | Physical
status | Copies/ng DNA | Physical
status |
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| CXCA samples
(HPV) | HPV16 E2 | HPV16 E6 | E2/E6 ratio
(0.79)a | Interpretation | HPV18 E2 | HPV18 E6 | E2/E6 ratio
(0.81)b | Interpretation | HPV16 E2 | HPV16 E6 | E2/E6 ratio
(0.78)c | Interpretation | HPV18 E2 | HPV18 E6 | E2/E6 ratio
(0.85)d | Interpretation |
|---|
| PCX 1 (16) | 5,139 | 498,762 | 0.01 | Mixed | – | – | – | – | 4,196 | 470,492 | 0.01 | Mixed | – | – | – | – |
| PCX 2 (16) | 312 | 32,190 | 0.01 | Mixed | – | – | – | – | 203 | 30,613 | 0.01 | Mixed | – | – | – | – |
| PCX 3 (16) | 5,815 | 6,087 | 0.96 | Episomal | – | – | – | – | 5644.39 | 6196.23 | 0.91 | Episomal | – | – | – | – |
| PCX 4 (18) | – | – | – | – | – | 191,065 | 0 | Integrated | – | – | – | – | – | 153,540 | 0 | Integrated |
| PCX 5 (18) | – | – | – | – | – | 9,380 | 0 | Integrated | – | – | – | – | – | 11,897 | 0 | Integrated |
| CX 1 (16) | 118,584 | 157,522 | 0.75 | Mixed | – | – | – | – | 108,968 | 133,776 | 0.81 | Episomal | – | – | – |
|
| CX2 (16) | 30,353 | 26,353 | 1.15 | Episomal | – | – | – | – | 32,588 | 24,118 | 1.35 | Episomal | – | – | – |
|
| CX 3 (16) | – | 24,237 | 0 | Integrated | – | – | – | – | – | 26,814 | 0 | Integrated | – | – | – |
|
| CX 4 (16) | 38,041 | 44,122 | 0.86 | Episomal | – | – | – | – | 38,788 | 48,108 | 0.81 | Episomal | – | – | – |
|
| CX 5 (16) | 6,209 | 6,721 | 0.92 | Episomal | – | – | – | – | 6,879 | 7,395 | 0.93 | Episomal | – | – | – |
|
| CX 6 (18) | – | – | – | – | 15,649 | 45,455 | 0.34 | Mixed | – | – | – | – | 16,857 | 42,240 | 0.4 | Mixed |
| CX 7 (18) | – | – | – | – | – | 73,929 | 0 | Integrated | – | – | – | – | – | 70,912 | 0 | Integrated |
| CX 8 (18) | – | – | – | – | 74,237 | 56,271 | 1.32 | Episomal | – | – | – | – | 71,243 | 54,039 | 1.32 | Episomal |
| CX 9 (16+18) | 855 | 3,468 | 0.25 | Mixed | 326 | 521 | 0.63 | Mixed | 1,014 | 3,919 | 0.26 | Mixed | 205 | 420 | 0.49 | Mixed |
| CX 10 (16+18) | 39,350 | 123,420 | 0.32 | Mixed | 266 | 410 | 0.65 | Mixed | 36,000 | 155,429 | 0.23 | Mixed | 113 | 237 | 0.48 | Mixed |
| CX 11 (16+18) | 2,834,887 | 6,008,765 | 0.47 | Mixed | 4,090 | 3,087 | 1.32 | Episomal | 2,592,857 | 5,571,429 | 0.47 | Mixed | 3,064 | 2,414 | 1.27 | Episomal |
| CX 12 (16+18) | 4,122 | 14,979 | 0.28 | Mixed | 403 | 478 | 0.84 | Episomal | 3,305 | 17,360 | 0.19 | Mixed | 371 | 356 | 1.04 | Episomal |
| CX 13 (16+18) | 120 | 277 | 0.43 | Mixed | 219 | 415 | 0.53 | Mixed | 92 | 227 | 0.41 | Mixed | 197 | 339 | 0.58 | Mixed |
| CX 14 (16+18) | 9,124 | 24,510 | 0.37 | Mixed | – | 1,034 | 0 | Integrated | 8,733 | 20,600 | 0.42 | Mixed | – | 853 | 0 | Integrated |
| CX 15 (16+18) | – | 512,980 | 0 | Integrated | – | 981 | 0 | Integrated | – | 488,500 | 0 | Integrated | – | 741 | 0 | Integrated |
| Internal
control |
| HPV16; Caski | 0 | 95,859 | 0 | Integrated | 0 | 0 | 0 | – | 0 | 91,791 | 0 | Integrated | 0 | 0 | 0 | – |
| HPV18; HeLa | 0 | 0 | 0 | – | 0 | 102,825 | 0 | Integrated | 0 | 0 | 0 | – | 0 | 110,275 | 0 | Integrated |
| Whole plasmid
HPV16 | 109,334 | 102,675 | 1.06 | Episomal | – | – | – | – | 106,445 | 105,988 | 1.00 | Episomal | – | – | – | – |
| Whole plasmid
HPV18 | – | – | – | – | 99,545 | 98,9655 | 1.01 | Episomal | – | – | – | – | 100,599 | 99,453 | 1.01 | Episomal |
Discussion
Multiple HR-HPV infection, particularly HPV16 and
HPV18 co-infection, is now a concern due to its effects on cervical
neoplasia development. The failure rate of treatment was previously
