Introduction
Prostate carcinoma is currently one of the most
common types of non-cutaneous cancer among males and its incidence
increases with age. The diagnosis and staging of this cancer has
attracted a great deal of public interest. Significant advances in
its treatment have been made recently; however, there are
limitations regarding the screening tools used for this cancer
(1). Therefore, it is imperative to
identify novel potential biomarkers for the diagnosis and
prevention of the induction and progression of prostate
carcinoma.
Biomarkers serve an essential role in the detection
and treatment of disease. A biomarker is a characteristic that is
capable of being measured in a laboratory/clinical setting, is
associated with the pathological process, and has diagnostic and
prognostic utility (2). If the
disease is diagnosed in the early stages, the therapeutic success
rates for patients with prostate carcinoma are substantially
improved. Therefore, a successful therapy for this disease depends
on the biomarkers for detection of the presence and progression of
the disease. However, the current biomarkers for prostate carcinoma
are not ideal (3). Previously,
microRNAs (miRNAs) have exhibited potential as biomarkers in
prostate cancer (4). The aberrant
regulation of miRNAs has long been recognized as one of the
mechanisms that contribute to numerous types of tumors, including
prostate cancer (5). miRNAs are
non-coding small RNAs of 19–25 nucleotides in length that regulate
mRNA expression primarily by binding to the complementary sequences
in the 3′-untranslated region (3′-UTR) (6). miRNAs have been demonstrated to serve
essential roles in cancer research including tumor growth, invasion
and metastasis (7,8). MiR-26a and miR-26b have been
demonstrated to inhibit cell aggressiveness by regulating
fucosyltransferase 4 in colorectal cancer (9). In solid tumors, the expression of miRNAs
has been identified to be dysregulated and the alternation of miRNA
expression is causatively associated with the development of cancer
(10). Although much is known about
the profiles of miRNAs in numerous tumors and tissues, the function
of miRNAs in prostate cancer has not yet been completely
elucidated.
The phosphatase and tensin homolog (PTEN) gene was
initially identified as a tumor suppressor (11,12).
Recently, PTEN has been verified as a functional target of various
miRNAs and has served different biological functions in different
type of cancers (13). It has been
reported that miR-21 regulated growth and metastasis in non-small
cell lung cancer cells by targeting PTEN (14). Additionally, miR-32 has been
identified to promote growth, migration, and invasion via PTEN in
colorectal carcinoma cells (15).
However, the role of PTEN is not fully understood in prostate
carcinoma.
Herein, the present study demonstrates that miR-106a
is significantly upregulated in prostate cancer tumors, with
functional studies indicating that miR-106a promotes PC-3 cell
growth via PTEN. PTEN may potentially be exploited in a therapeutic
approach to prostate carcinoma.
Materials and methods
Cell culture and tumor tissues
The PC-3 cell line is considered to be a classical
human prostate cancer cell line for the study of human prostate
cancer (16). The PC-3 cell line was
purchased from the American Tissue Culture Collection (ATCC;
Manassas, VA, USA) and cultured according to the supplier's
recommendations. Cells were cultured in a ATCC-formulated F-12K
medium and supplemented with 10% fetal bovine serum (FBS) (Atlanta
Biologicals, Flowery Branch, GA, USA), 100 U/ml penicillin, and 100
mg/ml streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
at 37°C in a humidified atmosphere with 5% CO2.
Prostate cancer and noncancerous prostate tissues
were obtained from the tissue bank of The First Hospital of Jilin
University (Changchun, China). The age in the cancer group ranged
between 49 and 80 years with a median age of 69 years. In the
noncancerous group, the age ranged between 50 and 76 years, with a
median age of 65 years. Tissues were collected between June 2014
and June 2016, and frozen in liquid nitrogen immediately after
mincing (cutting into small pieces) on ice and were then stored at
−80°C until subsequent analysis. All patients were male. There was
no inclusion/exclusion criterion for patient selection in the
present study. Written informed consent was acquired from all
patients, and ethical approval was obtained from Institutional
Review Board of The First Hospital of Jilin University.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells were isolated by a
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol.
