In traditional cell lines, established cells are
cultured in artificial conditions that may not accurately simulate
complex biological conditions, including tumor progression in low
oxygen conditions (4), excessive
hypoxia-induced transcription factor activation (5), immune escape mechanism deficiency
(6) and angiogenesis (7). In addition, autocrine, paracrine and
endocrine mechanisms also serve key functions in tumor development,
particularly in breast, ovarian and prostate cancer (8–10).
Therefore, in vitro-based culture experiments may be
inappropriate for individualized medicine.
Patient-derived xenograft (PDX) mouse models were
initially proposed 40 years ago. With the development of host
animals, numerous academic organizations have renewed their
interest in PDX models. PDXs involve the direct transfer of fresh
tumor samples into immunodeficient mice following surgical
resection or other medical operations. Gene expression in tumors
may be maintained by serial passages of tumors from mouse to mouse
(11). PDXs aid research into tumor
biology and pharmacology without manual manipulation of cell
cultures in vitro. Multiple PDX studies have reported that,
compared with their corresponding parental tumors, PDXs retain
sufficient fidelity regarding histology, the transcriptome and
genome (12–16). The present study reviewed current
collections of PDXs and evaluated the key issues facing their
future application.
Numerous academic institutions have begun renewing
their interest in genetically engineered mouse models (GEMMs), cell
line-derived xenografts, and PDX models.
GEMMs are classified into two categories: Transgenic
and targeted. In transgenic GEMMs, exogenous oncogenes are
expressed through pronuclear injection of embryonic stem cells.
Targeted GEMMs involve homologous recombination in mouse embryonic
stem cells, in which targeting vectors modify the homologous arms
of a genomic locus to the accuracy of a single base. GEMMs have
been used to elucidate multiple aspects of cancer development.
Khaled et al (17) summarized
data from multiple studies. For example, GEMMs are particularly
suitable for deleting or overexpressing targeted genes in a
tissue-specific manner, are easy to produce and use endogenous
regulatory elements. However, establishing the model takes ~1 year.
Another major disadvantage of GEMMs is the low success rate between
two targeted sites. In addition, GEMMs may not be able to mimic the
individualized therapy associated with a tumor-specific gene.
Therefore, GEMMs may not represent the optimal preclinical trial
model.
Cell lines are transplanted into immunodeficient
mice to establish tumor models by numerous means, including
subcutaneous implantation and orthotopic, venial or peritoneal
injection. In 2007, Hajitou et al (18) established the first soft tissue
sarcoma cell line-derived tumor in the hind limb of rats.
Subsequently, the development of cell line-derived xenograft models
has decreased the influence of irrelevant cells when studying a
single factor (16), improved the use
of experimental cellular operations in researching tumor-associated
signaling pathways and molecular mechanisms (19,20),
established acute myeloid leukemia animal models to mimic the
progression of liquid tumors (21),
and aided research into metastatic mechanisms via intravenous or
intraperitoneal injection (22). In
addition, cell line-derived xenograft mouse models are easily
established, and generating tumors takes only 2–8 weeks (23).
However, since generating cancer cell lines may
irreversibly alter the biological properties of the derived cells,
cell line-derived xenografts have limited predictive value for
cancer therapy. The major disadvantages of this model are as
follows: Following the dissociation of tumor tissues into a single
cell suspension in vitro, selection pressure tends to result
in a decrease in the heterogeneous characteristics of the tumors;
not all cell lines are suitable for patients with certain types of
cancer (17) and cell lines may not
accurately reflect the complexity of tumor heterogeneity (24), which impacts patient-specific
responses to clinical therapy. Therefore, cell line-derived
xenograft models may not satisfy the requirements of individualized
medicine.
Compared with GEMMs and cell line-derived xenograft
models, PDX models have more predictive value for clinical outcomes
(25,26). Multiple studies have reported the high
fidelity of PDXs regarding histology, the transcriptome and genome
(12–16). Previous studies have obtained more
detailed data on tumor cell population dynamics using deep-genome
and single-cell sequencing techniques in PDXs for breast cancer
(27,28). Therefore, the present study suggests
that PDXs may be optimal models for studying tumor heterogeneity
and currently represent the most powerful tool for assessing
cancer-associated mechanisms. However, PDXs remain to be
popularized in clinical application. Thus, the present study
reviewed the progression of PDXs, particularly the challenges faced
by these models, and future directions for individualized medicine
(Table I).