reported to be increased by 5-fold in multiple infection (57%),
compared with single infection (12%) (24). Therefore, a suitable risk assessment
among patients with persistent multiple HR-HPV infection is
required. To assess the risk, viral load and viral physical status
may be used for cancer prognosis. In the present study, a one tube
qPCR assay for HPV16 and HPV18 co-infection was successfully
established with acceptable performance in terms of specificity,
accuracy and precision.
Upon performing a literature search for multiplex
qPCR of 4 genes in one tube, one study by Zhao et al
(25) was identified. The authors
reported detection of 4 viral DNAs: HPV16 (E6 gene), HPV18 (E6
gene), HSV1 and HSV2, with an improved detection limit at 10 copies
compared with the present study. The difference in the detection
limit may result from the different size of viral DNA standard.
Small fragments of viral DNA (66–139 bp) were used as standard in
the study undertaken by Zhao et al, whereas the whole HPV
genome (10728–12267 bp) was used in the present study. Accordingly,
the small size template has an advantage for amplification when
compared with the whole genome. Therefore, our established
technique better represents the real viral infection in a clinical
setting. To resolve this limitation, more DNA template may be
adjusted. The accuracy of interpretation, including possible cross
reactivity, was verified using DNA of known viral status from
cervical Caski and HeLa cell lines. HPV58 and HPV45 were selected
for cross reactivity according to the top 4 common HR-HPV types
(HPV 16, 18, 45 and 58), covering 90% of cases among Thai women
(26). Accurate quantification was
not only demonstrated via comparison with simplex qPCR, but it was
also revealed that there was no competitive effect of an unequal
mixed HPV DNA template from 10- to 1,000-fold (Tables III and IV).
Furthermore, the comparison of viral physical status
between simplex and multiplex qPCR in clinical samples achieved 95%
(19/20 samples) agreement in the results of viral physical status.
The different physical status of CX1 was caused by the variation of
E2/E6 ratio between simplex (0.75) and multiplex (0.81) which
closed to the cut-off value for episomal form (0.79 and 0.78)
resulting in discrepancy results as mixed form and episomal form,
respectively. This limitation of accuracy occurred at values close
to the cut-off values.
In conclusion, the increased incidence of HPV16 and
HPV18 co-infection is a high-risk factor for CXCA progression in
patients with persistent HR-HPV infection. Therefore, the
successful development of multiplex qPCR for detecting HPV16 and
HPV18 viral load and physical status in a single tube would provide
a significant benefit in terms of cost effectiveness and shorter
assay time in the clinic. To assess the potential of using this
assay as a risk assessment for cancer progression in patients with
single and co-infection with HPV16 and HPV18, a larger sample size
with clinical outcome data should be included in future studies. In
particular, pre-cancerous and early cancerous cases harboring high
risk factors should be followed up frequently with monitoring of
risk factors.
Acknowledgements
Clinical stage of samples were provided by Dr Metee
Wongsena (Chief of Gynecologic Cancer Department, Ubonratchathani
Cancer Center, Ubonratchathani, Thailand) and Dr Pissamai Yuenyao
(Srinagarind Hospital, Khon Kaen, Thailand).
