Introduction
Hepatocellular carcinoma (HCC) is the third most
common cause of cancer-associated mortality worldwide, and the
highest numbers of HCC cases have been detected in Asia and Africa
(1,2).
The long-term prognosis of HCC remains unsatisfactory, as indicated
by a low overall survival rate of 22–35% over the last 10 years
(3). Since hepatocarcinogenesis
involves numerous oncogenes and tumour suppressor genes (4), the molecular carcinogenic mechanisms and
the pathogenic biology of HCC have become an issue of great
interest (5,6).
MicroRNAs (miRNAs), highly conserved small
non-coding RNAs of 19–25 nucleotides, are known to regulate
numerous protein-coding genes (7,8). By
binding directly to the 3′ untranslated regions (3′ UTR) of target
mRNAs, miRNAs may cause posttranscriptional repression of protein
synthesis, leading to deadenylation and subsequent mRNA degradation
and/or translational inhibition (9,10). A
number of miRNAs have been implicated in various cellular processes
including apoptosis, cell proliferation, differentiation, stem cell
renewal, stress responses and metabolism (11–15). Their
marked impact on the regulation of carcinogenesis and tumour
suppression indicates that the aberration and regulation of the
miRNA biogenesis pathway may contribute to human diseases,
including cancer (8,16,17).
In a previous study it was identified that the
expression of the miR-197-3p was markedly decreased in HCC compared
with the corresponding adjacent non-tumour samples. Additionally,
previous studies indicate that miR-197-3p is of great interest in
cancer therapy due to its association with various malignancies,
including bladder (18) and thyroid
cancer (19). The present study
demonstrated that low miR-197-3p expression in HCC tissues
correlated with poor patient prognosis; thus the focus of the study
centred on the roles and corresponding mechanisms of miR-197-3p in
the progression of HCC.
Materials and methods
Cell lines and culture
Human hepatocellular carcinoma cell lines (Huh7,
HCC-LM3, and Hep3B) and a hepatocellular cell line (THLE-3) were
purchased from the Cell Resource Centre of the Shanghai Institute
of Life Sciences (Shanghai, China). All cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% foetal bovine
serum (FBS) and 1% penicillin/streptomycin (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). All cells were maintained at
37°C in a humidified atmosphere, at 5% CO2.
Patients, tumour tissues and serum
sample
A total of 197 pairs of snap-frozen HCC and
peritumoural tissues were obtained from the Eastern Hepatobiliary
Surgery Hospital (Shanghai, China) from 19 females and 178 males
(age range, 35–83 years) between January 2010 and October 2014. The
tissues were used for reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) analysis. Clinical tissue samples were
verified as tumour or non-tumour through a histopathological
examination and the Edmondson grading system (20). Micrometastases were defined as tumours
adjacent to the border of the primary tumour, as observed using a
microscope. Tumour staging was defined according to the sixth
edition of the tumour, node, metastasis (TNM) classification system
(21) published by the International
Union Against Cancer. The tissue samples were stored at −80°C until
further use. Tumour differentiation was defined according to the R
and Barcelona Clinic Liver Cancer (BCLC) staging systems (22). The study was approved by the
Institutional Review Board of the Eastern Hepatobiliary Surgery
Hospital. All patients gave written informed consent to participate
in the study. The data were anonymized. All clinical specimens were
approved by the Clinical Research Ethics Committee of Eastern
Hepatobiliary Surgery Hospital.
RNA extraction and RT-qPCR
Total RNA from tissues or cells was extracted using
the miRNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) according
to the manufacturer's protocol. mRNA and miRNA were
reverse-transcribed from total RNA using the Revert Aid First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. cDNA was amplified and
quantified on a CFX96 system (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) using iQ SYBR Green (Bio-Rad Laboratories, Inc.). U6 or
GAPDH were used as endogenous controls. Primers for miR-197-3p
expression analysis were as follows: miR-197-3p forward,
5′-CACCACCTTCTCCACCCA-3′, and reverse, 5′-GGGACTGGACTTGGAGTC-3′; U6
forward, 5′-CTCGCTTCGGCAGCACA-3′, and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. Primers for epithelial mesenchymal
transition (EMT) analysis were as follows: E-cadherin forward,
5′-TACACTGCCCAGGAGCCAGA-3′, and reverse,
5′-TGGCACCAGTGTCCGGATTA-3′; N-cadherin forward,
5′-TCAGGCGTCTGTAGAGGCTT-3′, and reverse,
5′-ATGCACATCCTTCGATAAGACTG-3′; Vimentin forward,
5′-GACGCCATCAACACCGAGTT-3′, and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; GAPDH forward,
5′-TGTGGGCATCAATGGATTTGG-3′, and reverse,
5′-ACACCATGTATTCCGGGTCAAT-3′. Relative fold expressions were
calculated via the comparative quantification cycle
(2−ΔΔCq) method (23).
Mature miR-197-3p expression was detected using a TaqMan miRNA
assay kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The RNU6B gene was used as a normalization
control. All experiments were performed three times in duplicate.
The GAPDH gene was used as an internal control. qPCR was conducted
using the following conditions: 30 cycles consisting of
denaturation at 94°C for 30 sec, annealing at 56°C (58°C for GAPDH)
for 30 sec, and extension at 72°C for 30 sec.
Wound healing migration assay
Wound healing migration assays were performed as
described previously (24). Briefly,
1×105 cells were plated in each well of a 6-well plate.
Once attached, cells were scraped to form a wound in the middle of
the plate, and the medium was replaced with serum-free medium.
Cells were incubated for 36 h in a humidified atmosphere containing
5% CO2 at 37°C, prior to the measurement of migration
across the wound line.
Cellular migration and invasion
assays
Cellular migration and invasion were assessed using
Transwell chambers with a pore size of 8 µm (Corning Inc., Corning,
NY, USA). Following transfection for 48 h, 5×104
transfected cells in 300 µl medium without FBS were seeded into the
upper chamber, while 500 µl medium supplemented with 20% FBS was
placed into the lower chamber. For the Matrigel invasion assay, the
Transwell chamber was coated with Matrigel (BD Biosciences, San
Jose, CA, USA). A total of 5×104 transfected cells in
300 µl medium without FBS were seeded into the upper chamber of the
Transwell, while 500 µl medium supplemented with 20% FBS was placed
into the lower chamber. Cells were incubated at 37°C for a further
24 h for the migration assay and 48 h for the invasion assay. In
the two assays, the cells were fixed with 100% methanol for 5 min
(Beyotime Institute of Biotechnology, Haimen, China) and stained
with 0.5% crystal violet (Beyotime Institute of Biotechnology) for
5 min. Subsequently, cells remaining on the upper surface of the
membranes were removed carefully using cotton swabs. The migrated
and invaded cells were then counted in five randomly selected
fields with an inverted microscope (magnification, ×200). Each
experiment was repeated three times.
Synthetic RNA oligonucleotides and
transient transfection
The predicted binding sites between miR-197-3p and
zinc finger protein interacted with K protein 1 (ZIK1) were
obtained using TargetScan Human online software 7.1 (http://www.targetscan.org/vert_71/). miRNA
inhibitors, mimics and an miRNA negative control (NC) were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
5′-3′ sequences of the three miRNAs were as follows: Mimics,
CGGGUAGAGAGGGCAGUGGGAGG and UUCACCACCUUCUCCACCCAGC; inhibitor,
AAGUGGUGGAAGAGGUGGGUCG; and NC, CAGUACUUUUGUGUAGUACAA. ZIK1
overexpression vectors and the NC vector were obtained from
Shanghai GeneChem Co., Ltd. Cells were seeded at
1–1.5×105/well in a 6-well plate, and transiently
transfected with miRNAs (5 µg/well) using JetPrime®
(Polyplus-transfection SA, Illkirch, France), according to the
manufacturer's protocol. The cells were collected for analysis 48 h
post-transfection.