PDXs may have originally failed to enter mainstream
cancer research due to the limitations of using host animals that
are not sufficiently immunodeficient and initiate xenograft
rejection. However, using animal models is now common, since the
number of immunodeficient host animal models has increased and the
cost of immunodeficient mice has decreased (29). At present, nude, nonobese diabetic
(NOD)/severe combined immunodeficiency (SCID) and NOD SCID γ (NSG)
mice represent the three most commonly used types of
immunodeficient mice. Nude mice are athymic, which results in a
congenital deficiency of T and normal B lymphocytes and enhanced
activity of natural killer (NK) cells (30). NOD/SCID mice are generated by crossing
C.B.-17-SCID mice with NOD mice. SCID and NOD/SCID mice exhibit a
congenital deficiency of T and B lymphocytes; the latter also
exhibits decreased NK cell activity, which aids the use of the
model since residual NK cells serve a key function in rejecting
human tissues. NSG mice exhibit a deficiency of the interleukin 2
receptor γ-chain via the genetic engineering of NOD/SCID mice. NSG
mice are the most severely immunodeficient since they exhibit T, B
and NK cell deficiency. Therefore, the engrafting success rate in
NSG mice is typically increased compared with that in nude or
NOD/SCID mice (31). Therefore, NSG
mice may be the most suitable of the three to generate tumors
(32). NSG mice have been
successfully utilized in multiple types of cancer and the present
study has summarized the current state of PDXs (Table I).
Circulating tumor cells (CTCs) are released from
primary tumors into peripheral blood vessels. Conditions
permitting, CTCs may infiltrate distant tissues and thereby induce
metastasis by providing ‘seeds’ to distant organs, in accordance
with the ‘seed and soil’ theory (33). CTCs represent a readily accessible
method for liquid biopsy. Numerous researchers over the past
decades have focused on identifying and improving technically
challenging methods of isolating CTCs (34). Currently, the CellSearch System is the
only Food and Drug Administration-approved technique for CTC
enumeration (35). However, the
CellSearch System depends on epithelial cell adhesion molecules
(EpCAMs) and therefore may only identify a small number of CTCs,
potentially resulting in false negative or positive readings
(36). Therefore, researchers have
previously used the brain metastasis-selected markers (BMSMs)
erb-b2 receptor tyrosine kinase 2 (ERBB2)+/epidermal
growth factor receptor
(EGFR)+/heparanase+/notch 1+ to
identify EpCAM− CTCs (37). The infiltration and metastasis of
these CTCs were analyzed using BMSMs and these cells were then
demonstrated to be invasive and capable of generating brain or lung
metastasis in nude mice. Researching EpCAM− CTCs may
also serve to improve the understanding of metastasis. Enrichment
steps are crucial to increase the isolation success rate. Although
enriching CTCs may reach 98% of total CTCs by using the CTC-Chip,
this chip has not become available on the market yet (38). Previously, the drug sensitivity of
cultured CTCs was tested among multiple CTC lines (39). Furthermore, another study demonstrated
that testing the drug sensitivity of CTC lines for clinical regimes
is in accordance with the clinical treatment response (40). Drug sensitivity testing of CTCs may
predict the donor patient's response and direct appropriate therapy
for individualized medicine. In addition, CTC-derived xenografts
have been used in the laboratory for studying certain types of
cancer. One institution reported that the PDXs for small cell lung
cancer (SCLC) may reflect the responses of patients to clinical
regimes (34). Therefore, CTC-derived
xenograft models (CDXs) may be used to detect drug resistance
mechanisms.
PDXs are maintained by passaging tumor tissues
directly from mouse to mouse. Heterotopic or orthotopic PDXs
involve implanting tissues into the subcutaneous flank or the
organs of mice. PDXs are considered to be superior to traditional
cell line xenografts since the former may be more similar to
parental tumors compared with the latter, particularly in terms of
tumor, intratumor and intrametastasis heterogeneity (26). Traditional tumor models exhibit poor
predictive values due to the associated heterogeneity. The
development of PDXs satisfies the requirements of an effective
preclinical tool, and PDXs are a predictive model of carcinogenesis
physiology and clinical therapy. As genomic research develops,
further subtypes of cancer are predicted. For example, five
molecular subtypes of breast cancer have been characterized,
studied and categorized using the Prosigna Breast Cancer Prognostic
Gene Signature assay test: Luminal A, luminal B, ERBB2+, basal-like
and normal-like (41). Subtypes are
further divided into more detailed types for each individual in
clinical practice (42,43).
