Introduction
Of all pediatric bone malignant tumor types,
osteosarcoma has the highest incidence (1). Although surgical techniques and
chemotherapy have improved, the treatment effects remain unchanged
in detectable metastatic osteosarcoma (2,3). The rapid
growth of metastases and chemotherapy resistance are the primary
causes of not achieving complete remission (4); thus, it is vital to investigate an early
detection marker and molecular target in order to achieve complete
remission of osteosarcoma.
MicroRNAs (miRNAs) regulate protein expression
through complementary base pairing to specific mRNAs that act in
the 3′-untranslated regions (3′-UTRs) of target mRNAs (5). miRNAs serve an irreplaceable role in a
variety of biological processes in cancer development and
progression, including differentiation, proliferation metastasis
and apoptosis (6–9). Previous studies have demonstrated that
miR-218 is associated with the proliferation, migration and
invasion of osteosarcoma (10,11);
however, there is limited knowledge regarding the role that miR-218
serves in the proliferation of osteosarcoma. Therefore, the
molecular mechanism underlying the action of miR-218 in
osteosarcoma was investigated.
E2F2 is affiliated with a family of transcription
factors that includes E2F1-8 (12).
Studies have demonstrated that E2F factors are, not only involved
in the expression of cell cycle-associated genes, but also in the
expression of differentiation-, apoptosis-, autophagy- and
metabolism-associated genes (13–15).
Previously, E2F2 was determined to maintain G0 arrest by
regulating cell cycle-associated genes (13). In the present study, whether E2F2 was
essential for osteosarcoma tumorigenesis was investigated.
In brief, the present results demonstrated that
miR-218 inhibits the proliferation of human osteosarcoma cells
through E2F2, and that E2F2 promotes osteosarcoma
tumorigenesis.
Materials and methods
Antibodies
Anti-E2F2 (dilution, 1:500; cat. no. sc-633)
antibodies and β-actin (dilution, 1:1,000; cat. no. sc-58673) were
obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Secondary antibodies anti-mouse IgG, (dilution, 1:2,000; cat. no.
7076) were obtained from Cell Signaling Technology, Inc. (Danvers,
MA, USA).
Cell culture
293, Human osteoblast hFOB1.19 and human
osteosarcoma MG63 cell lines were obtained from the Shanghai
Institute of Cell Biology, Chinese Academy of Sciences (Shanghai,
China), and maintained at 37°C in an atmosphere containing 5%
CO2 in Dulbecco's modified Eagle's medium, D-MEM/F-12
medium and Minimum Eagle's medium, respectively, and with 10%
heat-inactivated fetal bovine serum (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA).
Constructs and production of
lentivirus
The miR-218 complementary DNA (cDNA) sequences were
inserted between the BamHI/MluI restriction sites in
the pGLV3/H1 vector (0.03 pmol) (Shanghai GenePharma Co., Ltd.,
Shanghai, China). The miR-218 short-hairpin RNA (shRNA) oligomer
(target sequence:
5′-GATCCACATGGTTAGATCAAGCACAACGATACATGGTTAGATCAAGCACAAACCGGTACATGGTTAGATCAAGCACAATCACACATGGTTAGATCAAGCACAATTTTTTG-3′)
was subcloned into the pGLV3/H1 construct between the
BamHI-MluI restriction sites. MG63 cells were
constructed using the three-plasmid lentivirus system, where the
second-generation lentiviral packaging, pspax2, and viral envelope,
pMD2G (Shanghai GenePharma Co., Ltd.) were co-transfected in 293
cells using PolyJet (SignaGen Laboratories, Rockville, MD, USA),
according to the manufacturer's protocols, to package the viruses.
The above three plasmids (pspax2: pMD2G: pGLV3/H1-miR-218=1 ug: 1
ug: 2 ug), opti-MEM (500 ul) and PolyJet (12 ul) were mixed and
allowed to stand for 20 min. It was subsequently added to a
six-well plate with MG63 cell density of 70%. After 12 h, the
medium was changed, and the supernatant, containing lentivirus, was
collected for 72 h.
Establishment of stable cell
lines
For overexpression of miR-218, the MG63 cells were
infected by green fluorescent protein (GFP) positive miR-218
viruses. Following 48 h of infection, the cells were cultured in
Minimum Eagle's Medium containing 2.5 lg/ml puromycin (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The surviving cells
were cultured into cell lines stably expressing miR-218. The
control, expressing GFP, and miR-218-sponge, expressing GFP, groups
were obtained according to the methods described above.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
cDNA was obtained using mRNA RT (Roche Diagnostics,
Basel, Switzerland), according to the manufacturer's protocol. qPCR
was performed using an ABI 7300 real-time PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with FastStart
Universal SYBR® Green Mix (Roche Diagnostics, Basel,
Switzerland), according to the manufacturer's protocols. The
primers for the amplification of E2F2 and β-actin were as follows:
E2F2 forward, 5′-AGCTGGATCTGGAGGGGATT-3′ and reverse,
5′-AGGACCCCATCCTCTGACTC-3′; and β-actin forward,
5′-CATGTACGTTGCTATCCAGGC-3′ and reverse,
5′-CGCTCGGTGAGGATCTTCATG-3′. The molecular weights were 196 and 195
bp, respectively. Thermocycling protocol included pre-denatured at
95°C for 3 min, denaturation at 95°C for 15 sec, annealing at 60°C
for 15 sec and extension at 72°C for 1 min for 40 cycles.
