Introduction
Lung cancer is one of the most common malignancies,
and the leading cause of cancer-associated mortality worldwide. The
mortality of lung cancer has decreased over the past decade, and
the current 5-year survival rate is ~18%. These alterations are
attributed to a decrease in tobacco use, the progress in lung
cancer screening with low-dose computed tomography, and
improvements in treatment in recent years (1). Novel targeted treatments for advanced
non-small-cell lung cancer (NSCLC) based on driver mutations and
immune checkpoints are continuously being developed (2–4).
Epidermal growth factor receptor (EGFR) mutations are the most
frequent driver mutations detected in lung adenocarcinomas and
exhibit clinically therapeutic implications. The occurrence of
these mutations varies between races, ranging from ~14% in
Caucasian patients to almost 50% in Asian patients, particularly
those with adenocarcinoma (5,6).
Currently, the detection of EGFR mutations relies mainly on tissue
specimens, and the identification of a mutation-positive tissue is
the premise of targeted therapy. Tests examining circulating tumor
DNA (ctDNA) are also used in the clinic. An early report detected
EGFR mutations in circulating tumor cells (CTCs) using a
microfluidic device, and achieved high rates of detecting
EGFR-activating mutation (7).
In the present study, a fast, simple CTC isolation
technique was established based on the CellSearch®
principle (8,9), which may simplify the verification of
the EGFR status in CTCs. The status of EGFR mutations was evaluated
by isolating blood epithelial cells from patients with advanced
NSCLC prior to treatment with tyrosine kinase inhibitors (TKIs).
The isolation and analysis of a few mixed CTCs significantly
improved the detection rate of EGFR mutations compared with
detection using single CTCs, and isolated CTCs were potentially
useful for analyzing the heterogeneity of mutations.
Materials and methods
Patients and ethics
The study was approved by the Beijing Chest Hospital
Committee on the Use of Humans as Experimental Subjects (Beijing
China). In addition, all participants provided written informed
consent. Patients were diagnosed with biopsy-validated lung cancer
in clinical phase III or IV. The 57 male and 34 female patients had
an age range of 32 to 87 years and a mean age of 62.3 years. Blood
samples (4.5 ml) were collected into Vacuette EDTA anticoagulant
tubes (Greiner Bio-One GmbH, Kremsmünster, Austria) from 91
untreated patients with lung cancer at the Beijing Chest Hospital,
and processed within 2 h using an autoMACS Pro instrument (Miltenyi
Biotec GmbH, Bergisch Gladbach, Germany).
CTC enrichment and single-cell
retrieval
Erythrocyte lysates were prepared from blood samples
using BD FACS Lysing Solution (BD Biosciences, San Jose, CA, USA).
Following red blood cell depletion, the Fc receptor reagent (cat.
no. 120-000-442; Miltenyi Biotec GmbH) and cluster of
differentiation CD326 microbeads (cat. no. 130061101; Miltenyi
Biotec GmbH) were used according to the manufacturer's protocol.
The cell suspension was subsequently loaded onto an autoMACS Pro
instrument, and CD326+ and CD326- fractions were obtained. The
CD326+ cell fraction was stained with anti-epithelial cell adhesion
molecule (EpCAM)-phycoerythrin (cat. no. 565399; BD Biosciences),
three types of anti-cytokeratin-fluorescein isothiocyanate antibody
(cat. no. ab870010, Abcam, Cambridge, UK; cat. no. ab72813, Abcam;
cat. no. 53-9898, eBioscience; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), anti-CD45allophycocyanin antibodies (cat. no.
ab28106; Abcam), and Hoechst 33342 (cat. no. C0030; Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) to
distinguish cancerous cells from healthy leukocytes. Individual
CTCs (EpCAM+, Hoechst+, anti-cytokeratin+, anti-CD45-) were
isolated from leukocytes (anti-CD45+, Hoechst+, anti-cytokeratin-,
EpCAM-) using a fluorescence microscope (Nikon Corporation, Tokyo,
Japan) with a robotic micromanipulator (CellEctor; Molecular
Machines & Industries GmbH, Eching, Germany). A total of three
commercially available lung cancer adenocarcinoma cell lines
(NCI-H2009, NCI-H1975, and HCC-827) were purchased from The
American Type Culture Collection (Manassas, VA, USA) for spiking
experiments and mutation analyses. For spiking experiments, CTCs
were picked up by applying CellEctor and spiked into 4.5 ml
blood.
DNA amplification
The selected CTCs were used for whole-genome
amplification (WGA) and quantification using a REPLI-g®
Single Cell kit (cat. no. 150343; Qiagen GmbH, Hilden, Germany).