Cell proliferation assay
Cell proliferation was measured using a Cell
Counting Kit-8 (CCK-8) assay (Sangon Biotech, Co., Ltd., Shanghai,
China). A total of 48 h post-transfection, 3×103
cells/well were seeded into a 96-well plate. The CCK-8 reagent was
added 72 h post-seeding, and optical density was measured using a
microplate reader at 450 nm.
Western blotting analysis
Cells were lysed with ice-cold lysis buffer
supplemented with protease inhibitor cocktail (Pierce; Thermo
Fisher Scientific, Inc.). Total protein concentration was
determined using the Enhanced BCA Protein Assay kit (Beyotime
Institute of Biotechnology). Protein samples (20 µg) were separated
on a 10% SDS-PAGE gel and then transferred to a polyvinylidene
fluoride membrane (Roche Diagnostics GmbH, Mannheim, Germany).
Following blocking with 5% skimmed milk at room temperature for 2
h, the membranes were incubated overnight at 4°C with rabbit
anti-human ZIK1 monoclonal primary antibody (1:1,000; Abcam,
Cambridge, UK; cat. no. 107918) and mouse anti-human β-actin
monoclonal primary antibody (1:1,000; Abcam; cat. no. 8226) primary
antibodies. Subsequently, the membranes were incubated with goat
anti-mouse horseradish peroxidase-conjugated secondary antibody
(1:3,000; Abcam; cat. no. ab6789) for 1 h at room temperature. The
membranes were visualized with Super Signal West Pico
Chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific,
Inc.). β-actin was used as a loading control. The Lab Works Image
Acquisition and Analysis Software (version 7.0; UVP, LLC, Phoenix,
AZ, USA) was used to quantify the band intensities.
Luciferase activity assay
The 3′ UTR of ZIK1 was amplified and cloned
downstream of the pGL3/Luciferase (Luc) vector (Shanghai GenePharma
Co., Ltd.). The mutant 3′ UTR of ZIK1 was amplified using the
pGL3/Luc-ZIK1 3′ UTR as the template and was cloned downstream of
the pGL3/Luc vector. For the luciferase reporter assay, either 100
nM of miR-197-3p mimic or control and 2 µg pGL3/Luc-ZIK1 3′ UTR or
the mutant 3′ UTR, in addition to the controls, were co-transfected
into HCCLM3 cells at 70% confluence using Lipofectamine®
2000 reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 48 h post-transfection, the cells were
lysed using radioimmunoprecipitation assay buffer (cat. no. P0013C;
Beyotime Institute of Biotechnology, China) according to the
manufacturer's protocol. Luciferase intensity was measured using an
F-4500 Fluorescence Spectrophotometer (Hitachi, Ltd., Tokyo, Japan)
and normalized to that of Renilla luciferase, according to
the manufacturer's protocol.
Cell proliferation (MTT) assay
HCCLM3 cells transfected with miR-197-3p mimics or
control and with the miR-197-3p inhibitor or control were plated
into 96-well plates at a density of 5×104 cells/well,
under 5% CO2 at 37°C for 48 h. Following incubation for
24, 48 or 72 h, the culture plates were incubated with 20 µl of
MTT/well in 100 µl medium at a final concentration of 5 mg/ml for a
further 4 h. The culture plates were centrifuged at 2,000 × g for 5
min at room temperature. The supernatant was carefully removed, and
100 µl dimethyl sulfoxide was added to each well to end the
reaction. Blue-violet formazan particles were dissolved for ~10 min
in the dark at 25°C. The absorbance at 570 nm was detected using a
Quant Universal microplate spectrophotometer (BioTek Instruments,
Inc., Winooski, VT, USA). The experiment was repeated three
times.
Statistical analysis
All values are presented as the mean ± standard
deviation. Significant differences were determined using GraphPad
5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The difference
between the HCC tissues and their corresponding adjacent non-tumour
samples was analysed for statistical significance using the paired
Student's t-test. Student's t-test was used to determine
significant differences between two groups. One-way analysis of
variance (ANOVA) was used to determine significant differences
between multiple testing. Student-Newman-Keuls test was used as a
post hoc test following ANOVA. The χ2 test was used to
analyse the association between miR-197-3p expression and
clinicopathological characteristics. Survival curves were plotted
using the Kaplan-Meier method and were compared via the log-rank
test. P<0.05 was considered to indicate a statistically
significant difference. To collect dependent variables,
multivariate regression analysis was conducted using the
significant variables identified in the univariate regression
analysis. All experiments were repeated three times.
Results
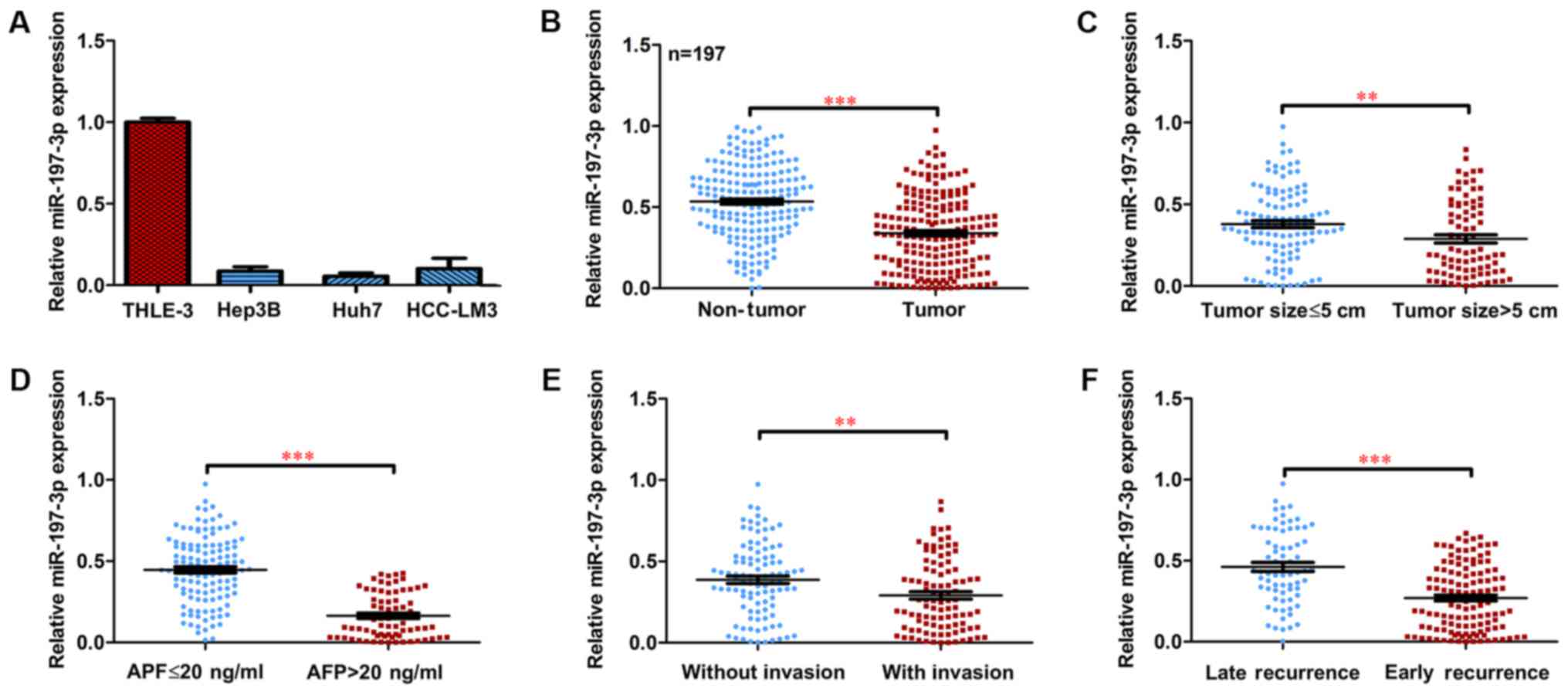
Expression of miR-197-3p is markedly
downregulated in HCC
To investigate the expression and clinical
significance of miR-197-3p in HCC, the expression of miR-197-3p in
HCC cell lines was examined. Decreased miR-197-3p expression was
observed in the HCC cells, compared with the normal liver cells
(Fig. 1A). The expression of
miR-197-3p was detected in a total of 197 paired primary HCC
tissues and their corresponding adjacent non-tumour samples.