Funding
The present study was supported by the Targeted
Research Fund of Khon Kaen University from the The National
Research Council of Thailand (Khon Kaen, Thailand) and the
Post-doctoral training program for MW (grant no. 58330) from the
Graduate School of Khon Kaen University, Thailand.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PJ supervised, designed and assisted in the
coordination of the present study. PP performed the research, the
analysis and prepared data for the first draft of manuscript. TL
and CL made substantial contributions in the conception and
experimental design. MW was involved in data analysis,
interpretation of data and drafting the final manuscript. PT
assisted with data interpretation and revising the final
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Ubonratchathani Cancer Hospital (approval no.
CE012/2013; Ubonratchathani, Thailand) and Khon Kaen University
(approval no. HE562296; Kho Kaen, Thailand).
Patient consent for publication
All patients provided written informed consent for
the publication of the present study.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CXCA
|
cervical cancer
|
|
HR-HPV
|
high risk human papillomavirus
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
CV
|
coefficient of variation
|
References
|
1
|
Bruni L, Barrionuevo-Rosas L, Albero G,
Serrano B, Mena M, Gómez D, Muñoz J, Bosch FX and de Sanjosé S: ICO
information centre on HPV and cancer (HPV Information Centre).
Human papillomavirus and related diseases in the World. Summary
Report. 30–July;2017.
|
|
2
|
Bruni L, Barrionuevo-Rosas L, Albero G,
Serrano B, Mena M, Gómez D, Muñoz J, Bosch FX and de Sanjosé S: ICO
information centre on HPV and cancer (HPV Information Centre).
Human papillomavirus and related diseases in Asia. Summary Report.
27–July;2017.
|
|
3
|
Li N, Franceschi S, Howell-Jones R,
Snijders PJ and Clifford GM: Human papillomavirus type distribution
in 30,848 invasive cervical cancers worldwide: Variation by
geographical region, histological type and year of publication. Int
J Cancer. 128:927–935. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu EQ, Yu XH, Zha X, Zhang GN, Wang JH,
Fan Y, Tang YY, Zhao ZX, Wu YG and Kong W: Distribution of human
papillomavirus genotypes in archival cervical lesions in eastern
inner Mongolian autonomous region. China Int J Gynecol Cancer.
19:919–923. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Youssef MA, Abdelsalam L, Harfoush RA,
Talaat IM, Elkattan E, Mohey A, Abdella RM, Farhan MS, Foad HA,
Elsayed AM, et al: Prevalence of human papilloma virus (HPV) and
its genotypes in cervical specimens of Egyptian women by linear
array HPV genotyping test. Infect Agent Cancer. 11:1750–9378. 2016.
View Article : Google Scholar
|
|
6
|
Téguété I, Dolo A, Sangare K, Sissoko A,
Rochas M, Beseme S, Tounkara K, Yekta S, De Groot AS and Koita OA:
Prevalence of HPV 16 and 18 and attitudes toward HPV vaccination
trials in patients with cervical cancer in Mali. PLoS One.
12:e01726612017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siddiqa A, Zainab M, Qadri I, Bhatti MF
and Parish JL: Prevalence and genotyping of high risk human
papillomavirus in cervical cancer samples from Punjab, Pakistan.
Viruses. 6:2762–2777. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suthipintawong C, Siriaunkgul S,
Tungsinmunkong K, Pientong C, Ekalaksananan T, Karalak A, Kleebkaow
P, Vinyuvat S, Triratanachat S, Khunamornpong S and Chongsuwanich
T: Human papilloma virus prevalence, genotype distribution, and
pattern of infection in Thai women. Asian Pac J Cancer Prev.
12:853–856. 2011.PubMed/NCBI
|
|
9
|
Chaturvedi AK, Katki HA, Hildesheim A,
Rodríguez AC, Quint W, Schiffman M, Van Doorn LJ, Porras C,
Wacholder S, Gonzalez P, et al: Human papillomavirus infection with
multiple types: Pattern of coinfection and risk of cervical
disease. J Infect Dis. 203:910–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Trottier H, Mahmud S, Costa MC, Sobrinho
JP, Duarte- Franco E, Rohan TE, Ferenczy A, Villa LL and Franco EL:
Human papillomavirus infections with multiple types and risk of
cervical neoplasia. Cancer Epidemiol Biomark Prev. 15:1274–1280.