The procedure was performed according to the manufacturer's
protocol. DNA was quantified using a NanoDrop 2000c
spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA), according to the manufacturer's protocol. The amplified DNA
was verified by polymerase chain reaction (PCR) using the Multiplex
PCR kit (cat no. 206143; Qiagen GmbH) and eight pairs of primers
specific for housekeeping genes were designed. The primers used
were follows: CYB5A forward, 5′-GGCAACGCTTAGACTCTGTGTG-3′ and
reverse, 5′-CTGCCCTTGGCCTAACTAACCT-3′; PRPH forward,
5′-GTTCCTCAAGAAGCTGCACGAG-3′ and reverse,
5′-CGTTAGACTCTGGATCTGGCGT-3′; GABARAPL2 forward,
5′-CCAGCCAATTCATGAGTCGGTG-3′ and reverse,
5′-CCTGACAACTCGCAAGTAGCAC-3′; ACTG1 forward,
5′-GCTCAATGGGGTACTTCAGGGT-3′ and reverse,
5′-GTGGACGTTACGTAAAAGGCCC-3′; NDUFA7 forward,
5′-TGCTCTGGATGTGAAGATGCCA-3′ and reverse,
5′-TTCCAGGTAAATCCAGCCCAGG-3′; UQCRC1 forward,
5′-CAGCCAGTCAGCATCATCCAAC-3′ and reverse,
5′-GAAAGCCGGATTGCGGTAACAT-3′; MYC forward,
5′-GGATAGCTCTGCAAGGGGAGAG-3′ and reverse,
5′-TCGTCGCAGTAGAAATACGGCT-3′; and MIF forward,
5′-AGAAGTCAGGCACGTAGCTCAG-3′ and reverse,
5′-GGCACGTTGGTGTTTACGATGA-3′.
Sequence and copy number variation
(CNV) analyses
For the analysis of CNVs, DNA was quantitatively
amplified from 2.5 ng/ml WGA products for library preparation and
barcoded with a Nextera Index kit (Illumina, Inc., San Diego, CA,
USA). The low-quality reads were removed from the obtained data
using the Illumina procedure following data quality control, and
Burrows-Wheeler Alignment Tool (version 0.7.15) (10,11) was
used to compare the sequences to the human genome (hg19). The data
normalized by unique reads were considered CNVs. Our test samples
used white blood cells from the same patient as a reference for
comparison.
EGFR mutation detection
EGFR mutations were detected using a quantitative
fluorescence PCR instrument (Cobas z480; Roche Diagnostics, Basel,
Switzerland) and an EGFR detection kit (human EGFR gene mutations
fluorescence PCR diagnostic kit; Amoy Diagnostics Co., Ltd.,
Xiamen, China) according to the manufacturers' protocols. EGFR
mutations were also analyzed by digital PCR (dPCR; ABI-QuantStudio
3D, Applied Biosystems; Thermo Fisher Scientific, Inc.), according
to the protocols for the Dual Flat Block GeneAmp PCR System 9700
(Thermo Fisher Scientific, Inc.); dPCR primers and probes (cat no.
A44177; Assay ID, HS000000026_rm; Thermo Fisher Scientific, Inc.)
for EGFR mutation detection were purchased from Applied Biosystems.
QuantStudio™ 3D AnalysisSuite™ software version 3.1.2 (Thermo
Fisher Scientific, Inc.) was used for data acquisition and
analysis.
Statistical analyses
Counts were compared using the χ2 test.
Continuous data were compared using Student's t-test if the
distribution of samples was normal, or nonparametric tests (the
Mann-Whitney U test) for comparisons between two groups. For each
test, two-sided P<0.05 was considered to indicate a
statistically significant difference. All statistical manipulations
were performed using the SPSS version 18 for Windows software
system (SPSS, Inc., Chicago, IL, USA).