RT-qPCR indicated that the average expression level of miR-197-3p
was significantly lower in the cancerous tissues compared with that
in the adjacent non-cancerous tissues (Fig. 1B; P<0.001). Moreover, the
miR-197-3p expression declined in HCC patients with larger tumours
(>5 cm), a higher α-fetoprotein (AFP) serum level (>20
ng/ml), vascular invasion and early recurrence (Fig. 1C-F).
Association between miR-197-3p
expression and clinicopathological characteristics
To evaluate the correlations between miR-197-3p and
the selected clinicopathological variables, the cohort of 197
patients were divided into subgroups with low and high miR-197-3p
expression. The median value of miR-197-3p expression in all 197
cases was selected as the cut-off value. Consistent with the
criteria above, the correlation of miR-197-3p expression with
clinicopathological characteristics and the prognosis of HCC were
analysed. As presented in Table I,
the lo expression group was associated with higher AFP levels (≥20
µg/l; P<0.001), larger tumour size (≥5 cm; P=0.012), multiple
tumours (n≥2; P<0.001), absence of capsule (P<0.001) and
vascular invasion (P=0.013).
 | Table I.Clinical characteristics of 197
patients with hepatocellular carcinoma, according to miR-197-3p
expression levels. |
Table I.
Clinical characteristics of 197
patients with hepatocellular carcinoma, according to miR-197-3p
expression levels.
|
| miR-197-3p |
|
|
|---|
|
|
|
|
|
|---|
| Feature | High (n=99) | Low (n=98) | χ2
value | P-value |
|---|
| Age in years |
|
| 0.610 | 0.435 |
|
≥55 | 44 | 49 |
|
|
|
<55 | 55 | 49 |
|
|
| Sex |
|
| 0.070 | 0.791 |
|
Male | 90 | 88 |
|
|
|
Female | 9 | 10 |
|
|
| HBsAg |
|
| 0.134 | 0.714 |
|
Positive | 83 | 84 |
|
|
|
Negative | 16 | 14 |
|
|
| AFP, µg/l |
|
| 56.840 | <0.001 |
|
Positive | 12 | 63 |
|
|
|
Negative | 87 | 35 |
|
|
| Cirrhosis |
|
| 1.845 | 0.174 |
|
Present | 47 | 56 |
|
|
|
Absent | 52 | 42 |
|
|
| Tumour size,
cm |
|
| 6.288 | 0.012 |
| ≥5 | 34 | 51 |
|
|
|
<5 | 65 | 47 |
|
|
| Tumour number |
|
| 0.063 | <0.001 |
|
Multiple | 34 | 32 |
|
|
|
Single | 65 | 66 |
|
|
| Capsule |
|
| 13.438 | <0.001 |
|
Present | 65 | 86 |
|
|
|
Absent | 34 | 12 |
|
|
| Vascular
invasion |
|
| 6.214 | 0.013 |
|
Present | 40 | 57 |
|
|
|
Absent | 59 | 41 |
|
|
| TNM stage |
|
| 0.412 | 0.521 |
|
I–II | 36 | 40 |
|
|
|
III–IV | 63 | 58 |
|
|
| BCLC stage |
|
| 0.414 | 0.520 |
|
A-B | 35 | 39 |
|
|
|
C-D | 64 | 59 |
|
|
Association between miR-197-3p
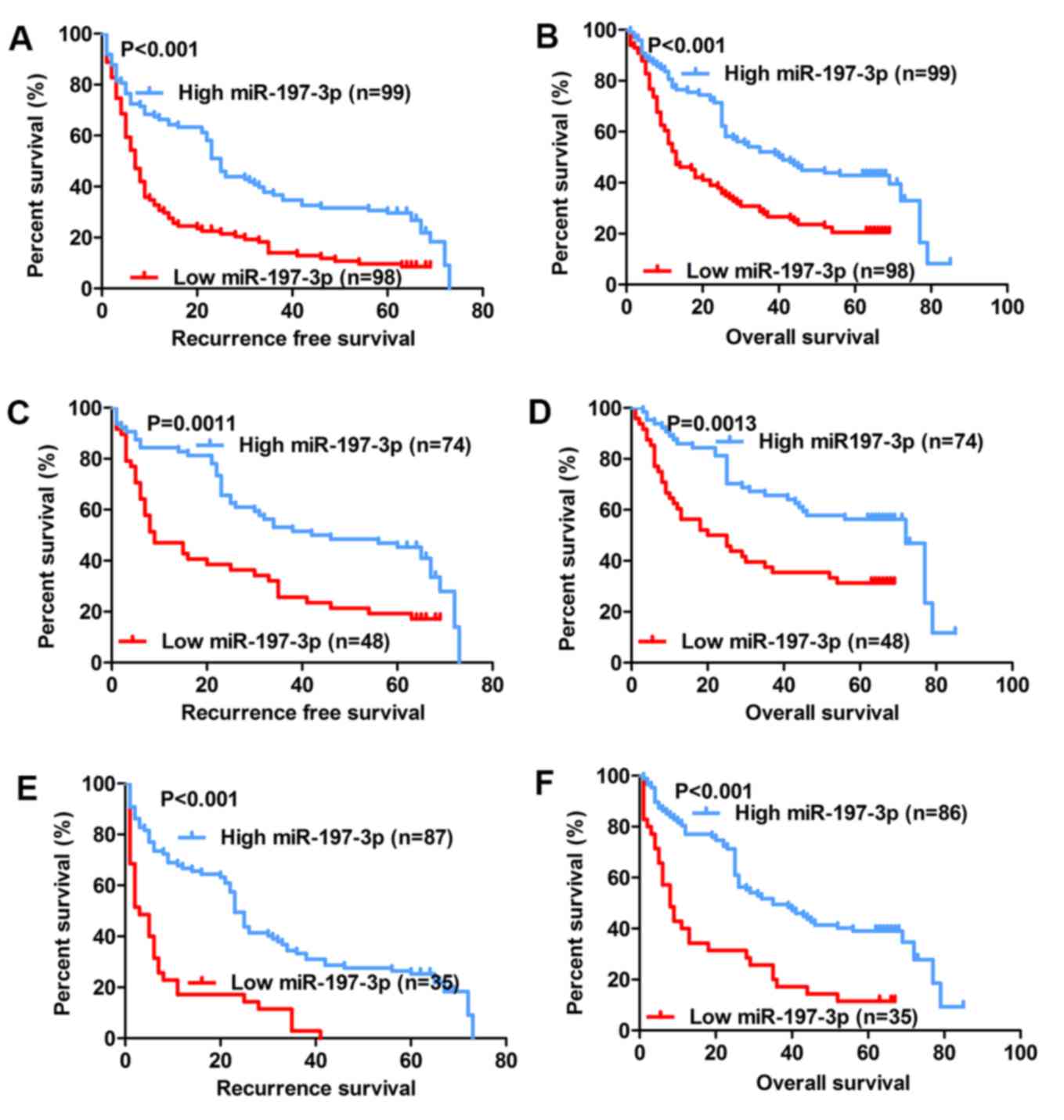
expression and HCC prognosis
The association between miR-197-3p expression and
the prognosis of patients with HCC following hepatectomy was
further analysed. It was determined that the group with low
miR-197-3p expression exhibited significantly poorer relapse-free
survival (RFS) (P<0.001; Fig. 2A)
and poorer overall survival (OS) (P<0.001; Fig. 2B). Previous studies indicated that
AFP-negative patients (AFP serum level <20 µg/l) are generally
considered to have a better prognosis (25,26);
nevertheless, many of these patients exhibit early recurrence and
poor OS. Therefore, a precise biomarker is required to predict the
prognosis of AFP-negative patients. Subgroup analysis revealed that
among the 122 AFP-negative patients (AFP serum level <20 µg/l),