Residual RNAs were reverse transcribed into cDNA with M-MLV reverse
transcriptase (Thermo Fisher Scientific, Inc.). Briefly, the
following components were added to a nuclease-free microcentrifuge
tube: Oligo (dT) (500 µg/ml) (1 µl), total RNA (1 µg), 10 mM dNTP
mix (1 µl) and distilled water to 12 µl. The mixture was heated to
65°C for 5 min and chilled on ice (0°C). Subsequently, the 5X
first-strand buffer (4 µl), 0.1 M DTT (2 µl) and distilled water (1
µl) were added into the tube. Following a brief centrifugation (1
min at 1,000 × g, at room temperature), the mixture was incubated
at 37°C for 2 min. Following the addition of M-MLV reverse
transcriptase (1 µl), the mixture was incubated at 37°C for 50 min.
The reaction was inactivated by heating at 70°C for 15 min. All
components were purchased from Thermo Fisher Scientific. The
residual DNA was removed by DNA-free DNase (Thermo Fisher
Scientific, Inc.). For mature miRNA determination in tissues and
cells, the TaqMan microRNA assay was performed in accordance with
the instruction of the TaqMan microRNA reverse transcription kit
(Thermo Fisher Scientific, Inc.).
For specific gene quantitation, RNAs were reverse
transcribed into cDNA with M-MLV reverse transcriptase (Gibco;
Thermo Fisher Scientific, Inc.). The Taqman RT-qPCR was used for
PTEN gene expression analysis. GAPDH was used as a reference gene
to normalization (17). All specific
Taqman probes are commercially available from Applied Biosystems
(Thermo Fisher Scientific, Inc.; PTEN cat no. Hs02621230_s1 and
GAPDH cat no. Hs02786624_g1). All assays were performed in
triplicate on an ABI 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Hold stage, 95°C for 20 sec; PCR stage, 95°C for 1 sec, 60°C for 20
sec for 1 cycle, 40 cycles total. The data are presented as the
mean ± standard error of the mean (SE) (n=3).
Transfection of microRNA mimic
A total of 50 nM miR-106a mimic or miRNA vector
control (miR-con) were reverse transfected in to PC-3 cells by
Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.) as described
by the study protocol of Reid et al (18). miR-106a mimic and miR-con were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The
miR-106a mimic sequence was as follows:
5′-GAUGGACGUGACAUUCGUGAAAA-3′. After 48 h transfection, the cells
were available for subsequent assays.
3′-UTR dual luciferase assay
PC-3 cells were seeded in a 96-well plate at a
density of 2×104 cells/well. The cells were
co-transfected with 100 ng miR-106 mimic (Shanghai GenePharma Co.,
Ltd.) and 10 ng PTEN 3′-UTR luciferase reporter construct
(GeneCopoeia, Inc., Rockville, MD, USA) with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The empty vector was used as a control (VC). The
dual-luciferase activities were measured after 24 h in accordance
with the instructions of the Duo-Luciferase kit 2.0 (GeneCopoeia,
Inc.). Results were normalized by comparison with a Renilla
luciferase activity.
miR-106a inhibition
Inhibition of miR-106a expression was performed by a
method previously described (19).
Briefly, 60 nM antisense inhibitor miR-106a or scrambled control
antisense inhibitor (GE Healthcare Dharmacon, Inc., Lafayette, CO,
USA) were transfected into PC-3 cells (2×104 cells/well)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). Cells were harvested for subsequent experimentation after 48
h incubation.
PTEN overexpression
For the PTEN overexpression, a PTEN lentiviral
system (lenti-PTEN) was used to stably overexpress PTEN in PC-3
cells. Lentiviral control vector (lenti-Con) was used as a negative
control. These are commercially available from Applied Biological
Materials Inc., (Richmond, Canada). PC-3 cells were seeded into a
24 well plate (at 0.5×105 cells/well). The next day,
PC-3 cells were infected with lenti-PTEN or lenti-Con at a
multiplicity of infection of 10 using polybrene (8 µg/ml; Thermo
Fisher Scientific, Inc.) for 24 h. Subsequently, the culture medium
was replaced with 1 ml complete medium (ATCC-formulated F-12K
medium and supplemented with 10% FBS, 100 U/ml penicillin, and 100
mg/ml streptomycin). The cells were incubated at 37°C with 5%
CO2 overnight for subsequent assays.