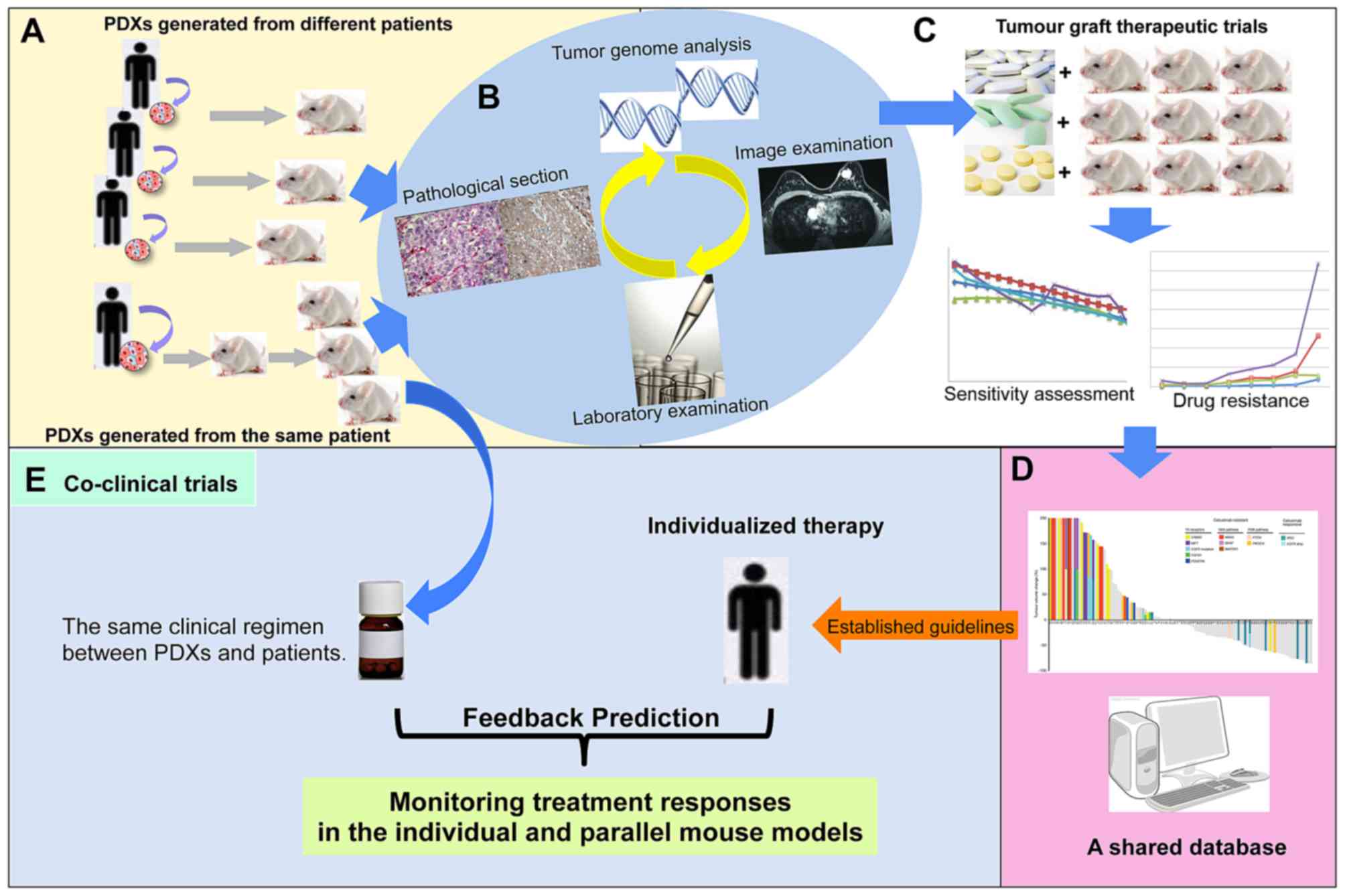
In addition, studies on targeted drug treatments
confirm the accurate predictive value, fidelity and stability of
PDXs and PDX-associated clinical prognosis (Fig. 1A, B and C).
The use of PDXs is advantageous in drug-screening
and resistance mechanism research. PDXs possess an effective
predictive value in targeted drug-screening for clinical
treatments. The development of optimal regimes, based on
drug-screening in PDXs, may improve the survival rate of patients
with cancer. Furthermore, gene mutations cause tumorigenesis,
particularly those in p53 and phosphatase and tensin homolog
(24,46). Although gene mutations may be
identified using whole-genome sequencing, identifying the
significant mutations with which specific diseases are associated
remains a challenge. Therefore, Berg et al (47) aimed to construct a comprehensive
dataset, including driver mutations, tumorigenicity variants and
clinical responses, but this was discovered to be time-consuming.
In addition, drug resistance mechanisms may consist of primary or
acquired resistance (48). PDXs may
represent intratumor and intrametastasis heterogeneity, and more
accurately predict resistance mechanisms to clinical treatments.
Therefore, PDXs may potentially represent a platform for evaluating
personalized resistance mechanisms. Personalized medicinal strategy
may become a future direction for personalized treatment and
translational medicine.
The poor predictive value of the preclinical models
used to select novel drugs is partly responsible for the low
success rate of novel agents in clinical application. PDXs are more
predictive of clinical outcomes and possess a vast development
foreground in the preclinical screening of novel anticancer drugs.