the difference in RFS and OS remained between the high- and
low-expression groups (P=0.0011 and P=0.0013, respectively;
Fig. 2C and D). Further analysis
indicated that of the 121 patients with tumour size <5 cm, the
group with low miR-197-3p expression exhibited significantly poorer
RFS and OS (P<0.001; Fig. 2E and
F, respectively). Univariate analysis indicated that among the
clinicopathological characteristics, RFS was associated with the
miR-197-3p expression level, presence of the hepatitis B virus
surface antigen, tumour size, tumour number, absence of capsule,
vascular invasion, TNM stage and BCLC stage, whilst the OS was
associated with the miR-197-3p expression level, tumour size,
tumour number, absence of capsule, vascular invasion, TNM stage and
BCLC stage (Table II). Furthermore,
multivariate Cox regression analysis revealed that independent risk
factors for HCC recurrence and survival included the miR-197-3p
expression level, tumour size, tumour number and vascular invasion,
while those for the OS in patients with HCC included the miR-197-3p
expression level, tumour size, tumour number, absence of capsule
and vascular invasion (Table III).
Based on the above results, it may be concluded that the expression
level of miR-197-3p may be adopted as an independent factor for
predicting the prognosis of HCC.
 | Table II.Univariable analysis for RFS and
OS. |
Table II.
Univariable analysis for RFS and
OS.
|
| RFS | OS |
|---|
|
|
|
|
|---|
| Variable | Hazard ratio | 95% CI | P-value | 95% CI | P-value |
|---|
| Age, years ≥55 vs.
<55 | 0.921 | 0.676–1.254 | 0.601 | 0.742–1.451 | 0.829 |
| Sex, male vs.
female | 0.925 | 0.559–1.531 | 0.763 | 0.845–3.070 | 0.147 |
| HBsAg, positive vs.
negative | 1.479 | 0.933–2.344 | 0.096 | 0.659–1.675 | 0.836 |
| AFP, µg/l, ≥20 vs.
<20 | 0.949 | 0.689–1.307 | 0.747 | 0.705–1.411 | 0.988 |
| Cirrhosis, present
vs. absent | 0.827 | 0.607–1.126 | 0.228 | 0.606–1.185 | 0.335 |
| Tumour diameter, cm
≥5 vs. <5 | 3.451 | 2.460–4.842 | <0.001 | 1.754–3.495 | <0.001 |
| Tumour number
multiple vs. solitary | 2.934 | 2.119–4.087 | <0.001 | 1.699–3.375 | <0.001 |
| Capsule absent vs.
present | 1.671 | 1.143–2.444 | 0.008 | 1.075–2.501 | 0.022 |
| Vascular invasion
present vs. absent | 3.602 | 2.578–5.033 | <0.001 | 2.526–5.229 | <0.001 |
| TNM stage III and
IV vs. I and II | 3.039 | 2.189–4.220 | <0.001 | 1.771–3.518 | <0.001 |
| BCLC stage C and D
vs. A and B | 3.129 | 2.250–4.352 | <0.001 | 1.755–3.486 | <0.001 |
| miR-197-3p
expression low vs. high | 1.940 | 1.418–2.656 | <0.001 | 1.388–2.759 | <0.001 |
 | Table III.Multivariable analysis of RFS and OS
in patients with hepatocellular carcinoma. |
Table III.
Multivariable analysis of RFS and OS
in patients with hepatocellular carcinoma.
|
| RFS | OS |
|---|
|
|
|
|
|---|
| Variable | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Tumour diameter cm,
≥5 vs. <5 | <0.001 | 2.286 | 1.580 | 3.306 | 0.053 | 1.466 | 0.995 | 2.161 |
| Tumour number
multiple vs. solitary | 0.001 | 1.877 | 1.317 | 2.676 | 0.001 | 1.874 | 1.275 | 2.753 |
| Vascular invasion
present vs. absent | <0.001 | 2.636 | 1.845 | 3.766 | <0.001 | 2.770 | 1.887 | 4.065 |
| miR-197-3p low vs.
high | 0.029 | 1.448 | 1.039 | 2.018 | 0.077 | 1.386 | 0.965 | 1.992 |
| Capsule absent vs.