Western blotting
Cells were lysed in RIPA Lysis and Extraction buffer
(Thermo Fisher Scientific, Inc.), and protein concentrations were
determined using a DC Protein Assay kit (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). A total of 15 µg protein from PC-3 cells
were separated using 4–20% gradient SDS-PAGE (Invitrogen; Thermo
Fisher Scientific, Inc.). Afterwards, the gel was
electrophoretically transferred onto a polyvinylidene difluoride
membrane and the membranes were incubated with the PTEN primary
antibody at 4°C overnight (cat no. ab31392; 1:500 dilution; Abcam,
Cambridge, MA, USA). Secondary antibodies (Goat anti-rabbit
antibody conjugated to horseradish peroxidase; cat no. 1662408EDU;
1:1,000 dilution; Bio-Rad Laboratories, Inc.) were added on the
next day after washing three times with TBST, 5 min each. After 2 h
incubation at room temperature, the proteins were visualized using
enhanced chemiluminescence (SuperSignal™ West Femto
Maximum Sensitivity Substrate; Thermo Fisher Scientific, Inc.) and
GAPDH (cat no. ab9485; 1:500 dilution; Abcam) from the same
membrane was used as a loading control. All experiments were
repeated three times.
Cell proliferation assay
CyQuant assay (CyQUANT™ Cell Proliferation Assay
kit; Thermo Fisher Scientific, Inc.), a fluorescence-based
microplate assay, was used to determine cell growth according to
the manufacturer's protocol. Briefly, 5×103 PC-3
cells/well were seeded in a 96-well tissue culture plate in 100 µl
media/well. The CyQuant solution was prepared immediately prior to
use at indicated time points. Following the removal of media, 100
µl of CyQuant solution was added to the wells and incubated in the
dark for 45 min at room temperature. The plate was read at
excitation at 497 nm and emission at 520 nm. Data was presented as
the mean ± SE of three independent experiments.
Statistical analysis
GraphPad Prism (version 6.07 for Windows; Graphpad
Software, Inc., La Jolla, CA, USA) was used to analyze data. Data
are presented as means ± SE. An unpaired Student's t-test was used
for comparison between two groups and one-way analysis of variance
test followed by Tukey's multiple comparison test was used to
compare the significance of differences between the means of
multiple groups. IBM SPSS Statistics 21 (IBM Corp., Armonk, NY,
USA) was used for statistical analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
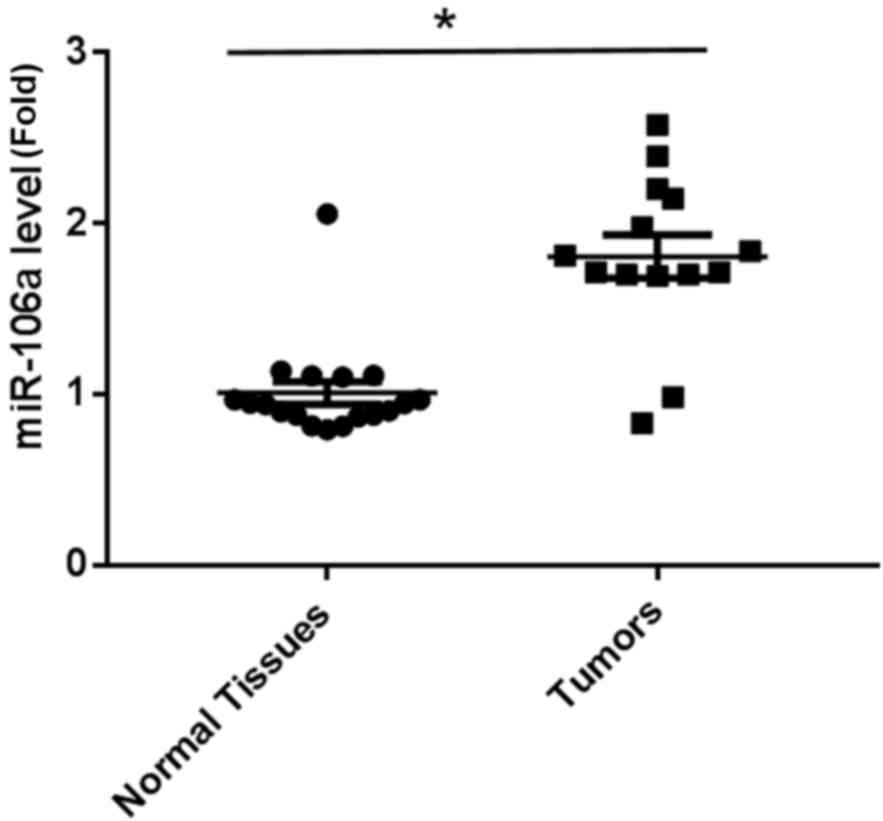
Levels of miR-106a are upregulated in
tumor tissues
Of microarray preliminary studies in the screening
of miRNAs from tumor tissues (14 cases) versus noncancerous
prostate tissues (18 cases), miR-106a was identified to have a
1.8-fold upregulation in prostate tumors, which is considered
significant (data not shown). In the present study, mature miR-106a
levels in prostatic tumors and normal tissues were compared using
RT-qPCR. As shown in Fig. 1, there
was a significantly increased expression of miR-106a in tumor
samples when compared with normal tissues, suggesting that miR-106a
may be responsible for the progression of prostate carcinoma.