Chiron et al (49) compared
the anticancer effect of aflibercept with that of bevacizumab by
using PDXs in multiple genetic backgrounds, demonstrating that
aflibercept had increased anticancer functions compared with
bevacizumab. Monsma et al (50) revealed that vemurafenib was effective
in PDXs of melanoma with B-Raf proto-oncogene, serine/threonine
kinase (BRAF)V600E or BRAFV600V. Furthermore,
the combination of mitogen-activated protein kinase inhibitors and
vemurafenib improved the effectiveness of anticancer therapies.
Consequently, screening of the susceptibility of drugs may be
effectively integrated into clinical translational medicine
(Fig. 1C and D).
Coclinical trials are characterized by parallel
studies between mouse models and patients, which may help determine
treatment strategies for patients and to identify underlying
cancer-associated mechanisms with the aid of PDXs (51,52). As
cancer progresses, the drug becomes less effective and novel
resistance mechanisms appear. When and how such mechanisms develop
is unpredictable during clinical treatment. Drug resistance may be
observed earlier with the aid of PDXs, which may assist in
determining subsequent treatment regimes. GEMMs have been used in
coclinical trials and exhibited positive results in parallel
clinical trails (53), including
those involving leukemia, melanoma, prostate cancer and non (N)SCLC
(53–56). Furthermore, coclinical trials may also
verify relevant hypotheses in a clinical setting and thereby affect
the design of future clinical studies (57). However, this may be associated with
increased costs (51). PDXs have not
been used in large scale coclinical trials (38). Bertotti et al (38) reported that 8 patients with metastatic
colorectal cancer were successfully treated using targeted
PDX-based therapies combining anti-ERBB2 with anti-EGFR to predict
resistance to anti-EGFR targeted therapy. Coclinical trials monitor
the responses of individuals and parallel mouse models
simultaneously and provide an in vivo model to research
suspicious resistance mechanisms and test combination strategies
for overcoming novel spontaneous resistance mechanisms (Fig. 1E).
Though the application of PDXs in tumor research is
associated with the aforementioned advantages, certain problems
with PDXs remain. PDXs depend on murine immunodeficiency models,
which lack functional elements of immune systems. Therefore, due to
the lack of stromal cells and degradation of tissue architecture,
the tumor microenvironment was virtually non-existent (58). In addition, murine fibroblasts differ
from those in humans (45). Morton
et al (59) successfully
isolated CD34+ cells from the blood of patients and
subsequently intravenously injected them into a mouse to
reconstruct a functional immune system in murine models that would
mimic that of patients. PDXs with patient-matched immune systems
may be valuable for study, particularly when screening immune
system-mediating agents.
Furthermore, less aggressive tumors exhibit
decreased implantation rates and more aggressive tumors exhibit
increased formation rates. For example, estrogen receptor-negative
types of breast cancer exhibit increased the rate of successful
tumor establishment compared with hormone-positive types of breast
cancer (60). Patients with initial
tumors may experience improved treatment outcomes compared with
those with more advanced tumors. However, constructing a PDX model
with initial tumor remains a challenge and tumor formation rate
remains low. Therefore, improvement of the implantation rate is
urgently required.
Developing PDXs delays treatment schedule and
increases costs. Typically, at least 3 months are required to
develop PDXs that may be used for preclinical study. This is a
major limiting factor for individualized medicine. Discovering the
most suitable conditions for certain subtypes of cancer may
decrease the duration of PDX generation. Another critical factor is
cost, comprising cloned animal and whole-genome analysis cost and
experimental preclinical expenses. Not all patients may be able to
afford these costs. Therefore, PDXs remain technically challenging,
time-consuming and costly.
The Human Genome Project was launched in 1990 and
has improved the understanding of the genome. However, gene
function remains to be fully understood, including regulatory
mechanisms and gene interactions. Under the complex background of
individual differences, achieving accurate clinical use of the
results of genomic analysis is challenging. Therefore, predictive
models are crucial. PDXs may provide evidence to personalize
clinical treatments for patients.
As aforementioned, the process of generating PDXs
differs among researchers. Low engrafting rates are a major factor
that restricts the personalized medicine development of PDXs. The
aim of the next phase of PDX development is to identify the most
appropriate conditions and methods to maximize tumor formation
rates. There is an increasing trend in industry and academia to
help develop PDXs. Hidalgo et al (61) have proposed the concept of the
‘EurOPDX Consortium’, which aims to establish a network of
clinically relevant models of human tumors, particularly PDXs, and
share the characteristics of currently available models.
Successfully establishing this shared database globally may help to
acquire analogical PDXs quickly by comparing data pertaining to
certain patients, thereby altering traditional concepts of clinical
treatment. Individualized therapy may be converted into
programmatic therapy. By comparing clinical samples with samples in
the database, the optimal treatment plan may be identified from the
shared database in cases where genomic characteristics are similar
or consistent between patients. Thus, treatment schedule delays and
high costs may cease to be limiting factors in the clinical
application of PDXs. The present study suggests that PDXs may be
commonly used in treating patients with cancer in the future.