Results
A simplified technique to yield
high-purity epithelial tumor cells from blood
An H2009 lung cancer cell line that stably expressed
both EpCAM and cytokeratins (CKs) was selected for spiking
experiments to establish an approach for isolating tumor cells from
blood. The aim was to optimize the collection of pure CTCs for
further molecular characterization at the individual cell level. An
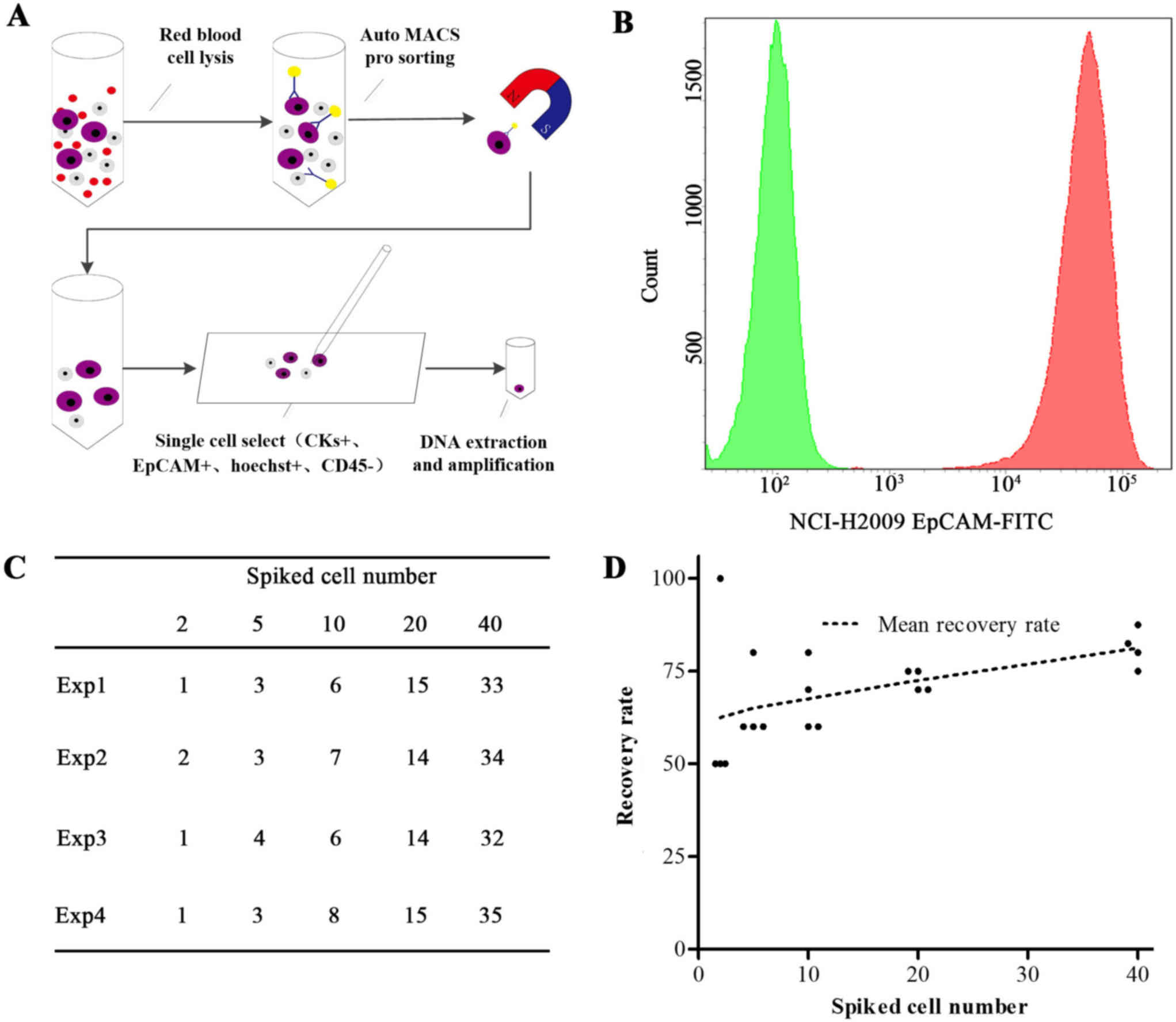
illustration of the CTC capture method is displayed in Fig. 1A and includes the following steps: i)
Ammonium chloride-based red blood cell lysis; ii) enrichment of
disseminated cancer cells using magnetically coated anti-EpCAM
beads; and iii) micromanipulation to obtain single cells. In the
present study, two epithelium-specific staining procedures (using
EpCAM and CK antibodies) were performed to identify cancer cells,
including EpCAM bead enrichment followed by staining with a
monoclonal antibody against EpCAM that bound well to the cancer
cell surface (Fig. 1B). The recovery
rates of H2009 cells (≥1 cell) ranged from 62.5–83.8% in four
independent experiments (Fig. 1C and
D) and increased when increasing the number of spiked cancer
cells via the precise recovery of cells using micromanipulation.
Similar capture efficiencies were achieved using cell lines
exhibiting relatively low EpCAM expression levels, including the
lung cancer cell lines H1975 and HCC-827. Using EpCAM and CK double
staining, epithelial marker-positive cells were not detected in
samples from 20 healthy subjects, from 10 patients with
tuberculosis and from 10 patients with other nonmalignant lung
diseases. This protocol was subsequently used to obtain rare CTCs
from clinical samples.
Efficient real-time isolation of CTCs
from patients with advanced lung cancer
A total of 91 patients with advanced lung cancer
were enrolled for the CTC analysis as described in this study,
between March 2016 and October 2017. Patient characteristics were
recorded, and ages ranged from 32–83 years; 63% were male and 66%
had adenocarcinoma. Patients with TNM stage III and IV diseases
represented 18 and 82% of the lung cancer cohort, respectively.