Expression levels were quantified using the 2−ΔΔCq
method (16).
EdU experiment
The EdU incorporation assay
(5-ethynyl-2′-deoxyuridine; Guangzhou RiboBio Co., Ltd., Guangzhou,
China) was used to evaluate the number of proliferating cells. A
total of 1×104 cells, MG63/control, MG63/miR-218 and
MG63/miR-218-sponge cells, were plated in 96-well plates and
cultured for an additional 24 h. Cells were treated with 50 µM EdU
for 2 h at 37°C and then fixed with 4% formaldehyde for 30 min at
room temperature. Subsequently, 2 mg/ml glycine antagonized
formaldehyde and 0.5% Triton X-100 was added and incubated for 15
min at room temperature. Following this, 100 µl 1X Apollo reaction
cocktail (Guangzhou RiboBio Co., Ltd.) and DAPI dye were added to
the cells and incubated for 30 min at room temperature,
respectively. Images were obtained using the fluorescent Olympus
IX-71 inverted microscope (magnification, ×200; Olympus
Corporation, Tokyo, Japan).
Flow cytometry
Cells (1×105) were plated in 6-well
plates and cultured for 24 h at 37°C. The cells were digested using
trypsin and single suspension cells were collected in 1.5 ml
Eppendorf tubes. Subsequently, the cells were treated with 70%
ethanol for 15 min at 0°C and then incubated with RNase for 30 min
at 37°C. Following this, the cells were dyed with propidium iodide
(10 ug/ml; Thermo Fisher Scientific, Inc.) and protected from light
for 30 min at room temperature. Cells were then subjected to flow
cytometry with a flow cytometer and the results were analyzed
through CellQuest Pro 4.0.2 software (BD Biosciences, Franklin
Lakes, NJ, USA).
Plate colony formation
A total of 2×102 cells, MG63/control,
MG63/miR-218 and MG63/miR-218-sponge cells, were plated in 6-cm
dishes and cultured for 14 days at 37°C. Cells were fixed with 4%
formaldehyde for 30 min and then incubated with 0.05% crystal
violet for 30 min at room temperature. Images were captured using a
Canon 70D camera (Canon, Inc., Tokyo, Japan). Colonies with >50
cells were counted.
Bioinformatics prediction
The miR-218 target prediction was performed using
bioinformatics algorithms from TargetScan 6.2 (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/).
Western blot analysis
Cell protein was extracted from the osteosarcoma
cell lines by bicinchoninic acid protein assay kit (Beyotime
Institute of Biotechnology Co., Ltd., Shanghai, China), according
to the manufacturer's protocols. Equivalent amounts (40 ug) of
protein were added to 10% SDS-PAGE and then transferred to
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica,
MA, USA) using a wet transfer method, where the PAGE gel and PVDF
membranes are placed in a thermostat covered with ice with a
constant current of 225 mA for 2.5 h at 0°C. Following blocking
with 5% skimmed milk powder overnight at 4°C, the primary
antibodies (E2F2 and β-actin) were added and incubated at 4°C
overnight. Subsequently, the secondary antibody was added and
incubated for 2 h at room temperature. The Pierce enhanced
chemiluminescent Plus Western Blotting Substrate (Thermo Fisher
Scientific, Inc.) was added to PVDF membranes and images were
obtained using Image Lab 5.2 (Bio-Rad, Laboratories, Inc. Hercules,
CA, USA). Results were analyzed using ImageJ 1.8.0 software
(National Institutes of Health, Bethesda, MD, USA).
Luciferase reporter experiments
E2F2 3′UTR was subcloned between the
SacI/XboI restriction sites in the pmirGLO vector
(Shanghai GenePharma Co., Ltd.). The pmirGLO-MT-BMI1 3′UTR plasmid
was constructed by removing the binding site for miR-218
(UUCGUGUU). PmirGLO-WT-E2F2 3′UTR/pmirGLO-MT-E2F2 3′UTR and phRL-TK
were co-transfected into MG63 cells using Polyjet. Luciferase
activity was assessed using the Dual-Luciferase Reporter Assay
system (Promega Corporation, Madison, WI, USA) 36 h after
transfection. Luciferase activity was normalized to Renilla
luciferase activity.
Animal experiments
Female BALB/c nude mice (n=6; ~20 g), 4–5 weeks of
age, were purchased from Charles River Laboratories, Inc.