The authors would like to thank Professor Hailing
Cheng and Chen Song (Department of Oncology and Cancer Institute,
The Second Hospital of Dalian Medical University, Dalian China) for
their guidance.
The present study was supported by grants from the
Provincial Natural Science Foundation of Liaoning (grant no.
2015020316) and National Natural Science Foundation of China (grant
no.81650018).
Not applicable.
PL, ML and FL designed the review and revised the
manuscript. XL and CX were responsible for manuscript drafting. All
authors have reviewed the final version of the manuscript and
approved it for publication.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
DiMasi JA, Reichert JM, Feldman L and
Malins A: Clinical approval success rates for investigational
cancer drugs. Clin Pharmacol Ther. 94:329–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kola I and Landis J: Can the
pharmaceutical industry reduce attrition rates? Nat Rev Drug
Discov. 3:711–715. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rosfjord E, Lucas J, Li G and Gerber HP:
Advances in patient-derived tumor xenografts: From target
identification to predicting clinical response rates in oncology.
Biochem Pharmacol. 91:135–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chawla SP, Cranmer LD, Van Tine BA, Reed
DR, Okuno SH, Butrynski JE, Adkins DR, Hendifar AE, Kroll S and
Ganjoo KN: Van tine Phase II study of the safety and antitumor
activity of the hypoxia-activated prodrug TH-302 in combination
with doxorubicin in patients with advanced soft tissue sarcoma. J
Clin Oncol. 32:3299–3306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schödel J, Grampp S, Maher ER, Moch H,
Ratcliffe PJ, Russo P and Mole DR: Hypoxia-inducible transcription
factors, and renal cancer. Eur Urol. 69:646–657. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Spranger S, Bao R and Gajewski TF:
Melanoma-intrinsic β-catenin signalling prevents anti-tumour
immunity. Nature. 523:231–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rivera LB and Bergers G: Cancer. Tumor
angiogenesis from foe to friend. Science. 349:694–695. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mungenast F and Thalhammer T: Estrogen
biosynthesis and action in ovarian cancer. Front Endocrinol
(Lausanne). 5:1922014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nwabo Kamdje AH, Seke Etet PF, Vecchio L,
Muller JM, Krampera M and Lukong KE: Signaling pathways in breast
cancer: Therapeutic targeting of the microenvironment. Cell Signal.
26:2843–2856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Attard G, Parker C, Eeles RA, Schröder F,
Tomlins SA, Tannock I, Drake CG and de Bono JS: Prostate cancer.
Lancet. 387:70–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang M, Shen A, Ding J and Geng M:
Molecularly targeted cancer therapy: Some lessons from the past
decade. Trends Pharmacol Sci. 35:41–50. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharpless NE and Depinho RA: The mighty
mouse: Genetically engineered mouse models in cancer drug
development. Nat Rev Drug Discov. 5:741–754. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang R, Jin S, Rao W, Song F, Yin Q, Wang
Y, Wang L, Xi Y, Zhang X, Wang M and Ge H: OVA12, a novel tumor
antigen, promotes cancer cell growth and inhibits
5-fluorouracil-induced apoptosis. Cancer Lett. 357:141–151. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park H, Kim Y, Sul JW, Jeong IG, Yi HJ,
Ahn JB, Kang JS, Yun J, Hwang JJ and Kim CS: Synergistic anticancer
efficacy of MEK inhibition and dual PI3K/mTOR inhibition in
castration-resistant prostatecancer. Prostate. 75:1747–1759. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Girotti MR, Lopes F, Preece N,
Niculescu-Duvaz D, Zambon A, Davies L, Whittaker S, Saturno G,
Viros A, Pedersen M, et al: Paradox-breaking RAF inhibitors that
also target SRC are effective in drug-resistant BRAF mutant
melanoma. Cancer Cell. 27:85–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wykosky J, Hu J, Gomez GG, Taylor T, Villa
GR, Pizzo D, VandenBerg SR, Thorne AH, Chen CC, Mischel PS, et al:
A urokinase receptor-Bim signaling axis emerges during EGFR
inhibitor resistance in mutant EGFR glioblastoma. Cancer Res.
75:394–404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khaled WT and Liu P: Cancer mouse models:
Past, present and future. Semin Cell Dev Biol. 27:54–60. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hajitou A, Lev DC, Hannay JA, Korchin B,
Staquicini FI, Soghomonyan S, Alauddin MM, Benjamin RS, Pollock RE,
Gelovani JG, et al: A preclinical model for predicting drug
response in soft-tissue sarcoma with targeted AAVP molecular
imaging. Proc Natl Acad Sci USA. 105:4471–4476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu B, Nandhu MS, Sim H, Agudelo-Garcia PA,
Saldivar JC, Dolan CE, Mora ME, Nuovo GJ, Cole SE and Viapiano MS:
Fibulin-3 promotes glioma growth and resistance through a novel
paracrine regulation of Notch signaling. Cancer Res. 72:3873–3885.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kung PP, Martinez R, Zhu Z, Zager M,
Blasina A, Rymer I, Hallin J, Xu M, Carroll C, Chionis J, et al:
Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a
critical role in triple-negative breast cancer. Mol Cancer Ther.