present |
|
|
|
| 0.082 | 1.476 | 0.952 | 2.288 |
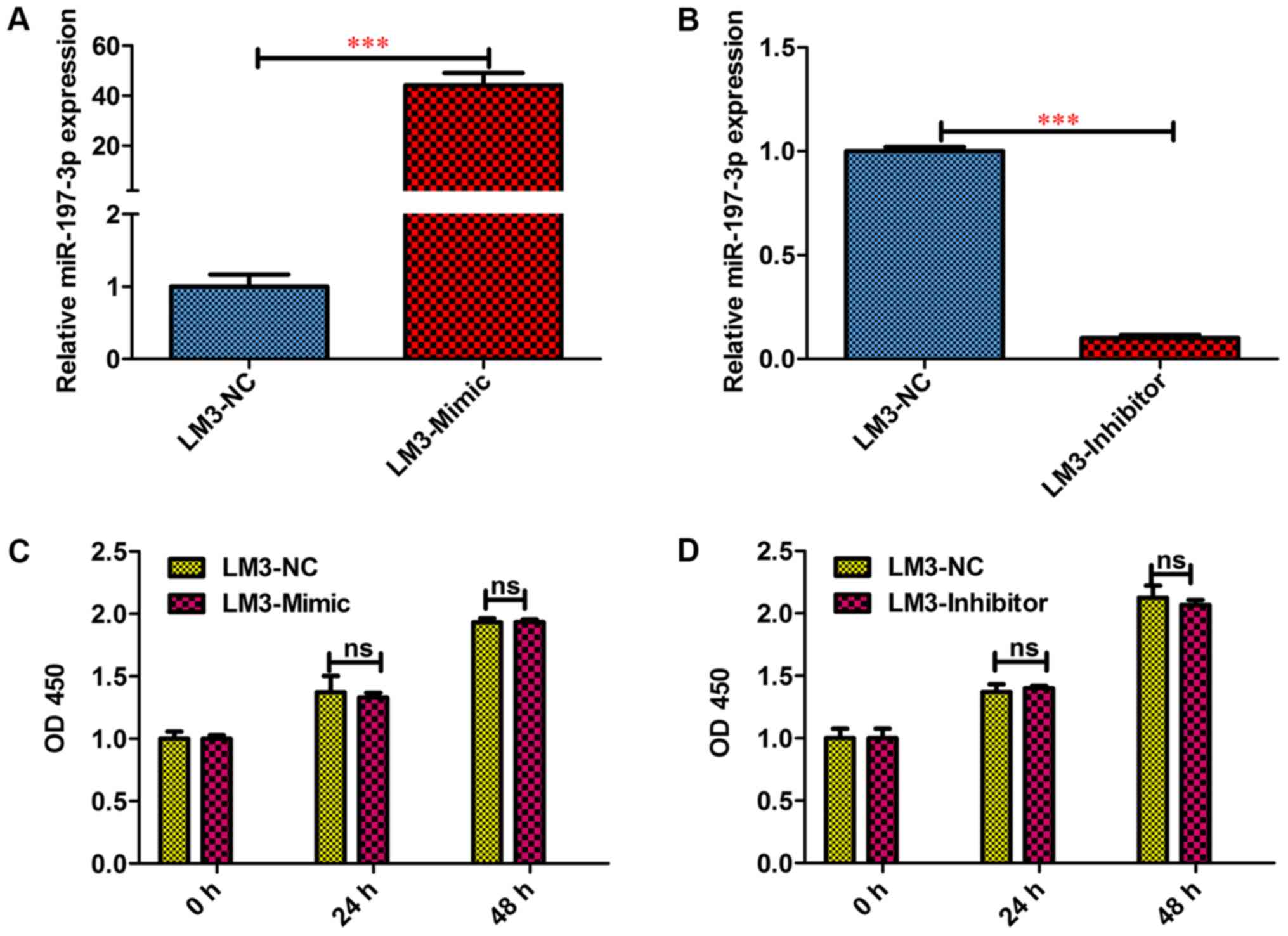
Inhibitory effects of miR-197-3p on
the metastasis and invasion of HCC cells in vitro
To functionally characterise miR-197-3p in HCC,
HCCLM3 cells were selected for a gain- of-function and
loss-of-function study. To this end, miR-197-3p
stably-overexpressing HCC-LM3-Mimic, and stably downregulated
HCC-LM3-Inhibitor cells were generated; the difference in the
miR-197-3p expression levels was detected via RT-qPCR (Fig. 3A and B). The MTT assay revealed that
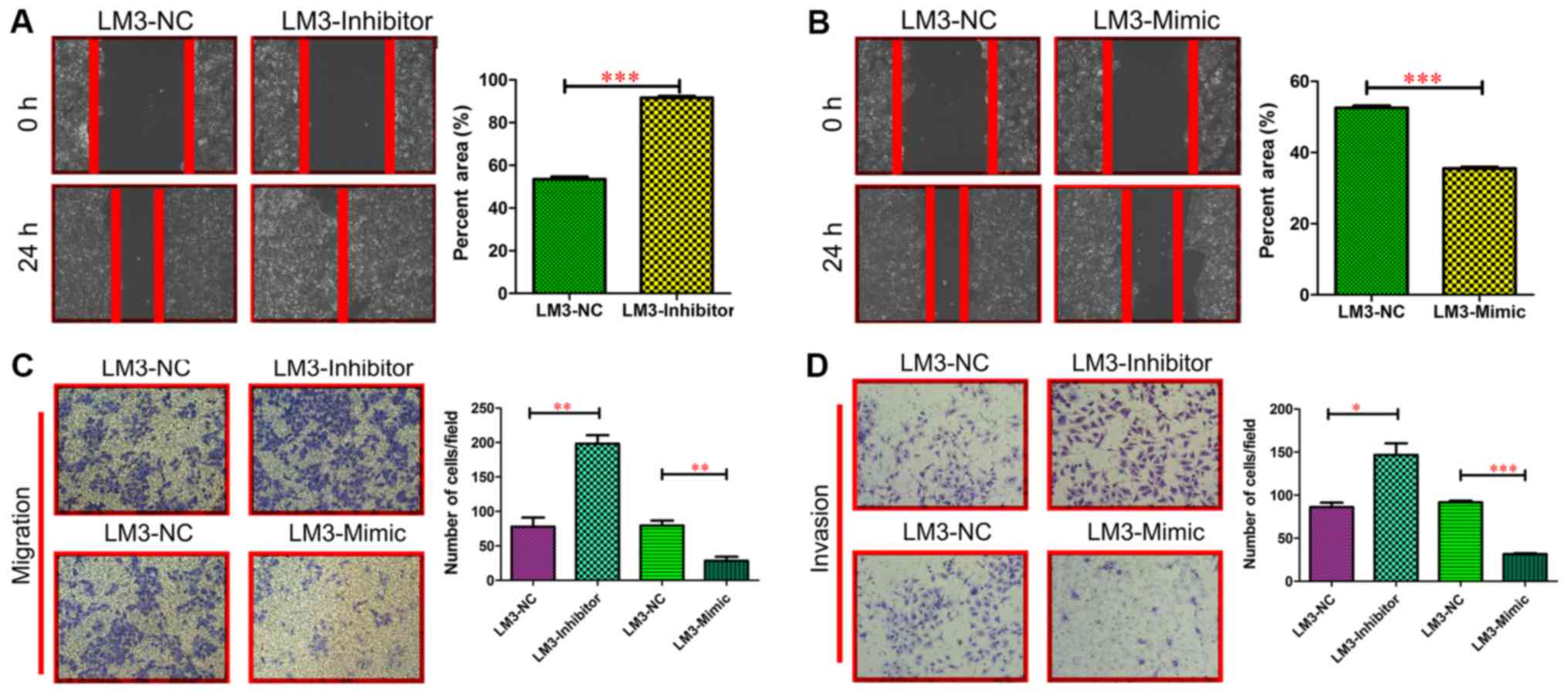
miR-197-3p did not affect the growth of HCC cells (Fig. 3C and D). In the wound-healing
migration assay, microscopic examination at 0 and 24 h revealed
that HCC-LM3-Inhibitor cell migration was significantly enhanced
compared with HCC-LM3-NC cell migration (P<0.001; Fig. 4A). By contrast, HCC-LM3-Mimic cell
migration was significantly delayed compared with HCC-LM3-NC cell
migration and invasiveness (P<0.001; Fig. 4B). The Transwell assay also revealed
that the HCC-LM3-Inhibitor cells displayed increased migration and
invasiveness, compared with the other cell types, whereas the
HCC-LM3-Mimic cells displayed decreased migration and invasiveness
compared with the other cell types (Fig.
4C and D). These data indicated that miR-197-3p may promote HCC
cell metastasis in vitro.