Among the total population, 56 patients (61.5%)
exhibited positive CTC counts at baseline, with the detection of at
least one CTC in 4.5 ml of peripheral blood (range, 1 to 29; median
2; mean 4.60). All identified cells were single CTCs, and no
clusters were detected. Representative images of CTCs from
individual patients are illustrated in Fig. 2. The prevalence of CTCs prior to
treatment and the association with patient characteristics are
listed in Table I. i) Stage IV NSCLC
was significantly associated with higher CTC numbers (Mann-Whitney
test, P=0.0081). At least one CTC was detected in 52 (69.3%) of the
75 patients with stage IV disease. In addition, at least one CTC
was detected in four (25.0%) of the 16 patients with stage III
NSCLC (Fig. 3), and CTCs were also
detected in one of the four patients with stage IIIa NSCLC, in whom
two CTCs were captured. Detectable CTCs, albeit at lower numbers,
positively correlated with bone and brain metastases compared with
metastatic disease at other sites. Among 56 patients with CTCs,
patients with bone, brain, liver and kidney metastasis accounted
for 31/56 (55.3%), 25/56 (44.6%), 15/56 (26.8%) and 9/56 (16.1%) of
the patients, respectively. Furthermore, the number of CTCs
detected was specifically associated with bone metastasis, and the
mean number of CTCs detected in patients with bone metastases was
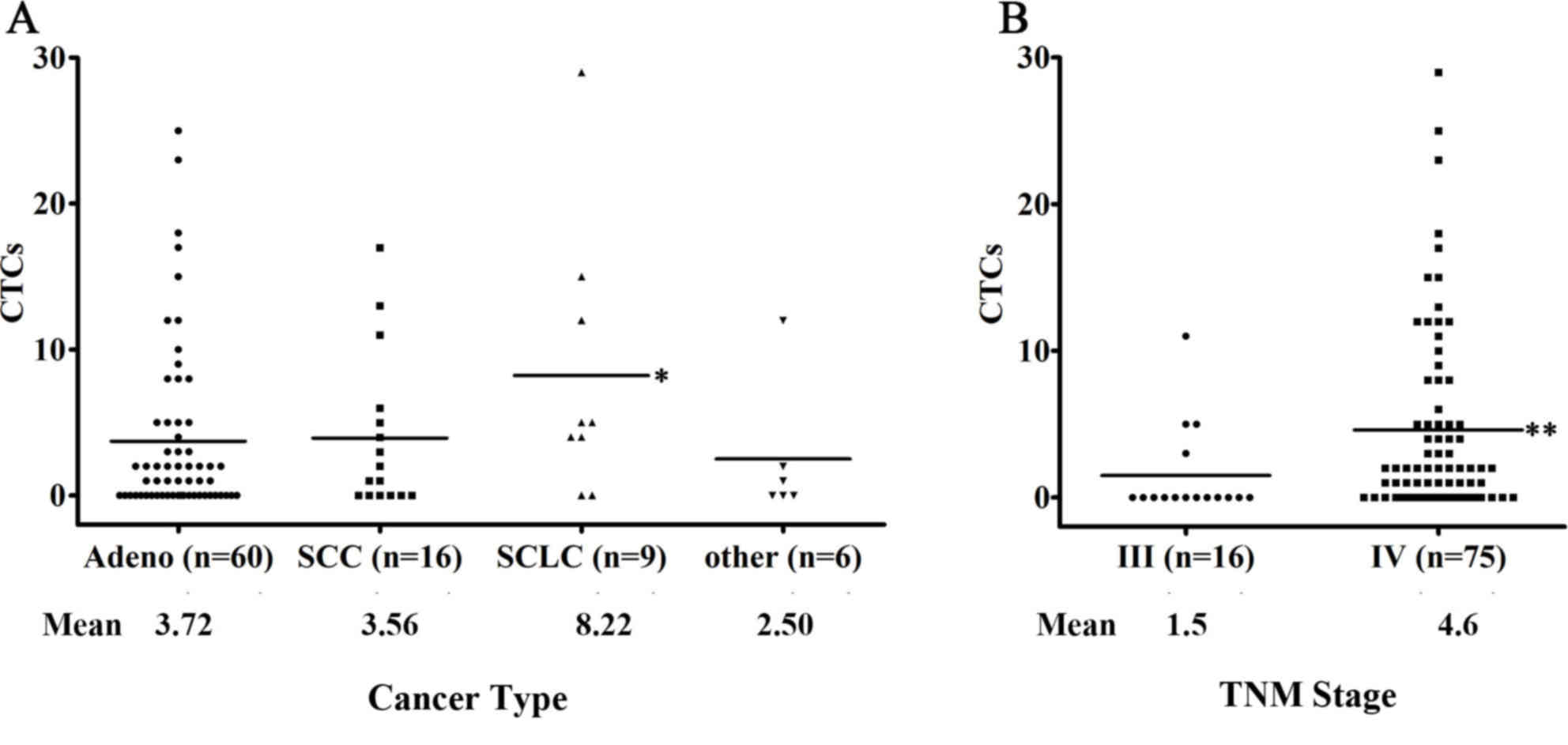
5.7. ii) According to the histological features, patients with
small-cell lung cancer (SCLC) displayed the highest CTC counts and
a CTC detection rate of 77.8% (7 of 9 patients; median, 5; mean,
8.22) compared with patients with NSCLC or other types of
metastatic diseases (P>0.05; Fig.