13:2104–2115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saland E, Boutzen H, Castellano R, Pouyet
L, Griessinger E, Larrue C, de Toni F, Scotland S, David M,
Danet-Desnoyers G, et al: A robust and rapid xenograft model to
assess efficacy of chemotherapeutic agents for human acute myeloid
leukemia. Blood Cancer J. 5:e2972015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Santel A, Aleku M, Röder N, Möpert K,
Durieux B, Janke O, Keil O, Endruschat J, Dames S, Lange C, et al:
Atu027 prevents pulmonary metastasis in experimental and
spontaneous mouse metastasis models. Clin Cancer Res. 16:5469–5480.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee SH, Hong JH, Park HK, Park JS, Kim BK,
Lee JY, Jeong JY, Yoon GS, Inoue M, Choi GS and Lee IK: Colorectal
cancer-derived tumor spheroids retain the characteristics of
original tumors. Cancer Lett. 367:34–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X and Lewis MT: Establishment of
patient-derived xenograft (PDX) models of human breast cancer. Curr
Protoc Mouse Biol. 3:21–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chou J, Fitzgibbon MP, Mortales CL,
Towlerton AM, Upton MP, Yeung RS, McIntosh MW and Warren EH:
Phenotypic and transcriptional fidelity of patient-derived colon
cancer xenografts in immune-deficient mice. PLoS One. 8:e798742013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cho YB, Hong HK, Choi YL, Oh E, Joo KM,
Jin J, Nam DH, Ko YH and Lee WY: Colorectal cancer patient-derived
xenografted tumors maintain characteristic features of the original
tumors. J Surg Res. 187:502–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eirew P, Steif A, Khattra J, Ha G, Yap D,
Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al: Dynamics of
genomic clones in breast cancer patient xenografts at single-cell
resolution. Nature. 518:422–426. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Emes RD, Goodstadt L, Winter EE and
Ponting CP: Comparison of the genomes ofhuman and mouse lays the
foundation of genome zoology. Hum Mol Genet. 12:701–709. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pelleitier M and Montplaisir S: The nude
mouse: A model of deficient T-cell function. Methods Achiev Exp
Pathol. 7:149–166. 1975.PubMed/NCBI
|
|
31
|
Lapidot T, Fajerman Y and Kollet O:
Immune-deficient SCID and NOD/SCID mice models as functional assays
for studying normal and malignant human hematopoiesis. J Mol Med.
75:664–673. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shultz LD, Lyons BL, Burzenski LM, Gott B,
Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al:
Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R
gamma null mice engrafted with mobilized human hemopoietic stem
cells. J Immunol. 174:6477–6489. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alunni-Fabbroni M and Sandri MT:
Circulating tumour cells in clinical practice: Methods of detection
and possible characterization. Methods. 50:289–297. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin
W, Kumar D, Goodman JC, Groves MD and Marchetti D: The
Identification and characterization of breast cancer CTCs competent
for brain metastasis. Sci Transl Med. 5:180ra482013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu M, Bardia A, Aceto N, Bersani F, Madden
MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al:
Cancer therapy. Ex vivo culture of circulating breast tumor cells
for individualized testing of drug susceptibility. Science.
345:216–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bertotti A, Migliardi G, Galimi F, Sassi
F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti
C, et al: A molecularly annotated platform of patient-derived
xenografts (‘xenopatients’) identifies HER2 as an effective
therapeutic target in cetuximab-resistant colorectal cancer. Cancer
Discov. 1:508–523. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hughes AD, Marshall JR, Keller E, Powderly
JD, Greene BT and King MR: Differential drug responses of
circulating tumor cells within patient blood. Cancer Lett.
352:28–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hodgkinson CL, Morrow CJ, Li Y, Metcalf
RL, Rothwell DG, Trapani F, Polanski R, Burt DJ, Simpson KL, Morris
K, et al: Tumorigenicity and genetic profiling of circulating
tumorcells in small-cell lung cancer. Nat Med. 20:897–903. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peters LJ: Radiation therapy tolerance
limits. For one or for all?-Janeway Lecture. Cancer. 77:2379–2385.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van Dijk LK, Boerman OC, Kaanders JH and
Bussink J: PET Imaging in head and neck cancer patients to monitor
treatment response: A future role for EGFR-targeted imaging. Clin
Cancer Res. 21:3602–3609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stebbing J, Paz K, Schwartz GK, Wexler LH,
Maki R, Pollock RE, Morris R, Cohen R, Shankar A, Blackman G, et
al: Patient-derived xenografts for individualized care in advanced
sarcoma. Cancer. 120:2006–2015. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garralda E, Paz K, López-Casas PP, Jones
S, Katz A, Kann LM, López-Rios F, Sarno F, Al-Shahrour F, Vasquez
D, et al: Integrated next-generation sequencing and avatar mouse
models for personalized cancer treatment. Clin Cancer Res.