2006. View Article : Google Scholar
|
|
11
|
Salazar KL, Zhou HS, Xu J, Peterson LE,
Schwartz MR, Mody DR and Ge Y: Multiple human papilloma virus
infections and their impact on the development of high-risk
cervical lesions. Acta Cytol. 59:391–398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pett M and Coleman N: Integration of
high-risk human papillomavirus: A key event in cervical
carcinogenesis? J Pathol. 212:356–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cricca M, Morselli-Labate AM, Venturoli S,
Ambretti S, Gentilomi GA, Gallinella G, Costa S, Musiani M and
Zerbini M: Viral DNA load, physical status and E2/E6 ratio as
markers to grade HPV16 positive women for high-grade cervical
lesions. Gynecol Oncol. 106:549–557. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu EQ, Zha X, Yu XH, Zhang GN, Wu YG, Fan
Y, Ren Y, Kong LQ and Kong W: Profile of physical status and gene
variation of human papillomavirus 58 genome in cervical cancer. J
Gen Virol. 90:1229–1237. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qureshi MN, Bolick D, Ringer PJ, Spagler
FL and Zimmerman G: HPV testing in liquid cytology specimens:
Comparison of analytic sensitivty and specificity for in situ
hybridization and chemiluminescent nucleic acid testing. Acta
Cytol. 49:120–126. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fujii T, Masumoto N, Saito M, Hirao N,
Niimi S, Mukai M, Ono A, Hayashi S, Kubushiro K, Sakai E, et al:
Comparison between in situ hybridization and real-time PCR
technique as a means of detecting the integrated form of human
papillomavirus 16 in cervical neoplasia. Diagn Mol Pathol.
14:103–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaiwongkot A, Pientong C, Ekalaksananan
T, Vinokurova S, Kongyingyoes B, Chumworathayi B, Patarapadungkit
N, Siriaunkgul S and von Knebel Doeberitz M: Detection of the human
papillomavirus 58 physical state using the amplification of
papillomavirus oncogene transcripts assay. J Virol Methods.
189:290–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wanram S, Limpaiboon T, Leelayuwat C,
Yuenyao P, Guiney DG, Lulitanond V, Lulitanond V and Jearanaikoon
P: The use of viral load as a surrogate marker in predicting
disease progression for patients with early invasive cervical
cancer with integrated human papillomavirus type 16. Am J Obstet
Gynecol. 201:79.e1–e7. 2009. View Article : Google Scholar
|
|
19
|
Sotlar K, Diemer D, Dethleffs A, Hack Y,
Stubner A, Vollmer N, Menton S, Menton M, Dietz K, Wallwiener D, et
al: Detection and typing of human papillomavirus by E6 nested
multiplex PCR. J Clin Microbiol. 42:3176–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peitsaro P, Johansson B and Syrjanen S:
Integrated human papillomavirus type 16 is frequently found in
cervical cancer precursors as demonstrated by a novel quantitative
real-time PCR technique. J Clin Microbiol. 40:886–891. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Damay A, Didelot-Rousseau MN, Costes V,
Konate I, Ouedraogo A, Nagot N, Foulongne V, Van de Perre P, Mayaud
P and Segondy M: Viral load and physical status of human
papillomavirus (HPV) 18 in cervical samples from female sex workers
infected with HPV 18 in Burkina Faso. J Med Virol. 81:1786–1791.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Broeders S, Huber I, Grohmann L, Berben G,
Taverniers I, Mazzara M, Roosens N and Morisset D: Guidelines for
validation of qualitative real-time PCR methods. Trends Food Sci
Technol. 37:115–126. 2014. View Article : Google Scholar
|
|
24
|
Munagala R, Donà MG, Rai SN, Jenson AB,
Bala N, Ghim SJ and Gupta RC: Significance of multiple HPV
infection in cervical cancer patients and its impact on treatment
response. Int J Oncol. 34:263–271. 2009.PubMed/NCBI
|
|
25
|
Zhao Y, Cao X, Tang J, Zhou L, Gao Y, Wang
J, Zheng Y, Yin S and Wang Y: A novel multiplex real-time PCR assay
for the detection and quantification of HPV16/18 and HSV1/2 in
cervical cancer screening. Mol Cell Probes. 26:66–72. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bruni L, Barrionuevo-Rosas L, Albero G,
Serrano B, Mena M, Gómez D, Muñoz J, Bosch FX and de Sanjosé S: ICO
Information Centre on HPV and Cancer (HPV information centre).
Human papillomavirus and related diseases in Thailand. Summary
Report. 27–July;2017.
|