Direct binding of miR-197-3p to the 3′
UTR of ZIK1
To investigate the molecular mechanisms underlying
the miR-197-3p-mediated decrease in HCC metastasis, the involvement
of miR-197-3p in manipulating EMT was assessed, which is regarded
as a key process underlying cell metastasis. It was observed that
miR-197-3p exhibited no significant effect on the expression levels
of EMT-associated genes including E-cadherin, N-cadherin, and
vimentin (Fig. 5). To identify the
genes potentially involved in enhancing miR-197-3p expression in
HCC cell metastasis, TargetScan was used to predict the genes that
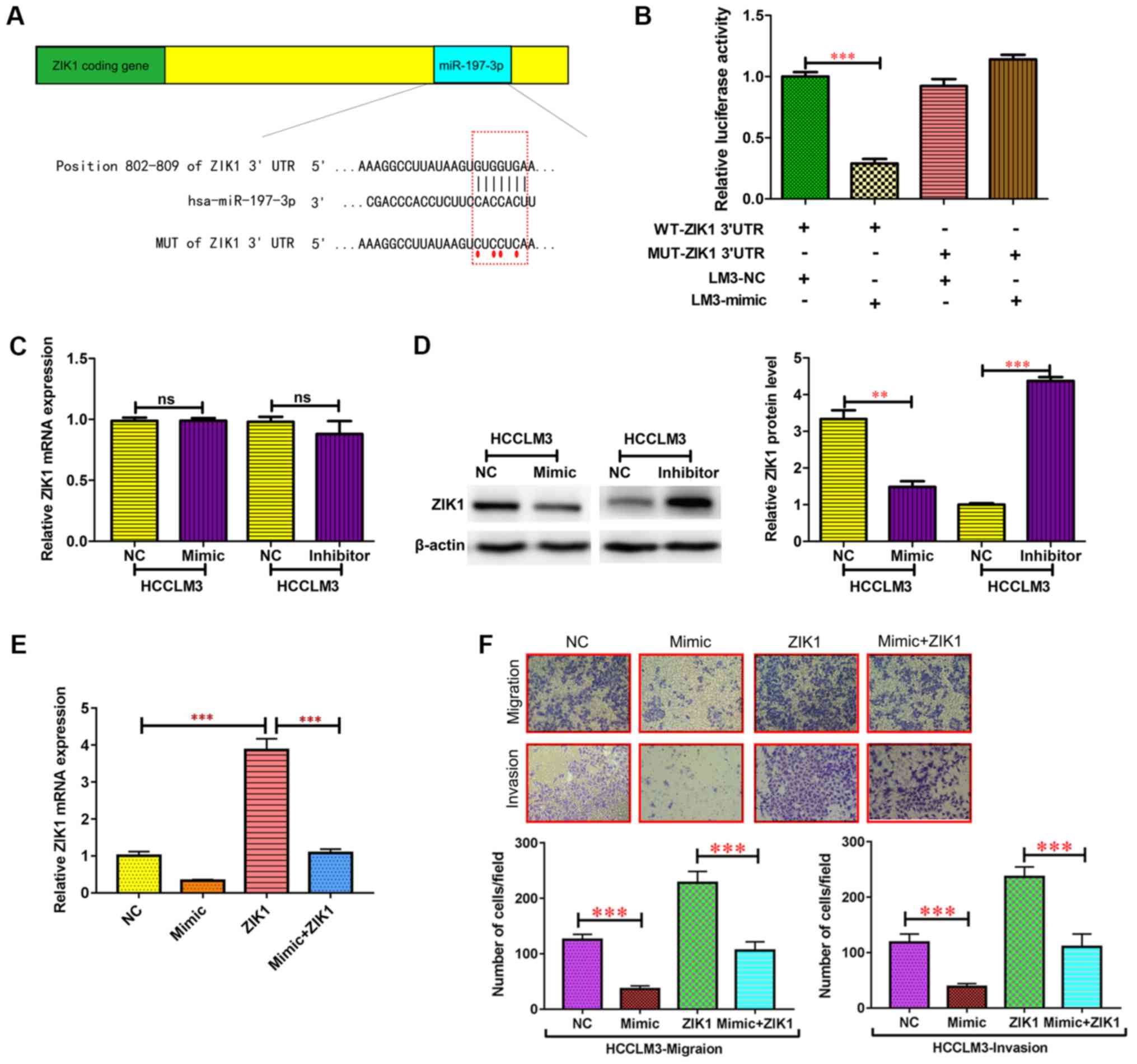
miR-197-3p may target. A putative miR-197-3p-binding site was
identified within the 3′UTR of ZIK1 (Fig.
6A), and a luciferase reporter assay was used to validate
whether ZIK1 was a direct target of miR-197-3p. wild-type (WT) and
mutant (MUT) versions of the ZIK1 3′ UTR, the latter containing
site-directed mutations in the putative miR-197-3p target sites,
were cloned into reporter plasmids. The miR-197-3p overexpression
markedly suppressed luciferase activity from the wild-type, though
not the mutant reporter. This suggested that the 3′ UTR of ZIK1 was
targeted by miR-197-3p, and that the point mutations in this
sequence abolished this effect in HCCLM3 (Fig. 6B). In addition, the protein expression
levels of ZIK1 were markedly reduced following miR-197-3p
overexpression, while the protein expression level of ZIK1 was
increased following miR-197-3p knockdown in HCCLM3 cells (Fig. 6D). By contrast, no significant
differences were observed in the ZIK1 mRNA expression levels
(Fig. 6C). These results indicated
that miR-197-3p may suppress ZIK1 expression through translational
repression.
To determine whether the ZIK1 gene is required for
the effect of miR-197-3p on HCC cell metastasis, ectopic
overexpression of ZIK1 was performed to conduct functional studies
in HCCLM3 cells (Fig. 6E). The
migration and invasion capacities in HCC cells overexpressing ZIK1
were significantly inhibited, while overexpression of ZIK1
abolished the effect of miR-197-3p on migration and invasion of HCC
cells (Fig. 6E). Therefore,
overexpression of ZIK1 abolished the effects of miR-197-3p on
specific HCC cell phenotypes.
Discussion
Although numerous growth factors, oncogenes and
tumour suppressor genes have been associated with
hepatocarcinogenesis (27,28), the molecular mechanisms and the
pathogenic biology of HCC remain unclear. In addition to reports
that miRNAs participates in the regulation of almost every cellular
process investigated to date, there have been reports that they are
involved in the pathogenesis of HCC (29,30).
miR-197-3p was recognized as an oncogene in prostate cancer
(31); however, the characterisation
of miR-197-3p in HCC and its association with cancer progression
and development remain largely unknown. In the present study,
miR-197-3p was downregulated in HCC tumour tissues and HCC cell
lines. miR-197-3p downregulation correlated with higher AFP levels
(≥20 µg/l), larger tumour sizes (≥5 cm), multiple tumours (n ≥2),
absence of capsule and vascular invasion. AFP is a well-established
diagnostic biomarker in the screening of HCC (32–35); an
online literature search revealed that the positive rate of AFP in
HCC patients was reportedly between 50 (36) and 53% (37). which declines to 47% for patients with
small hepatocellular carcinoma (38).
In the present study, 61% of the patients were in the early stages
of HCC (TNM stage I or II), and many had a small tumour volume;
therefore, the incidence of AFP appeared to be lower compared with
that in other reports. The correlation between the expression of
miR-197-3p and AFP levels indicated that miR-197-3p may be a
potential biomarker in HCC diagnosis. Additionally, its association
with tumour number and size, presence of capsule and vascular
invasion suggests a potential role of miR-197-3p in HCC.
MicroRNAs can function as tumour suppressors or
oncogenes by targeting their respectively associated genes
(39,40). miR-197-3p targets that may contribute
to miR-197-3p-mediated inhibition of cell metastasis in HCC were
further investigated. Using TargetScan bioinformatics, the ZIK1
gene was potentially identified as a direct target for miR-197-3p.