20:2476–2484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jones N, Bonnet F, Sfar S, Lafitte M,
Lafon D, Sierankowski G, Brouste V, Banneau G, Tunon de Lara C,
Debled M, et al: Comprehensive analysis of PTEN status in breast
carcinomas. Int J Cancer. 133:323–334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Berg JS, Amendola LM, Eng C, Van Allen E,
Gray SW, Wagle N, Rehm HL, DeChene ET, Dulik MC, Hisama FM, et al:
Processes and preliminary outputs for identification of actionable
genes as incidental findings in genomic sequence data in the
Clinical Sequencing Exploratory Research Consortium. Genet Med.
15:860–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Garraway LA and Jänne PA: Circumventing
cancer drug resistance in the era of personalized medicine. Cancer
Discov. 2:214–226. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chiron M, Bagley RG, Pollard J, Mankoo PK,
Henry C, Vincent L, Geslin C, Baltes N and Bergstrom DA:
Differential antitumor activity of aflibercept and bevacizumab in
patient-derived xenograft models of colorectal cancer. Mol Cancer
Ther. 13:1636–1644. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Monsma DJ, Cherba DM, Eugster EE, Dylewski
DL, Davidson PT, Peterson CA, Borgman AS, Winn ME, Dykema KJ, Webb
CP, et al: Melanoma patient derived xenografts acquire distinct
Vemurafenib resistance mechanisms. Am J Cancer Res. 5:1507–1518.
2015.PubMed/NCBI
|
|
51
|
Clohessy JG and Pandolfi PP: Mouse
hospital and co-clinical trial project-from bench to bedside. Nat
Rev Clin Oncol. 12:491–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lunardi A and Pandolfi PP: A co-clinical
platform to accelerate cancer treatment optimization. Trends Mol
Med. 21:1–5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen Z, Akbay E, Mikse O, Tupper T, Cheng
K, Wang Y, Tan X, Altabef A, Woo SA, Chen L, et al: Co-clinical
trials demonstrate superiority of crizotinib to chemotherapy in
ALK-rearranged non-small cell lungcancer and predict strategies to
overcome resistance. Clin Cancer Res. 20:1204–1211. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kwong LN, Boland GM, Frederick DT, Helms
TL, Akid AT, Miller JP, Jiang S, Cooper ZA, Song X, Seth S, et al:
Co-clinical assessment identifies patterns of BRAF inhibitor
resistance in melanoma. J Clin Invest. 125:1459–1470. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Malaney P, Nicosia SV and Davé V: One
mouse, one patient paradigm: New avatars of personalized cancer
therapy. Cancer Lett. 344:1–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nardella C, Lunardi A, Patnaik A, Cantley
LC and Pandolfi PP: The APL paradigm and the ‘co-clinical trial’
project. Cancer Discov. 1:108–116. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen Z, Cheng K, Walton Z, Wang Y, Ebi H,
Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, et al: A murine lung
cancer co-clinical trial identifies genetic modifiers of
therapeutic response. Nature. 483:613–617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cassidy JW, Caldas C and Bruna A:
Maintaining tumor heterogeneity in patient-derived tumor
xenografts. Cancer Res. 75:2963–2968. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Morton JJ, Bird G, Keysar SB, Astling DP,
Lyons TR, Anderson RT, Glogowska MJ, Estes P, Eagles JR, Le PN, et
al: XactMice: Humanizing mouse bone marrow enables microenvironment
reconstitution in a patient-derived xenograft model of head and
neck cancer. Oncogene. 35:290–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Julien S, Merino-Trigo A, Lacroix L,
Pocard M, Goéré D, Mariani P, Landron S, Bigot L, Nemati F,
Dartigues P, et al: Characterization of a large panel of
patient-derived tumor xenografts representing the clinical
heterogeneity of human colorectal cancer. Clin Cancer Res.
18:5314–5328. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hidalgo M, Amant F, Biankin AV, Budinská
E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo
GM, et al: Patient-derived xenograft models: An emerging platform
for translational cancer research. Cancer Discov. 4:998–1013. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cottu P, Marangoni E, Assayag F, de
Cremoux P, Vincent-Salomon A, Guyader Ch, de Plater L, Elbaz C,
Karboul N, Fontaine JJ, et al: Modeling of response to endocrine
therapy in a panel of human luminal breast cancer xenografts.
Breast Cancer Res Treat. 133:595–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
DeRose YS, Wang G, Lin YC, Bernard PS,
Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, et al:
Tumor grafts derived from women with breast cancer authentically
reflect tumor pathology, growth, metastasis and disease outcomes.