(Wilmington, MA, USA). The mice were housed in a pathogen-free
plastic cage with a sealed air filter, at room temperature
(26–28°C) and relative humidity of 40–60%. Mice had access to
drinking water and fed with an autoclaved diet and were under a
10/14 h light/dark cycle, respectively. Following corresponding
lentivirus infection of MG63 cells, the corresponding cell lines,
MG63/control, MG63/miR-218 and MG63/miR-218-sponge cells, was
obtained. A total of 2×106 cells/0.1 ml MG63/control,
MG63/miR-218 and MG63/miR-218-sponge cells in phosphate saline were
injected subcutaneously into the lower back of nude mice. After
eight weeks, nude mice were sacrificed by intraperitoneal injection
of pentobarbital sodium (150 to 200 mg/kg), in accordance with the
guidelines for laboratory animals established by and approved by
the Xuzhou Children's Hospital Animal Care and Use Committee. The
tumors were measured by mean as follows: Volume=(length × width
2)/2).
Immunohistochemistry
Immunohistochemistry experiments were performed
using the S-P immunohistochemistry kit (OriGene Technologies, Inc.,
Rockville, MD, USA). The 4% paraformaldehyde fixed paraffin
sections (4 micron) were deparaffinized in xylene for 5 min and
rehydrated in graded ethanol (100, 95, 85 and 75%) at room
temperature for 5 min per step. Subsequently, antigen retrieval was
conducted by heating sections in citric buffer (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) in a pressure cooker at 120°C.
Endogenous peroxidase activity was blocked in 3% hydrogen peroxide
at room temperature for 30 min. Sections were incubated with 2.5%
bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA) to reduce
non-specific binding at room temperature for 10 min.
Anti-proliferating cell nuclear antigen antibodies at 1:100
dilution in 2.5% BSA (PBS as control) were incubated at room
temperature for 1 h and then incubated with biotin-conjugated goat
anti-rabbit IgG (cat. no. ZB-2010; Zhongshan Goldenbridge Biotech
Co.) and horseradish peroxidase-conjugated streptavidin (cat. no.
AR100017; OriGene Technologies, Inc.) at room temperature for 20
min. Staining was performed by incubating sections for 2–10 min
with the chromogenic reagent 3,3′-diaminobenzidine (DAB) followed
by quenching of the reaction with water. Sections were analyzed
according to the degree of color, as longer periods of incubation
resulted to darker background color for analysis. Hematoxylin was
used as the counterstain for cell nuclei for 2 min and then
sections were dehydrated using graded ethanol (100, 95, 85 and 75%)
and xylene at room temperature. Images were captured using an
fluorescent Olympus IX-71 inverted microscope (magnification,
×400).
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA)
was used to perform the statistical analyses. Results are shown as
the mean ± standard error of the mean. Statistical significance was
analyzed using the Student's t-test (two groups) and one-way
analysis of variance (multiple groups) and Dunnett's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-218 expression is attenuated in
osteosarcoma cells
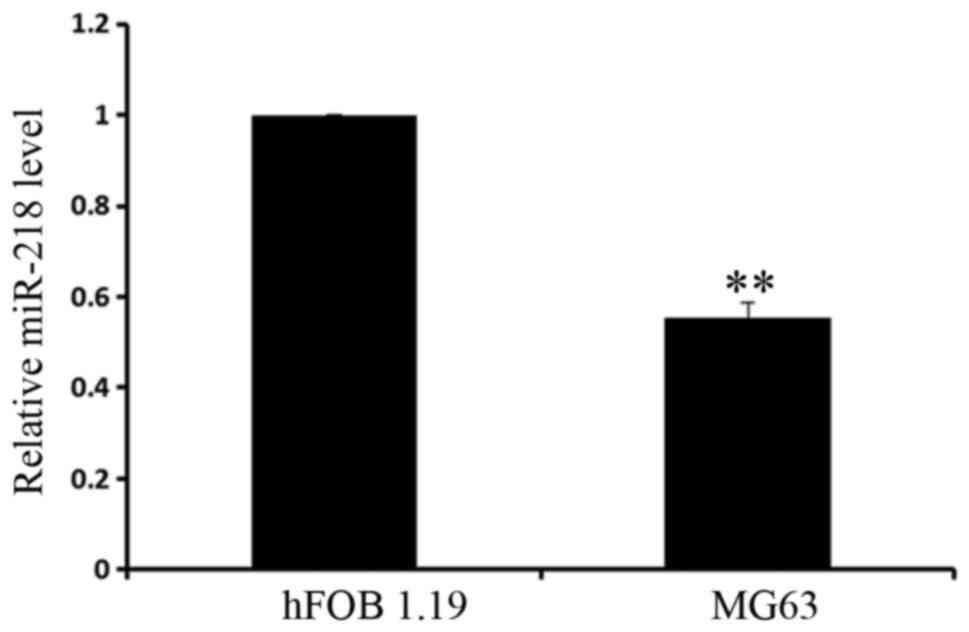
To determine the expression level of miR-218, it was
examined in the osteoblastic cell line hFOB1.19 and the
osteosarcoma cell line MG63 using RT-qPCR. It was determined that
miR-218 had significantly increased expression in osteoblasts,
compared with osteosarcoma cells (Fig.