ZIK1 was first proposed as a transcriptional repressor that binds
to the nuclear ribonucleoprotein particle K protein (41), and was subsequently identified by
methylation sensitive representational difference analysis as being
hypermethylated in the intestinal metaplasia of the stomach
(42). In support of this finding,
Mihara et al (43) reported
that ZIK1 is hypermethylated in 100% of human gastric intestinal
metaplasia samples, which suggests that epigenetic silencing of
ZIK1 may contribute to the pathogenesis of gastric neoplasia. The
expression levels of ZIK1 are also reportedly associated with the
progression of renal cell carcinoma (44), diffuse large B-cell lymphoma (45), oesophageal cancer (46) and colorectal cancer (47,48);
Borinstein et al (48)
assessed the role of aberrant DNA methylation in the azoxymethane
rodent model of colon cancer and revealed that ZIK1 displayed
aberrant DNA methylation in the tumours that were absent or present
at a low frequency in the normal colon, which suggested that ZIK1
may serve an important role in the transformation of
gastrointestinal mucosal cells Moreover, ZIK1 was significantly
upregulated in cluster of differentiation (CD) 133+
colorectal carcinoma cells compared with their CD133−
counterpart. CD133 expression, which identified a considerable
population of tumour-initiating cells in colorectal cancer,
reportedly correlated with tumour metastasis (47). The level of ZIK1 expression was also
higher in renal cell carcinoma compared with the normal kidney
(44). However, despite the fact that
it is a zinc finger protein and a potential transcription factor,
little is known of its function. The present study verified that
miR-197-3p directly targets ZIK1 by interacting with its 3′UTR,
indicating that miR-197-3p may suppress HCC cell metastasis, in
part by targeting ZIK1.
In conclusion, the present study indicated that
miR-197-3p expression was frequently decreased in HCC tumour
tissues, and it may serve as a prognostic biomarker in patients
with HCC. The results indicated that miR-197-3p may inhibit HCC
cell metastasis by directly suppressing the expression of ZIK1,
which not only sheds new light on HCC progression and metastasis,
but provides a potential target for cancer prevention and
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
Basic Research Program of China, (grant no. 2014CB542102); The
Shanghai Health and Family Planning Commission Foundation (grant
no. 20164Y0189); The National Human Genetic Resources Sharing
Service Platform (grant no. 2005DKA21300); The Science Fund for
Creative Research Groups, NSFC, China (grant no. 81521091); and The
State Key Infection Disease Project of China (grant no.
2017ZX10203208).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WPZ and YY conceived the study and designed the
experiments. ZGW and MCW provided the experimental materials. JSN,
HZ, ZPH, YLO and YPT performed the experiments with the help of
YGH. JSN and HZ performed the data analysis. ZPH and YPT wrote the
manuscript. All authors contributed to the interpretation and
discussion of the results and reviewed the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the Eastern Hepatobiliary Surgery Hospital. All
patients gave written informed consent to participate in the study
and the data were anonymised.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RFS
|
relapse-free survival
|
|
OS
|
overall survival
|
|
BCLC
|
Barcelona Clinic Liver Cancer
|
|
EMT
|
epithelial mesenchymal transition
|
|
HCC
|
hepatocellular carcinoma
|
|
ZIK1
|
zinc finger protein interacting with K
protein 1
|
|
TNM
|
tumour node metastasis
|
|
AFP
|
alpha-fetoprotein
|
References
|
1
|
Ye JZ, Wang YY, Bai T, Chen J, Xiang BD,
Wu FX and Li LQ: Surgical resection for hepatocellular carcinoma
with portal vein tumor thrombus in the Asia-Pacific region beyond
the Barcelona Clinic Liver Cancer treatment algorithms: A review
and update. Oncotarget. 8:93258–93278. 2017.PubMed/NCBI
|
|
2
|
Orcutt ST and Anaya DA: Liver resection
and surgical strategies for management of primary liver cancer.
Cancer Control. 25:10732748177446212018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ren FH, Yang H, He RQ, Lu JN, Lin XG,
Liang HW, Dang YW, Feng ZB, Chen G and Luo DZ: Analysis of
microarrays of miR-34a and its identification of prospective target
gene signature in hepatocellular carcinoma. BMC Cancer. 18:122018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geng M, Xin X, Bi LQ, Zhou LT and Liu XH:
Molecular mechanism of hepatitis B virus X protein function in
hepatocarcinogenesis. World J Gastroenterol. 21:10732–10738. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Novikova MV, Khromova NV and Kopnin PB:
Components of the hepatocellular carcinoma microenvironment and
their role in tumor progression. Biochemistry (Mosc). 82:861–873.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoo S, Wang W, Wang Q, Fiel MI, Lee E,
Hiotis SP and Zhu J: A pilot systematic genomic comparison of
recurrence risks of hepatitis B virus-associated hepatocellular
carcinoma with low- and high-degree liver fibrosis. BMC Med.
15:2142017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang G, Fang X, Han M, Wang X and Huang Q:
MicroRNA-493-5p promotes apoptosis and suppresses proliferation and
invasion in liver cancer cells by targeting VAMP2. Int J Mol Med.
41:1740–1748. 2018.PubMed/NCBI
|
|
8
|
Lin H, Ewing LE, Koturbash I, Gurley BJ
and Miousse IR: MicroRNAs as biomarkers for liver injury: Current
knowledge, challenges and future prospects. Food Chem Toxicol.
110:229–239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loosen SH, Schueller F, Trautwein C, Roy S
and Roderburg C: Role of circulating microRNAs in liver diseases.
World J Hepatol. 9:586–594. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashmawy AM, Elgeshy KM, Abdel Salam ET,
Ghareeb M, Kobaisi MH, Amin HAA, Sharawy SK and Abdel Wahab AHA:
Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway
in hepatocellular carcinoma patients. Arab J Gastroenterol.
18:144–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Singaravelu R, Quan C, Powdrill MH, Shaw
TA, Srinivasan P, Lyn RK, Alonzi RC, Jones DM, Filip R, Russell RS
and Pezacki JP: MicroRNA-7 mediates cross-talk between metabolic
signaling pathways in the liver. Sci Rep. 8:3612018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang
Y, Ma R, Jiang YH and Sun NF: MicroRNA-199b-5p attenuates
TGF-β1-induced epithelial-mesenchymal transition in hepatocellular
carcinoma. Br J Cancer. 117:233–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu D, Hu Y, Xu W, Yu H, Yang N, Ni S and
Fu R: miR-203 inhibits the expression of collagen-related genes and
the proliferation of hepatic stellate cells through a
SMAD3-dependent mechanism. Mol Med Rep. 16:1248–1254. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Riazalhosseini B, Mohamed R, Apalasamy YD,
Langmia IM and Mohamed Z: Circulating microRNA as a marker for
predicting liver disease progression in patients with chronic
hepatitis B. Rev Soc Bras Med Trop. 50:161–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang S, Wang JQ and Lv XW: Exosomal miRNAs
as biomarkers in the diagnosis of liver disease. Biomark Med.
11:491–501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Slaby O, Laga R and Sedlacek O:
Therapeutic targeting of non-coding RNAs in cancer. Biochem J.
474:4219–4251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vettukattil JJ: Is the hepatic factor a
miRNA that maintains the integrity of pulmonary microvasculature by
inhibiting the vascular endothelial growth factor? Curr Cardiol
Rev. 13:244–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu
S and Meng JS: LINC00312 inhibits the migration and invasion of
bladder cancer cells by targeting miR-197-3p. Tumor Biol.