Nat Med. 17:1514–1520. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang X, Claerhout S, Prat A, Dobrolecki
LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano
M, et al: A renewable tissue resource of phenotypically stable,
biologically and ethnically diverse, patient-derived human breast
cancer xenograft models. Cancer Res. 73:4885–4897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kabos P, Finlay-Schultz J, Li C, Kline E,
Finlayson C, Wisell J, Manuel CA, Edgerton SM, Harrell JC, Elias A
and Sartorius CA: Patient-derived luminal breast cancer xenografts
retain hormone receptor heterogeneity and help define unique
estrogen-dependent gene signatures. Breast Cancer Res Treat.
135:415–432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Petrillo LA, Wolf DM, Kapoun AM, Wang NJ,
Barczak A, Xiao Y, Korkaya H, Baehner F, Lewicki J, Wicha M, et al:
Xenografts faithfully recapitulate breast cancer-specific gene
expression patterns of parent primary breast tumors. Breast Cancer
Res Treat. 135:913–922. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Moro M, Bertolini G, Tortoreto M,
Pastorino U, Sozzi G and Roz L: Patient-derived xenografts of non
small cell lung cancer: Resurgence of an old model for
investigation of modern concepts of tailored therapy and cancer
stem cells. J Biomed Biotechnol. 2012:5685672012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nakajima T, Geddie W, Anayama T, Ko HM, da
Cunha Santos G, Boerner S, Wang T, Wang YH, Li M, Pham NA, et al:
Patient-derived tumor xenograft models established from samples
obtained by endobronchial ultrasound-guided transbronchial needle
aspiration. Lung Cancer. 89:110–114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dangles-Marie V, Pocard M, Richon S,
Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N,
Validire P, et al: Establishment of human colon cancer cell lines
from fresh tumors versus xenografts: Comparison of success rate and
cell line features. Cancer Res. 67:398–407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Puig I, Chicote I, Tenbaum SP, Arqués O,
Herance JR, Gispert JD, Jimenez J, Landolfi S, Caci K, Allende H,
et al: A personalized preclinical model to evaluate the metastatic
potential of patient-derived colon cancer initiating cells. Clin
Cancer Res. 19:6787–6801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng S, Creighton CJ, Zhang Y, Sen B,
Mazumdar T, Myers JN, Lai SY, Woolfson A, Lorenzi MV, Bell D, et
al: Tumor grafts derived from patients with head and neck squamous
carcinoma authentically maintain the molecular and histologic
characteristics of human cancers. J Transl Med. 11:1982013.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Priolo C, Agostini M, Vena N, Ligon AH,
Fiorentino M, Shin E, Farsetti A, Pontecorvi A, Sicinska E and Loda
M: Establishment and genomic characterization of mouse xenografts
of human primary prostate tumors. Am J Pathol. 176:1901–1913. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wetterauer C, Vlajnic T, Schüler J,
Gsponer JR, Thalmann GN, Cecchini M, Schneider J, Zellweger T,
Pueschel H, Bachmann A, et al: Early development of human lymphomas
in a prostate cancer xenograft program using triple knock-out
immunocompromised mice. Prostate. 75:585–592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Boone JD, Dobbin ZC, Straughn JM Jr and
Buchsbaum DJ: Ovarian and cervical cancer patient derived
xenografts: The past, present, and future. Gynecol Oncol.
138:486–491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bankert RB, Balu-Iyer SV, Odunsi K, Shultz
LD, Kelleher RJ Jr, Barnas JL, Simpson-Abelson M, Parsons R and
Yokota SJ: Humanized mouse model of ovarian cancer recapitulates
patient solid tumor progression, ascites formation, and metastasis.
PLoS one. 6:e244202011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Han C, Shen J, Wang H, Yu L, Qian X, Liu B
and Guan W: Personalized primary tumor xenograft model established
for the pre-clinical trial to guide postoperative chemotherapy. Med
Hypotheses. 79:705–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xue A, Julovi SH, Samra JS, et al:
Establishment of patient-derived subrenal capsule xenograft of
pancreatic cancersin NOD/SCID mice: Potential models for drug
responses of personalized chemotherapy. Proceedings of the
Australian Health and Medical Research Congress (AHMRC). 2012.
|
|
78
|
Pavía-Jiménez A, Tcheuyap VT and
Brugarolas J: Establishing a human renal cell carcinoma tumorgraft
platform for preclinical drug testing. Nat Protoc. 9:1848–1859.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mohseni MJ, Amanpour S, Muhammadnejad S,
Sabetkish S, Muhammadnejad A, Heidari R, Haddadi M, Mazaheri Z,
Vasei M and Kajbafzadeh AM: Establishment of a patient-derived
Wilms' tumor xenograft model: A promising tool for individualized
cancer therapy. J Pediatr Urol. 10:123–129. 2014. View Article : Google Scholar : PubMed/NCBI
|