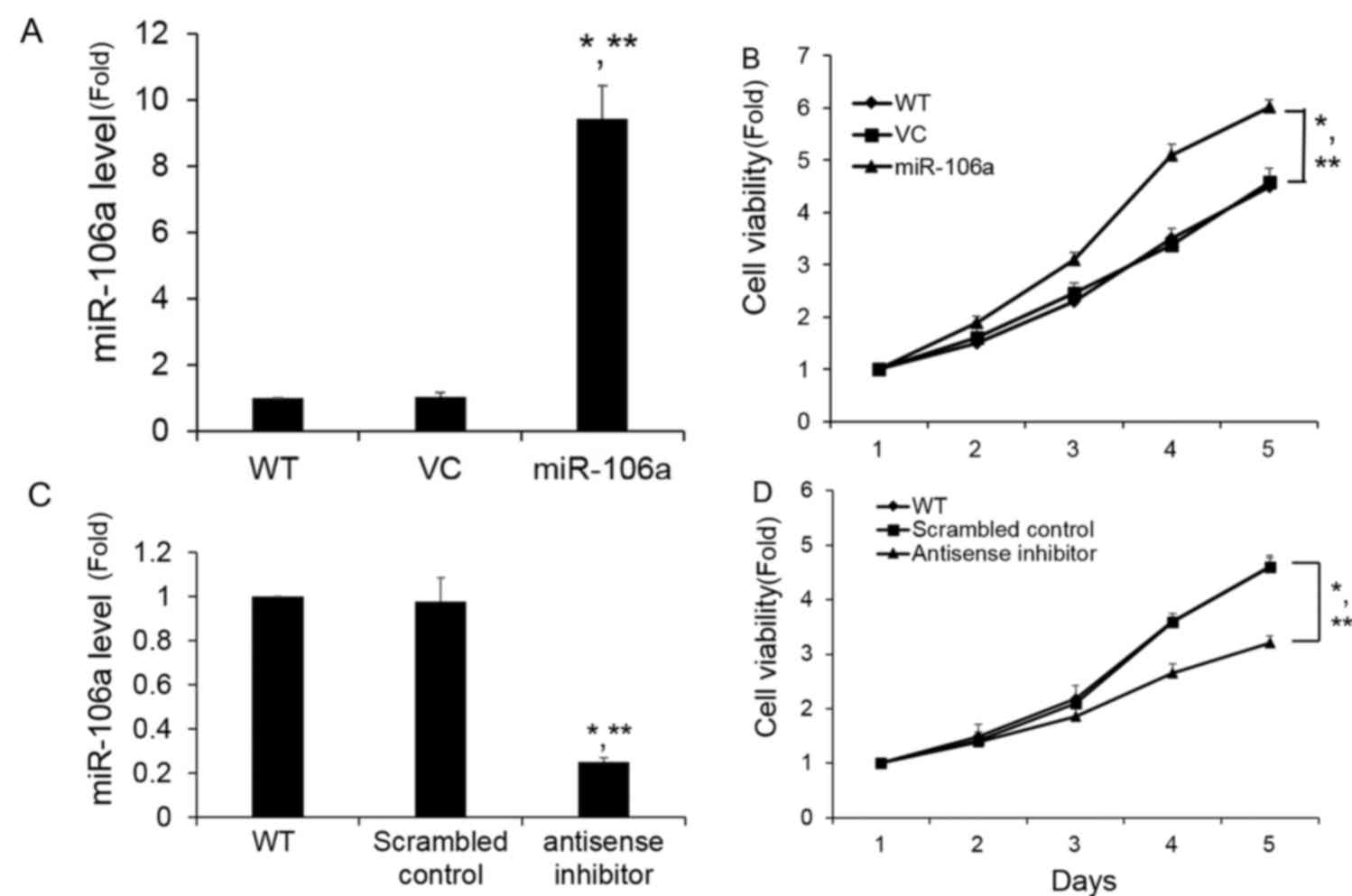
miR-106a promotes cell growth in
vitro
To decipher the functional effects of miR-106a, a
synthetic miR-106a mimic was transfected into PC-3 cells, and the
miR-106a expression level following transfection was determined to
be significantly increased compared with the wild-type and VC
control groups (Fig. 2A). As shown in
Fig. 2B, the miR-106a mimic
significantly promoted cell growth in PC-3 cells compared with a
control mimic, which did not affect growth. To further confirm
these results, a reverse study was performed to confirm the role of
miR-106a in the PC-3 cells following the inhibition of miR-106a.