1). The present results demonstrated that miR-218 serves an
important role in osteosarcoma tumorigenesis, which provides
preliminary evidence for future research.
miR-218 inhibits proliferation of
osteosarcoma cells
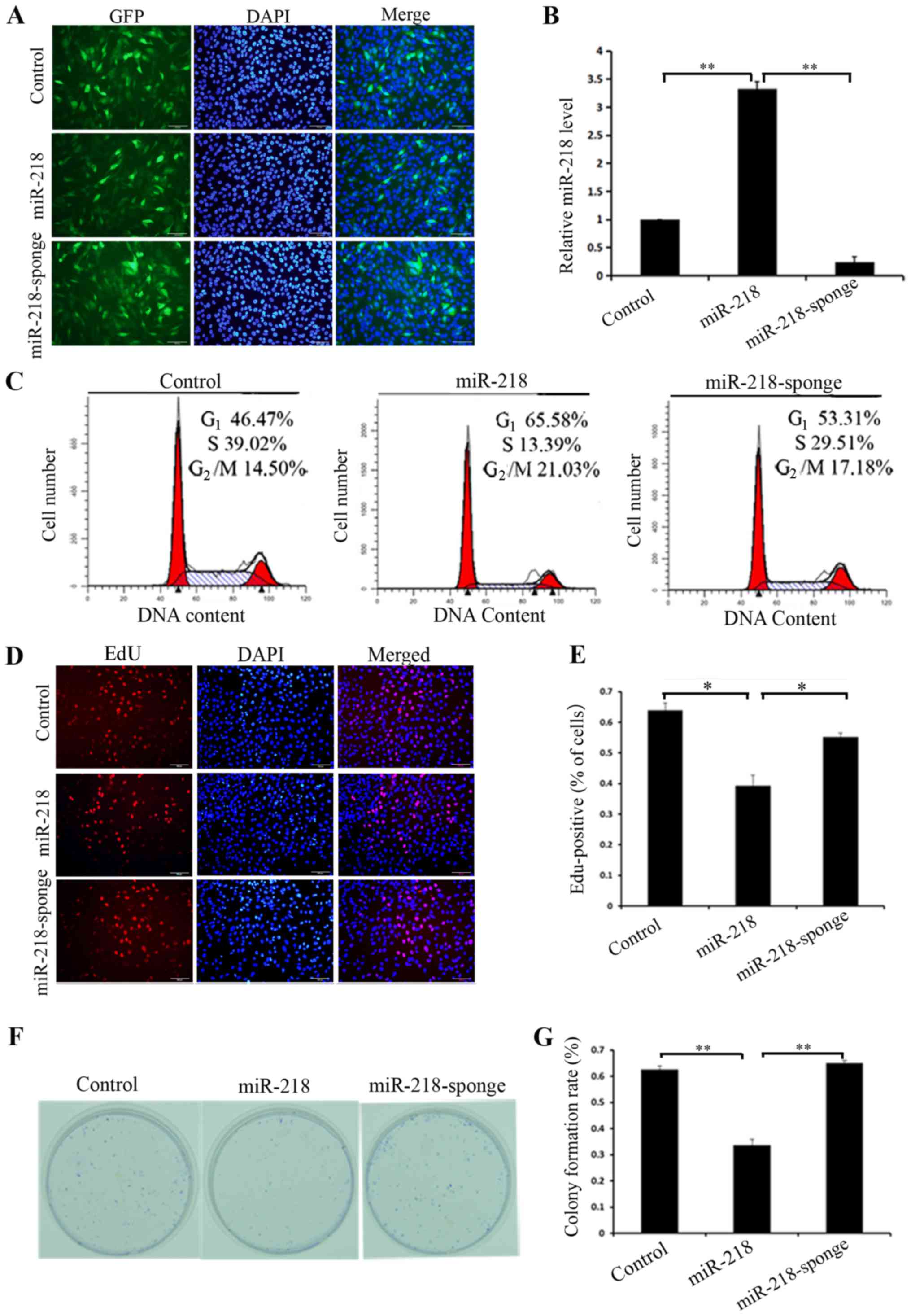
To further investigate the molecular mechanism
underlying miR-218 in osteosarcoma cells, a miR-218 overexpressing
cell line was constructed using lentiviral infection and also
miR-218 was knocked down in MG63 cells using shRNA specific for
miR-218 (Fig. 2A). miR-218 expression
was verified using RT-qPCR (Fig. 2B).
To observe the results of altering the expression of miR-218 in
MG63 cells, the cell cycle was analyzed using flow cytometry.
Control cells had the following distribution of cell cycle phases:
46.47% in G1; 39.02% in S and 14.50% in G2/M.
miR-218-overexpressing cells had the following distribution of cell
cycle phases: 65.58% in G1, 13.39% in S and 21.03% in
G2/M. miR-218-knockdown cells had the following
distribution of the cell cycle phases: 53.31% in G1,
29.51% in S and 17.18% in G2/M (Fig. 2C). Similar results were obtained using
EdU proliferation (P=0.016) and colony formation assays (P=0.001),
and demonstrated that overexpression of miR-218 inhibited
proliferation and silencing of miR-218 promoted proliferation
(Fig. 2D-G).