3A). iii) No significant association was observed between a
higher CTC number and tumor burden or smoking status; however, a
clear trend toward a correlation between an increased CTC detection
rate and increased tumor diameter was observed (<3 cm, 48.7% vs.
≥3 cm, 71.1%; P=0.078).
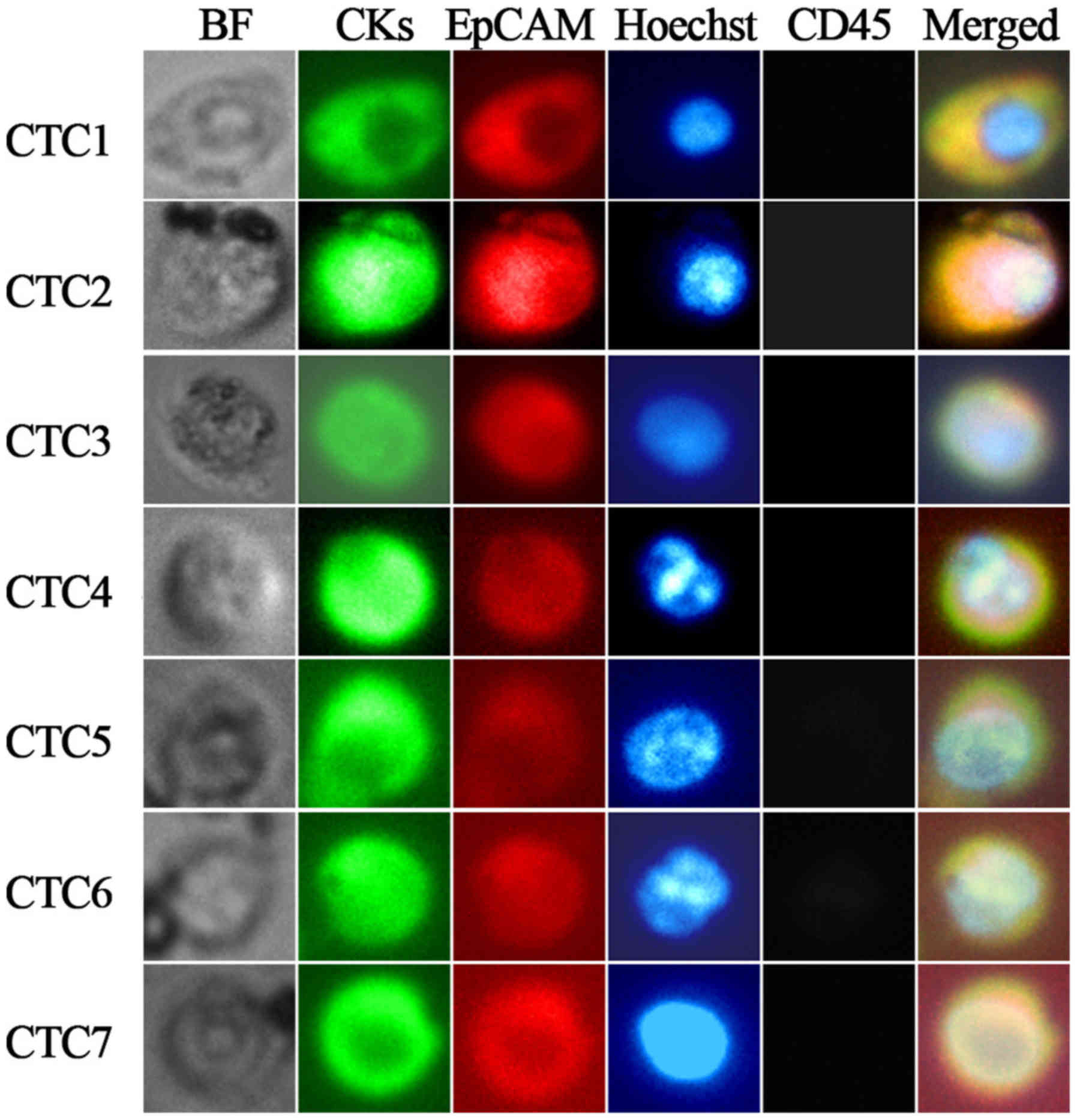 | Figure 2.Representative images of individual
CTCs isolated from 4.5 ml of blood from patients with advanced lung
cancer (magnification, ×400). CTCs 1–4 were captured from four
different patients with lung cancer; CTCs 5–7 were captured from
the same patient with lung cancer. Tumor cells were identified as
EpCAM-positive (red-PE), pan-CK-positive (green-FITC),
CD45-negative (gray-APC) and Hoechst 33342-positive (blue) cells.