Transfection of miR-106a antisense inhibitor into PC-3 cells
produced an ~70% decrease in miR-106a levels as measured by RT-qPCR
compared with the scrambled control (Fig.
2C). As shown in Fig. 2D, the
miR-106a inhibitor significantly suppressed PC-3 cell growth, which
is in line with the effects observed following overexpression of
miR-106 in PC-3 cells. Therefore, the next step was to evaluate the
potential mechanisms of miR-106a on cell proliferation.
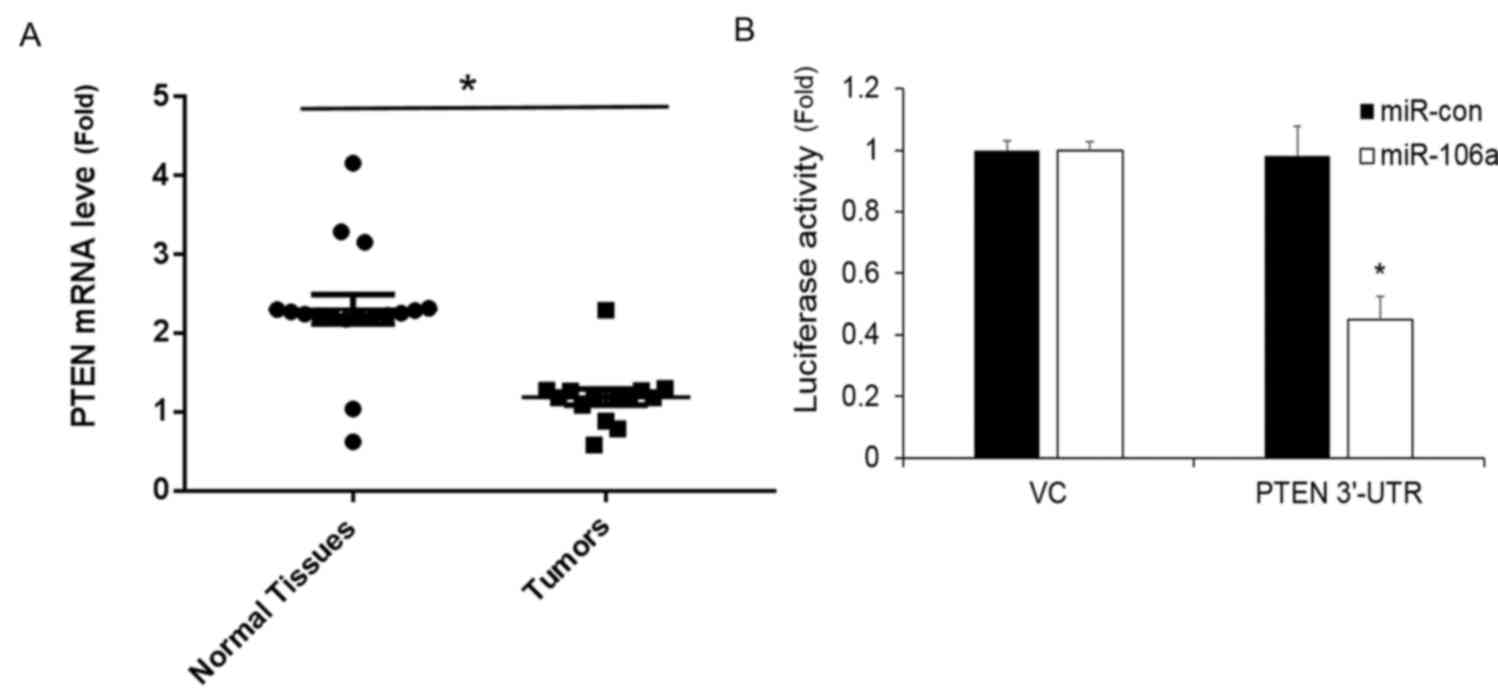
PTEN expression is repressed by
miR-106a
A previous study identified an association between
miR-106a and PTEN (20). To determine
whether the cell growth promotive effects observed with miR-106a in
PC-3 cells was also associated with the downregulation of PTEN, the
expression of PTEN mRNA in tumor and normal tissues was analyzed.