E2F2 is a direct target of
miR-218
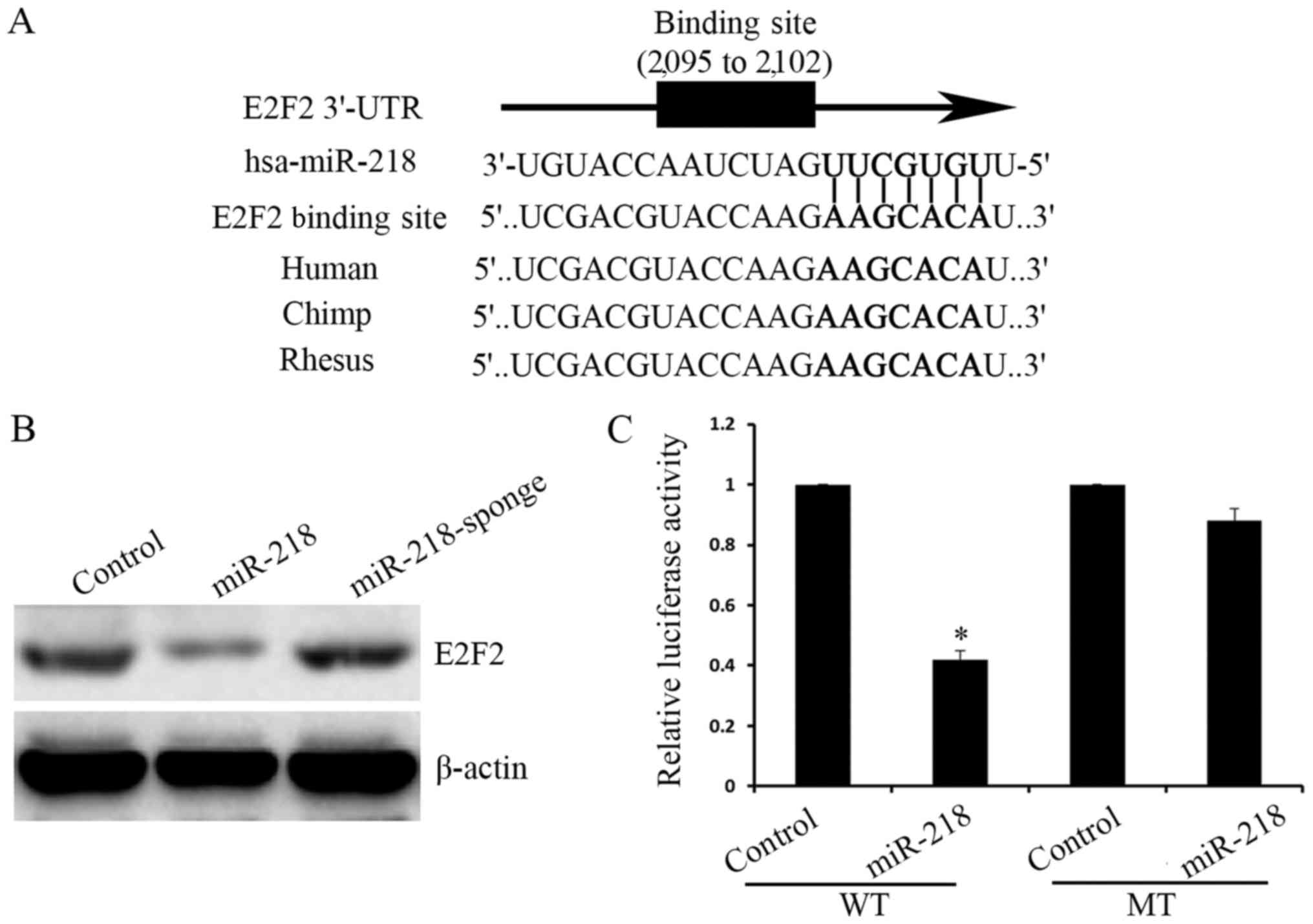
To study the mechanism underlying miR-218 inhibition
of osteosarcoma proliferation, a bioinformatics software was used
to demonstrate that miR-218 directly targeted the E2F2 3′UTR
(Fig. 3A). The expression of E2F2 was
significantly increased following overexpression of miR-218, as
indicated by western blot analysis, and expression of E2F2 was
increased following silencing of miR-218 (Fig. 3B). Furthermore, pmirGLO-WT-E2F2 3′UTR
and pmirGLO-MT-E2F2 3′UTR plasmids were constructed and the
luciferase activity in the pmirGLO-WT-E2F2 3′UTR group was
determined to be significantly decreased, compared with the control
group, while there was no difference between the control and the
pmirGLO-MT-E2F2 3′UTR groups (Fig.