CTC, circulating tumor cell; EpCAM, epithelial cell adhesion
molecule; CK, cytokeratin; PE, phycoerythrin; FITC, fluorescein
isothiocyanate; APC, allophycocyanin; BF, bright field; CD, cluster
of differentiation. |
 | Table I.Correlation between captured CTC
counts and clinical characteristics among patients with lung cancer
prior to treatment. |
Table I.
Correlation between captured CTC
counts and clinical characteristics among patients with lung cancer
prior to treatment.
|
|
| CTC count (%) |
|---|
|
|
|
|
|---|
| Factor | Patients with CTC
(Cutoff ≥1) (%) | 0 | 1 | 2-3 | 4–5 | 6–10 | >10 |
|---|
| All patients
(n=91) | 56 (61.5) | 35 (38.5) | 10 (10.99) | 15 (16.5) | 11 (12.1) | 6 (6.6) | 14 (15.4) |
| C-stage |
| III
(n=16) | 4 (25.0) | 12 (75) | 0 (0) | 1 (6.3) | 2 (12.4) | 0 (0) | 1 (6.3) |
| IV
(n=75) | 52 (69.3) | 23 (30.7) | 10 (13.3) | 14 (18.7) | 9 (12) | 6 (8) | 13 (17.3) |
| Histological cell
type |
| Adeno
(n=60) | 36 (60.0) | 24 (30) | 7 (11.7) | 12 (20) | 5 (8.3) | 5 (8.3) | 7 (11.7) |
| SCC
(n=16) | 10 (62.5) | 6 (37.5) | 2 (12.5) | 2 (12.5) | 2 (12.5) | 1 (6.3) | 3 (18.7) |
| SCLC
(n=9) | 7 (77.8) | 2 (22.2) | 0 (0) | 0 (0) | 4 (44.4) | 0 (0) | 3 (33.3) |
| Other
(n=6) | 3 (50.0) | 3 (50) | 1 (33.3) | 1 (33.3) | 0 (0) | 0 (0) | 1 (33.3) |
| Smoking
history |
| Yes
(n=61) | 36 (59.0) | 25 (41) | 3 (4.9) | 11 (18.0) | 8 (13.1) | 5 (8.2) | 9 (14.8) |
| No
(n=30) | 20 (66.7) | 10 (33.3) | 7 (23.3) | 4 (13.3) | 3 (10) | 1 (3.3) | 5 (16.7) |
| Tumor size |
| <3
cm (n=39) | 19 (48.7) | 20 (51.3) | 4 (10.3) | 5 (12.8) | 2 (2.2) | 3 (5.1) | 5 (12.8) |
| >3
cm (n=52) | 37 (71.1) | 15 (28.9) | 6 (11.5) | 10 (19.2) | 9 (17.3) | 3 (5.8) | 9 (17.3) |
CNV analysis and single-cell EGFR
mutation detection
The use of CTCs as a real-time liquid biopsy has
attracted increasing attention. Accurate and reliable techniques
for the capture of purified CTCs from peripheral blood are
therefore required to facilitate the clinical utility of CTC