PTEN mRNA levels were demonstrated to be significantly decreased in
the tumor tissues in comparison with normal tissues (Fig. 3A).
To further confirm this observation, PC-3 cells were
co-transfected with the PTEN 3′-UTR luciferase reporter and
miR-106a or miR-con. The results revealed that miR-106a
significantly repressed the activity of PTEN compared with the
miR-con (Fig. 3B).
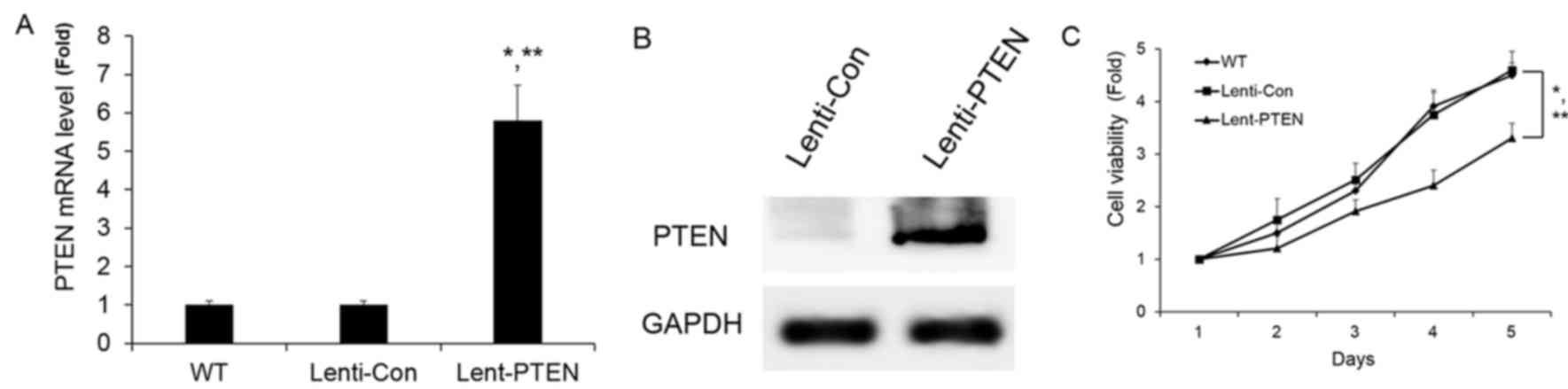
PTEN overexpression suppressed cell
growth in vitro
To assess whether PTEN overexpression caused a
similar response in PC-3 cell growth as was caused by miR-106a
downregulation, PTEN was stably overexpressed in PC-3 cells. PTEN
was overexpressed by lentiviral-PTEN. Lentiviral-PTEN significantly
increased the mRNA and protein expression of PTEN (Fig. 4A and B). The overexpression of PTEN
was observed to significantly repress PC-3 cell growth, which was
consistent with the effects of miR-106a inhibition (Fig. 4C). This data suggests that miR-106a
regulates prostate carcinoma progression through the involvement of
PTEN.
Discussion
The expression and roles of miRNAs are tissue and
cell specific. Dysregulated expression of miRNAs is a
well-recognized feature of cancer. The expression and functions of
miRNAs in the progression of prostate carcinoma are not fully
characterized or completely understood. In the present study,
miR-106a was observed to be significantly upregulated in prostate
cancer tissues. Furthermore, increased expression of miR-106a
significantly enhanced PC-3 cell proliferation, and loss of
miR-106a repressed PC-3 cell proliferation. In addition, the
present study also identified that PTEN miRNA and protein levels
are negatively associated with miR-106a expression. Furthermore,
PTEN overexpression significantly suppressed PC-3 cell growth in
vitro. Therefore, these data suggested that miR-106a
contributes to the prostate carcinoma progression with PTEN
involvement and miR-106a may be a promising biological biomarker
for prostate carcinoma.
miRNAs have previously exhibited promising results
as potential circulating biomarkers in prostate cancer (4). miR-125 has been identified to interact
with the apoptotic signaling pathway in prostate cells by targeting
P53, P21 and Puma (21). miR-21 has
been demonstrated to stimulate apoptosis through the P53 pathway
via targeting programmed cell death 4 and PTEN (22,23).