3C).
miR-218 inhibits tumor formation in
vivo
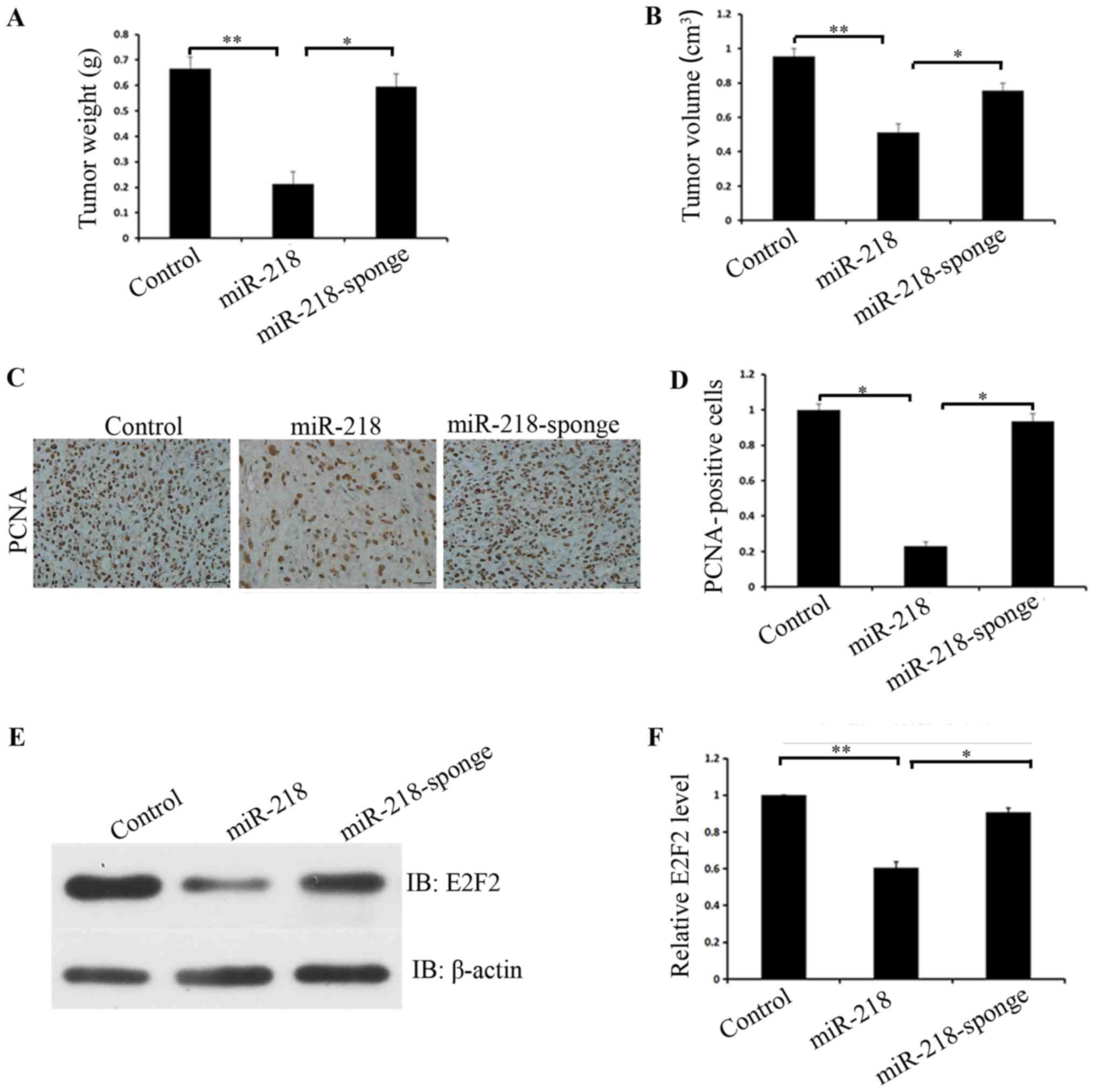
Finally, the in vivo role miR-218 serves in
osteosarcoma tumorigenicity in nude mice was investigated. After
eight weeks, nude mice were sacrificed and their tumors were
measured. Tumor sizes of mice injected with MG63 cells expressing
miR-218 were reduced (P=0.001) and had a reduced growth rate,
compared with mice injected with control or miR-218 sponge cells
(P=0.001) (Fig. 4A and B).
Immunohistochemistry also confirmed that overexpression of miR-218
inhibited proliferation and silencing of miR-218 promoted
proliferation (P=0.001) (Fig. 4C and
D). Western blot analysis of tumor tissues derived from mice
receiving different treatments also confirmed that E2F2 was
significantly downregulated in the miR-218 overexpressing group,
compared with the control group, and that E2F2 was significantly
upregulated in the miR-218 knockdown group, compared with the
miR-218 overexpressing group (P=0.02) (Fig. 4E and F).
In conclusion, the present results indicated that
miR-218 suppresses the proliferation of human osteosarcoma cells
through E2F2 and E2F2 is a direct target of miR-218.