37:14553–14563. 2016. View Article : Google Scholar
|
|
19
|
Liu K, Huang W, Yan DQ, Luo Q and Min X:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 37(pii):
BSR201701092017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pirisi M, Leutner M, Pinato DJ, Avellini
C, Carsana L, Toniutto P, Fabris C and Boldorini R: Reliability and
reproducibility of the edmondson grading of hepatocellular
carcinoma using paired core biopsy and surgical resection
specimens. Arch Pathol Lab Med. 134:1818–1822. 2010.PubMed/NCBI
|
|
21
|
Chansky K, Sculier JP, Crowley JJ, Giroux
D, Van Meerbeeck J and Goldstraw P: International Staging Committee
and Participating Institutions: The international association for
the study of lung cancer staging project: Prognostic factors and
pathologic TNM stage in surgically managed non-small cell lung
cancer. J Thorac Oncol. 4:792–801. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong JH, Xiang BD, Gong WF, Ke Y, Mo QG,
Ma L, Liu X and Li LQ: Comparison of long-term survival of patients
with BCLC stage B hepatocellular carcinoma after liver resection or
transarterial chemoembolization. PLoS One. 8:e681932013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang RY, Chen L, Chen HY, Hu L, Li L, Sun
HY, Jiang F, Zhao J, Liu GM, Tang J, et al: MUC15 inhibits
dimerization of EGFR and PI3K-AKT signaling and is associated with
aggressive hepatocellular carcinomas in patients. Gastroenterology.
145:1436–1448, e1-e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shiraki K, Takase K, Tameda Y, Hamada M,
Kosaka Y and Nakano T: A clinical study of lectin-reactive
alpha-fetoprotein as an early indicator of hepatocellular carcinoma
in the follow-up of cirrhotic patients. Hepatology. 22:802–807.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li D, Mallory T and Satomura S: AFP-L3: A
new generation of tumor marker for hepatocellular carcinoma. Clin
Chim Acta. 313:15–19. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lv X, Li J and Yang B: Clinical effects of
miR-101 on prognosis of hepatocellular carcinoma and carcinogenic
mechanism of anti-miR-101. Oncol Rep. 36:2184–2192. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qadir MI and Rizvi SZ: miRNA in
hepatocellular carcinoma: Pathogenesis and therapeutic approaches.
Crit Rev Eukaryot Gene Expr. 27:355–361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Almas I, Afzal S, Idrees M, Ashraf MU,
Amin I, Shahid M, Zahid K and Zahid S: Role of circulatory
microRNAs in the pathogenesis of hepatitis C virus. Virusdisease.
28:360–367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Daniel R, Wu Q, Williams V, Clark G,
Guruli G and Zehner Z: A panel of MicroRNAs as diagnostic
biomarkers for the identification of prostate cancer. Int J Mol
Sci. 18(pii): E12812017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tayob N, Richardson P, White DL, Yu X,
Davila JA, Kanwal F, Feng Z and El-Serag HB: Evaluating screening
approaches for hepatocellular carcinoma in a cohort of HCV related
cirrhosis patients from the Veteran's Affairs Health Care System.
BMC Med Res Methodol. 18:12018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ricco G, Cavallone D, Cosma C, Caviglia
GP, Oliveri F, Biasiolo A, Abate ML, Plebani M, Smedile A, Bonino
F, et al: Impact of etiology of chronic liver disease on
hepatocellular carcinoma biomarkers. Cancer Biomark. 21:603–612.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luma HN, Eloumou SAFB, Okalla C,
Donfack-Sontsa O, Koumitana R, Malongue A, Nko'Ayissi GB and Noah
DN: Prevalence and characteristics of hepatitis delta virus
infection in a tertiary hospital setting in cameroon. J Clin Exp
Hepatol. 7:334–339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu WF, Fei YM, Zhou JK, Shen HJ, Chen XF,
Lv QQ and Ding YY: Significance of serum golgi protein 73 (GP73),
alpha-fetoprotein (AFP) and lectin-reactive alpha-fetoprotein
(AFP-L3) expresssion in primary hepatic carcinoma. Zhonghua Shi Yan
He Lin Chuang Bing Du Xue Za Zhi. 25:286–288. 2011.(In Chinese).
PubMed/NCBI
|
|
36
|
Kamiyama T, Takahashi M, Nakagawa T,
Nakanishi K, Kamachi H, Suzuki T, Shimamura T, Taniguchi M, Ozaki
M, Matsushita M, et al: AFP mRNA detected in bone marrow by
real-time quantitative RT-PCR analysis predicts survival and
recurrence after curative hepatectomy for hepatocellular carcinoma.
Ann Surg. 244:451–463. 2006.PubMed/NCBI
|
|
37
|
Hartmann JT, Rick O, Oechsle K, Kuczyk M,
Gauler T, Schöffski P, Schleicher J, Mayer F, Teichmann R, Kanz L
and Bokemeyer C: Role of postchemotherapy surgery in the management
of patients with liver metastases from germ cell tumors. Ann Surg.
242:260–266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang GT, Lee PH, Tsang YM, Lai MY, Yang
PM, Hu RH, Chen PJ, Kao JH, Sheu JC, Lee CZ and Chen DS:
Percutaneous ethanol injection versus surgical resection for the
treatment of small hepatocellular carcinoma: A prospective study.
Ann Surg. 242:36–42. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Y, Yang Z, Du F, Yang Q, Hou J, Yan X,
Geng Y, Zhao Y and Wang H: Molecular mechanisms of pathogenesis in
hepatocellular carcinoma revealed by RNA-sequencing. Mol Med Rep.
16:6674–6682. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morimoto A, Kannari M, Tsuchida Y, Sasaki
S, Saito C, Matsuta T, Maeda T, Akiyama M, Nakamura T, Sakaguchi M,
et al: An HNF4α-microRNA-194/192 signaling axis maintains hepatic
cell function. J Biol Chem. 292:10574–10585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Denisenko ON, O'Neill B, Ostrowski J, Van
Seuningen I and Bomsztyk K: Zik1, a transcriptional repressor that
interacts with the heterogeneous nuclear ribonucleoprotein particle
K protein. J Biol Chem. 271:27701–27706. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mihara M, Yoshida Y, Tsukamoto T, Inada K,
Nakanishi Y, Yagi Y, Imai K, Sugimura T, Tatematsu M and Ushijima
T: Methylation of multiple genes in gastric glands with intestinal
metaplasia: A disorder with polyclonal origins. Am J Pathol.
169:1643–1651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao Q, Kun D, Hong B, Deng X, Guo S, Tang
X, Yang Y, Gong K, Li Q, Ye L, et al: Identification of novel
proteins interacting with vascular endothelial growth inhibitor 174
in renal cell carcinoma. Anticancer Res. 37:4379–4388.
2017.PubMed/NCBI
|
|
45
|
Liu P, Jiang W, Zhao J and Zhang H:
Integrated analysis of genome-wide gene expression and DNA
methylation microarray of diffuse large B-cell lymphoma with TET
mutations. Mol Med Rep. 16:3777–3782. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oka D, Yamashita S, Tomioka T, Nakanishi
Y, Kato H, Kaminishi M and Ushijima T: The presence of aberrant DNA
methylation in noncancerous esophageal mucosae in association with
smoking history: A target for risk diagnosis and prevention of
esophageal cancers. Cancer. 115:3412–3426. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim ST, Sohn I, Do IG, Jang J, Kim SH,
Jung IH, Park JO, Park YS, Talasaz A, Lee J and Kim HC:
Transcriptome analysis of CD133-positive stem cells and prognostic
value of survivin in colorectal cancer. Cancer Genomics Proteomics.
11:259–266. 2014.PubMed/NCBI
|
|
48
|
Borinstein SC, Conerly M, Dzieciatkowski
S, Biswas S, Washington MK, Trobridge P, Henikoff S and Grady WM:
Aberrant DNA methylation occurs in colon neoplasms arising in the
azoxymethane colon cancer model. Mol Carcinog. 49:94–103.
2010.PubMed/NCBI
|