Furthermore, downregulated miR-221 enhanced the
androgen-independent tumor growth and triggered a more aggressive
prostate cancer phenotype (24).
miR-106a has been demonstrated to serve an oncogenic role in
pancreatic cancer (25). miR-106a was
also identified to be upregulated in the prostate cancer group
(4,10). In the present study, miR-106a
expression was identified to be upregulated in prostate tumor and
that an increased miR-106a level was associated with the promotion
of PC-3 cell growth.
PTEN is well established as a key positive and
negative regulator, and as a mediator of cell growth, survival and
proliferation. Additionally, PTEN is one of the most frequently
disrupted tumor suppressors in cancer (26–30). Loss
of PTEN cooperates with aberrant ETS transcription factor
expression to promote cancer progression in the prostate (31). Furthermore, PTEN has been demonstrated
as a prognostic marker for patients with non-small cell lung cancer
(32). However, the association of
PTEN and miR-106a remains unclear in prostate carcinoma
progression. In the present study, PTEN expression was demonstrated
to be decreased in prostate tumor samples. Additionally, PTEN mRNA
and protein expression were increased following stable
overexpression by lentiviral transfection. Furthermore, PTEN
overexpression repressed PC-3 cell growth, similar to the effects
observed following miR-106a inhibition. However, the data was
derived from a small patient cohort. The present study generated a
hypothesis that miR-106a promotes prostate cancer cell growth
through regulating PTEN; however, further validation with larger
datasets is necessary.
In conclusion, the present study has demonstrated
that miR-106a regulates prostate carcinoma progression with PTEN
involvement. The effects of miR-106a on cell proliferation may
unveil its contribution to prostate cancer development, and as a
result, miR-106a may be used as a potential therapeutic target for
prostate cancer.
Acknowledgements
Not applicable.
Funding
This work is partly supported by National Natural
Science Foundation of China (grant no. 31501052), Bethune Program
of Jilin University (grant no. 2015338) and Science and Technology
Development Program of Jilin Province (grant no.
20160520163JH).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
KH designed the experiments. JL performed the
experiments. XM and QY derived the models and analyzed the data.
All authors contributed to the final version of the manuscript.
Ethics approval and consent to
participate
Written informed consent was acquired from all
patients, and ethical approval was obtained from Institutional
Review Board of The First Hospital of Jilin University (Changchun,
China).
Consent for publication
Informed consent was obtained from all patients for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Delongchamps Barry N: Prostate cancer:
Review in 2014. Diagn Interv Imaging. 95:739–742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lesko LJ and Atkinson AJ Jr: Use of
biomarkers and surrogate endpoints in drug development and
regulatory decision making: Criteria, validation, strategies. Annu
Rev Pharmacol Toxicol. 41:347–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Madu CO and Lu Y: Novel diagnostic
biomarkers for prostate cancer. J Cancer. 1:150–177. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jackson BL, Grabowska A and Ratan HL:
MicroRNA in prostate cancer: Functional importance and potential as
circulating biomarkers. BMC Cancer. 14:9302014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coppola V, De Maria R and Bonci D:
MicroRNAs and prostate cancer. Endocr Relat Cancer. 17:F1–F17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmad J, Hasnain SE, Siddiqui MA, Ahamed
M, Musarrat J and Al-Khedhairy AA: MicroRNA in carcinogenesis &
cancer diagnostics: a new paradigm. Indian J Med Res. 137:680–694.