analyses. WGA was performed for DNA amplification and to identify
the genomic features of CTCs. The DNA was subsequently subjected to
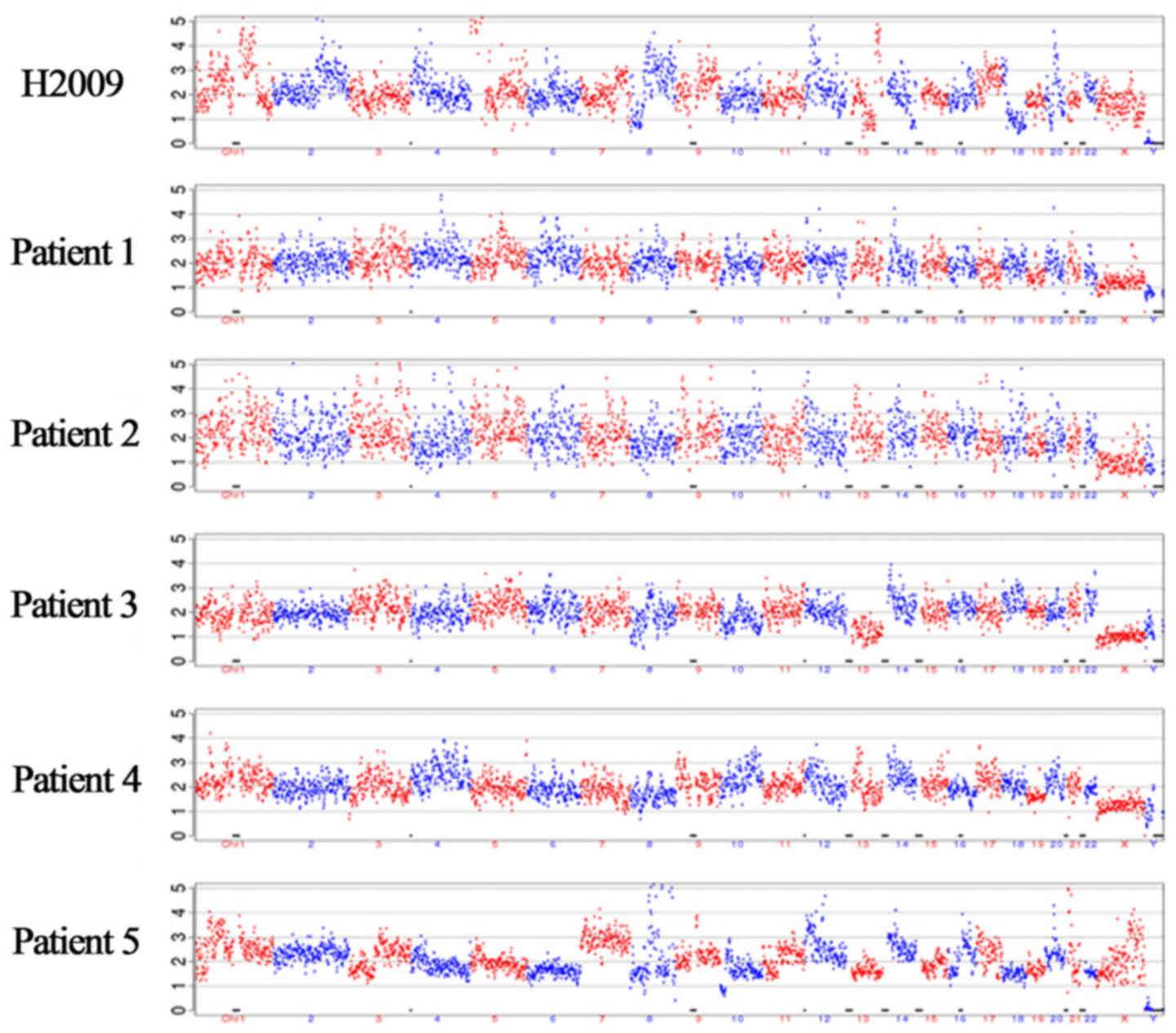
CNV analyses. Multiple chromosomal variations were observed in the
H2009 lung cancer cell line at the single-cell level, compared with
those in normal white blood cells. Representative CNV profiles of
five patients are illustrated in Fig.
4. Tumor-characteristic CNVs were detected in four patients
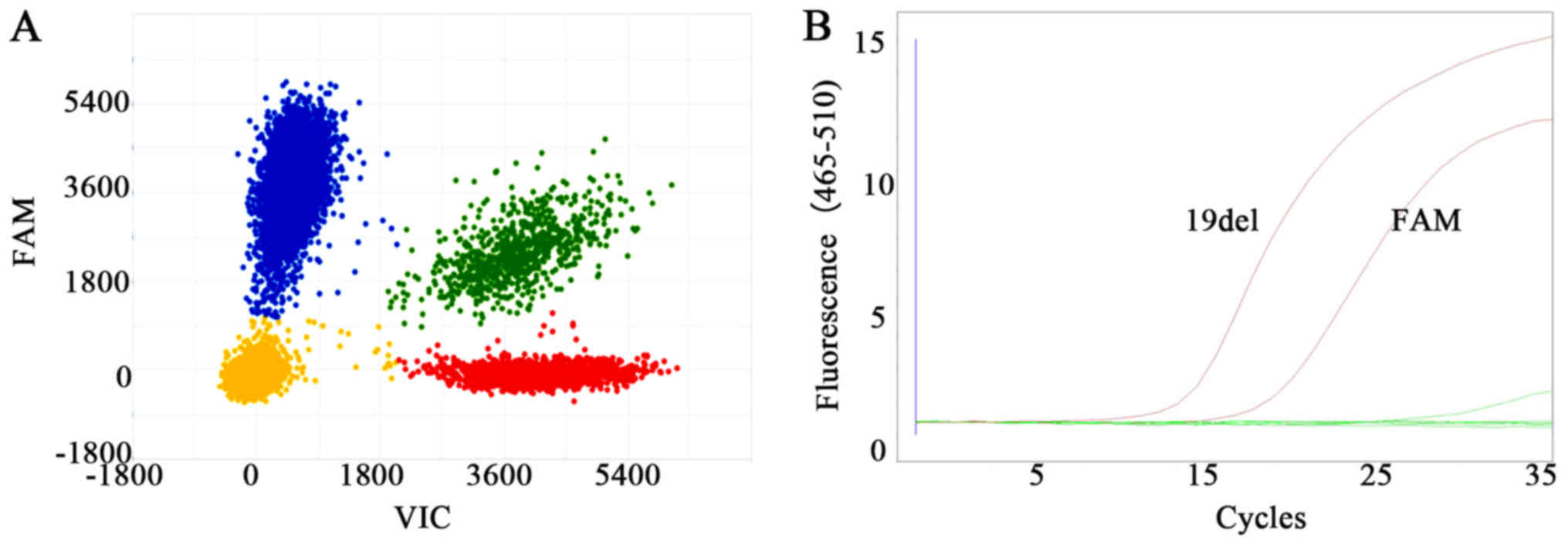
using 1–10 CTCs. Based on the sensitivity of 1/1,000 copies for
dPCR and 1/100 copies for the amplification refractory mutation
system (ARMS) procedures, our study confirmed that both dPCR- and
ARMS-based assays were suitable for analyzing mutations in single
H1975 or HCC-827 cells, as displayed in Fig. 5. A total of three other lung cancer
cell lines were also used to verify that dPCR- and ARMS-based
approaches were effective for the detection of expected EGFR
mutations (Table II).
 | Table II.Lung cancer cell lines with the
expected EGFR mutations and the EGFR mutations detected by digital
polymerase chain reaction or amplification refractory mutation
system. |
Table II.
Lung cancer cell lines with the
expected EGFR mutations and the EGFR mutations detected by digital
polymerase chain reaction or amplification refractory mutation
system.
|
| EGFR mutation |
|
|---|
|
|
|
|
|---|
| Cell lines | Expected | Detected | Histological cell
type |
|---|
| NCI-H2009 | Wild-type | Wild-type | Adeno |
| HCC-827 | 19del | 19del | Adeno |
| NCI-H1975 | L858RT790M | L858RT790M | Adeno |
Homogeneity analysis of CTC-EGFR
mutations
Initially, EGFR mutations were analyzed in single
CTCs following WAG; two single cells carried the same EGFR
mutations as the primary tumor among 12 single cells derived from
different patients using dPCR- and ARMS-based detection methods,
indicating a 16.67% sensitivity. When samples containing more CTCs
(10 CTCs from four patients) were analyzed, all the samples