Discussion
Previous studies demonstrated that miR-218
suppressed the migration and invasion in osteosarcoma cells
(11); however, research on the role
of miR-218 in proliferation has not been reported for osteosarcoma
cells. In the present study, it was determined that miR-218
exhibited reduced expression in osteosarcoma cells, compared with
osteoblasts. miR-218 may serve a tumor suppressor role in
osteosarcoma. To investigate the exact role of miR-218 in
osteosarcoma cells, vectors used to upregulate and downregulate the
expression of miR-218 in human osteosarcoma MG63 cells were
constructed using the three-plasmid lentivirus system. The results
demonstrated that miR-218 suppresses the proliferation of human
osteosarcoma cells through E2F2 in vitro and in vivo
and E2F2 is a direct target of miR-218.
Numerous studies have demonstrated that miR-218 can
be regarded as a tumor suppressor in a number of cancer types,
including esophageal carcinoma (17),
hepatocellular carcinoma (18), lung
carcinoma (19), pancreatic cancer
(20), glioma (21) and prostate cancer (22). It has been reported that miR-218 can
also be regarded as a tumor suppressor in osteosarcoma (11), which is consistent with the present
data. In a previous study, expression of miR-218 was significantly
reduced in osteosarcoma tissues, compared with non-tumorous tissues
(10). In agreement with that report,
it was determined that miR-218 was expressed at a reduced level in
osteosarcoma, compared with osteoblast cells. Previous studies
demonstrated that miR-218 may serve a role in tumor suppression
through targeting BMI1 (17,23), RET (18), tumor protein D52 (22), KIT (24)
and SH3 domain-containing growth factor receptor bound protein
2-like protein 1 (25). Furthermore,
it was determined that miR-218 directly targeted the E2F2 3′UTR in
osteosarcoma.
E2F2 is a member of the E2F transcription factor
family, which serves a central role in the regulation of cell cycle
progression (13,26,27). E2F2
may serve the role of a promoter of tumorigenesis. A number of
recent studies support this point, reporting that E2F2 is a pivotal
member in promoting the proliferation of osteosarcoma (26), human glioblastoma (28) and hepatocellular carcinoma cells
(29); however, in contrast to these
data, E2F2 suppresses the proliferation of renal carcinoma
(30) and colon carcinoma cells
(31). In the present study, it was
indicated that the downregulation of E2F2 suppressed the
proliferation of human osteosarcoma cells by overexpressing miR-218
and that upregulation of E2F2 promoted the proliferation of human
osteosarcoma cells by silencing miR-218. Thus, it was confirmed
that E2F2 promotes osteosarcoma tumorigenesis.
In conclusion, it was determined that miR-218 had
reduced expression in osteosarcoma cells, inhibiting the
proliferation through targeting E2F2 directly, and E2F2 can promote
osteosarcoma tumorigenesis. This result may be significant in the
early detection of osteosarcoma, which could have treatment
implications and could be a valuable protein marker in the early
detection of osteosarcoma in children.
Acknowledgements
Not applicable.
Funding
The present study was supported by Xuzhou Municipal
Bureau on Science and Technology (grant no. XM12B055).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CX, MJ, and YG proposed the concept and design of
the study. CX, YG, RH and QA performed experiments, prepared all
figures, tables and manuscript. SX applied biology software
predicts target genes for miRNAs. LW and YW performed the data and
figure analysis.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Xuzhou Children's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sakamoto A and Iwamoto Y: Current status
and perspectives regarding the treatment of osteo-sarcoma:
Chemotherapy. Rev Recent Clin Trials. 3:228–231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mori K, Rédini F, Gouin F, Cherrier B and
Heymann D: Osteosarcoma: Current status of immunotherapy and future
trends (Review). Oncol Rep. 15:693–700. 2006.PubMed/NCBI
|
|
3
|
Loeb DM: Is there a role for immunotherapy
in osteosarcoma? Cancer Treat Res. 152:447–457. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Habel N, Hamidouche Z, Girault I,
Patiño-García A, Lecanda F, Marie PJ and Fromigué O: Zinc
chelation: A metallothionein 2A's mechanism of action involved in
osteosarcoma cell death and chemotherapy resistance. Cell Death
Dis. 4:e8742013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yanokura M, Banno K, Kobayashi Y, Kisu I,
Ueki A, Ono A, Masuda K, Nomura H, Hirasawa A, Susumu N and Aoki D:
MicroRNA and endometrial cancer: Roles of small RNAs in human
tumors and clinical applications (Review). Oncol Lett. 1:935–940.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rogers K and Chen X: Biogenesis, turnover,
and mode of action of plant microRNAs. Plant Cell. 25:2383–2399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Yan W, Zhang W, Chen L, You G, Bao
Z, Wang Y, Wang H, Kang C and Jiang T: MiR-218 reverses high
invasiveness of glioblastoma cells by targeting the oncogenic
transcription factor LEF1. Oncol Rep. 28:1013–1021. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang HT, Liu AG, Luo DS, Zhou ZN, Lin HG,
Chen RZ, He JS and Chen K: miR-218 expression in osteosarcoma
tissues and its effect on cell growth in osteosarcoma cells. Asian
Pac J Trop Med. 7:1000–1004. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin J, Cai L, Liu ZM and Zhou XS:
miRNA-218 inhibits osteosarcoma cell migration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lukas J, Petersen BO, Holm K, Bartek J and
Helin K: Deregulated expression of E2F family members induces
S-phase entry and overcomes p16INK4A-mediated growth suppression.