2013.PubMed/NCBI
|
|
9
|
Li Y, Sun Z, Liu B, Shan Y, Zhao L and Jia
L: Tumor-suppressive miR-26a and miR-26b inhibit cell
aggressiveness by regulating FUT4 in colorectal cancer. Cell Death
Dis. 8:e28922017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Steck PA, Pershouse MA, Jasser SA, Yung
WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T,
et al: Identification of a candidate tumour suppressor gene, MMAC1,
at chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou
H and Jia L: MicroRNA-130b targets PTEN to mediate drug resistance
and proliferation of breast cancer cells via the PI3K/Akt signaling
pathway. Sci Rep. 7:419422017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu ZL, Wang H, Liu J and Wang ZX:
MicroRNA-21 (miR-21) expression promotes growth, metastasis, and
chemo- or radioresistance in non-small cell lung cancer cells by
targeting PTEN. Mol Cell Biochem. 372:35–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alimirah F, Chen J, Basrawala Z, Xin H and
Choubey D: DU-145 and PC-3 human prostate cancer cell lines express
androgen receptor: Implications for the androgen receptor functions
and regulation. FEBS Lett. 580:2294–2300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reid G, Pel ME, Kirschner MB, Cheng YY,
Mugridge N, Weiss J, Williams M, Wright C, Edelman JJ, Vallely MP,
et al: Restoring expression of miR-16: a novel approach to therapy
for malignant pleural mesothelioma. Ann Oncol. 24:3128–3135. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thomson DW, Bracken CP, Szubert JM and
Goodall GJ: On measuring miRNAs after transient transfection of
mimics or antisense inhibitors. PLoS One. 8:e552142013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu
FQ, Hu R and Wang MS: miR-106a promotes growth and metastasis of
non-small cell lung cancer by targeting PTEN. Int J Clin Exp
Pathol. 8:3827–3834. 2015.PubMed/NCBI
|
|
21
|
Amir S, Ma AH, Shi XB, Xue L, Kung HJ and
White Devere RW: Oncomir miR-125b suppresses p14(ARF) to modulate
p53-dependent and p53-independent apoptosis in prostate cancer.
PLoS One. 8:e610642013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang CH, Yue J, Fan M and Pfeffer LM: IFN
induces miR-21 through a signal transducer and activator of
transcription 3-dependent pathway as a suppressive negative
feedback on IFN-induced apoptosis. Cancer Res. 70:8108–8116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Spahn M, Kneitz S, Scholz CJ, Stenger N,
Rüdiger T, Ströbel P, Riedmiller H and Kneitz B: Expression of
microRNA-221 is progressively reduced in aggressive prostate cancer
and metastasis and predicts clinical recurrence. Int J Cancer.
127:394–403. 2010.PubMed/NCBI
|
|
25
|
Li P, Xu Q, Zhang D, Li X, Han L, Lei J,
Duan W, Ma Q, Wu Z and Wang Z: Upregulated miR-106a plays an
oncogenic role in pancreatic cancer. FEBS Lett. 588:705–712. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Lin G, Wu B, Zhou X and Zhou K:
Overexpression of PTEN induces cell growth arrest and apoptosis in
human breast cancer ZR-75-1 cells. Acta Biochim Biophys Sin
(Shanghai). 39:745–750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qin Y, Huo Z, Song X, Chen X, Tian X and
Wang X: mir-106a regulates cell proliferation and apoptosis of
colon cancer cells through targeting the PTEN/PI3K/AKT signaling
pathway. Oncol Lett. 15:3197–3201. 2018.PubMed/NCBI
|
|
28
|
Yu YX, Wang Y and Liu H: Overexpression of
PTEN suppresses non-small-cell lung carcinoma metastasis through
inhibition of integrin alphaVbeta6 signaling. Am J Transl Res.
9:3304–3314. 2017.PubMed/NCBI
|
|
29
|
de la Rosa J, Weber J, Rad R, Bradley A
and Cadinanos J: Disentangling PTEN-cooperating tumor suppressor
gene networks in cancer. Mol Cell Oncol. 4:e13255502017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li S, Shen Y, Wang M and Yang J, Lv M, Li
P, Chen Z and Yang J: Loss of PTEN expression in breast cancer:
Association with clinicopathological characteristics and prognosis.
Oncotarget. 8:32043–32054. 2017.PubMed/NCBI
|
|
31
|
Carver BS, Tran J, Gopalan A, Chen Z,
Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino
PT, et al: Aberrant ERG expression cooperates with loss of PTEN to
promote cancer progression in the prostate. Nat Genet. 41:619–624.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao J, Hu CP, He BX, Chen X, Lu XX, Xie
MX, Li W, He SY, You SJ and Chen Q: PTEN expression is a prognostic
marker for patients with non-small cell lung cancer: A systematic
review and meta-analysis of the literature. Oncotarget.
7:57832–57840. 2016. View Article : Google Scholar : PubMed/NCBI
|