Mol Cell Biol. 16:1047–1057. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Infante A, Laresgoiti U, Fernández-Rueda
J, Fullaondo A, Galán J, Díaz-Uriarte R, Malumbres M, Field SJ and
Zubiaga AM: E2F2 represses cell cycle regulators to maintain
quiescence. Cell Cycle. 7:3915–3927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Muller H, Bracken AP, Vernell R, Moroni
MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD and
Helin K: E2Fs regulate the expression of genes involved in
differentiation, development, proliferation, and apoptosis. Genes
Dev. 15:267–285. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang T, Chen T, Niu H, Li C, Xu C, Li Y,
Huang R, Zhao J and Wu S: MicroRNA-218 inhibits the proliferation
and metastasis of esophageal squamous cell carcinoma cells by
targeting BMI1. Int J Mol Med. 36:93–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sui C, Xu F, Shen W, Geng L, Xie F, Dai B,
Lu J, Zhang M and Yang J: Overexpression of miR-218 inhibits
hepatocellular carcinoma cell growth through RET. Tumour Biol.
36:1511–1518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song L, Li D, Zhao Y, Gu Y, Zhao D, Li X,
Bai X, Sun Y, Zhang X, Sun H, et al: miR-218 suppressed the growth
of lung carcinoma by reducing MEF2D expression. Tumour Biol.
37:2891–2900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Xu Y, Long J, Guo K, Ge C and Du R:
microRNA-218 suppresses the proliferation, invasion and promotes
apoptosis of pancreatic cancer cells by targeting HMGB1. Chin J
Cancer Res. 27:247–257. 2015.PubMed/NCBI
|
|
21
|
Jun GJ, Zhong GG and Ming ZS: miR-218
inhibits the proliferation of glioma U87 cells through the
inactivation of the CDK6/cyclin D1/p21 pathway. Oncol Lett.
9:2743–2749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han G, Fan M and Zhang X: microRNA-218
inhibits prostate cancer cell growth and promotes apoptosis by
repressing TPD52 expression. Biochem Biophys Res Commun.
456:804–809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng Y, Yang X, Deng X, Zhang X, Li P,
Tao J and Lu Q: MicroRNA-218 inhibits bladder cancer cell
proliferation, migration, and invasion by targeting BMI-1. Tumour
Biol. 36:8015–8023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fan R, Zhong J, Zheng S, Wang Z, Xu Y, Li
S, Zhou J and Yuan F: MicroRNA-218 inhibits gastrointestinal
stromal tumor cell and invasion by targeting KIT. Tumour Biol.
35:4209–4217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi J, Yang L, Wang T, Zhang J, Guo X, Huo
X and Niu H: miR-218 is downregulated and directly targets SH3GL1
in childhood medulloblastoma. Mol Med Rep. 8:1111–1117. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iwasaki T, Tanaka K, Kawano M, Itonaga I
and Tsumura H: Tumor-suppressive microRNA-let-7a inhibits cell
proliferation via targeting of E2F2 in osteosarcoma cells. Int J
Oncol. 46:1543–1550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pusapati RV, Weaks RL, Rounbehler RJ,
McArthur MJ and Johnson DG: E2F2 suppresses Myc-induced
proliferation and tumorigenesis. Mol Carcinog. 49:152–156.
2010.PubMed/NCBI
|
|
28
|
Zhang Y, Han D, Wei W, Cao W, Zhang R,
Dong Q, Zhang J, Wang Y and Liu N: MiR-218 inhibited growth and
metabolism of human glioblastoma cells by directly targeting E2F2.
Cell Mol Neurobiol. 35:1165–1173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong Y, Zou J, Su S, Huang H, Deng Y, Wang
B and Li W: MicroRNA-218 and microRNA-520a inhibit cell
proliferation by downregulating E2F2 in hepatocellular carcinoma.
Mol Med Rep. 12:1016–1022. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao Y, Ma X, Yao Y, Li H, Fan Y, Zhang Y,
Zhao C, Wang L, Ma M, Lei Z and Zhang X: miR-155 regulates the
proliferation and invasion of clear cell renal cell carcinoma cells
by targeting E2F2. Oncotarget. 7:20324–20337. 2016.PubMed/NCBI
|
|
31
|
Li T, Luo W, Liu K, Lv X and Xi T: miR-31
promotes proliferation of colon cancer cells by targeting E2F2.
Biotechnol Lett. 37:523–532. 2015. View Article : Google Scholar : PubMed/NCBI
|