displayed clear EGFR mutation status, consistent with the tumor
tissue, including two patients with the L858R mutation, one patient
with the 19del mutation, and one patient with a non-activating
mutation. The two samples with the L858R mutation were further
confirmed by dPCR.
Inconsistent rare EGFR mutations are
identifiable in CTCs
In addition to the four patients described above,
EGFR mutations were also detected in the remaining four samples,
with CTC numbers ranging between three and nine cells. Among them,
two samples yielded identical results to those of the tissue, while
one sample contained four CTCs from the patient (patient 2) that
were detected as wild type, which was a different result from the
tissue pathological examination that revealed a 19del mutation.
Another sample from patient 4 contained three CTCs in which a
single new G791× mutation was detected that differed from the L858R
mutation that was observed in this patient's tumor tissue (Table III).
 | Table III.Analysis of the consistency of EGFR
mutations among various CTCs and tissue specimens. |
Table III.
Analysis of the consistency of EGFR
mutations among various CTCs and tissue specimens.
|
|
|
| EGFR mutation
(ARMS) |
|
|---|
|
|
|
|
|
|
|---|
| Patient | CTC sample | CTC number | Tissue | CTC | Matched |
|---|
| 1 | 1 | 10 | L858R | L858R | Yes |
|
| 2 | 9 | L858R | L858R | Yes |
| 2 | 1 | 10 | 19del | 19del | Yes |
|
| 2 | 4 | 19del | N | No |
| 3 | 1 | 10 | N | N | Yes |
|
| 2 | 3 | N | N | Yes |
| 4 | 1 | 10 | L858R | L858R | Yes |
|
| 2 | 3 | L858R | G719X | No |
Discussion
A number of promising techniques, including the use
of microfluidic and microarray devices, may potentially enrich CTCs
with greater sensitivity. However, these techniques have not been
approved for clinical use, and their utility in the discovery of
new CTC markers is in the early stages of development. The
CellSearch® system has undergone extensive development
and is an accredited method for analyzing CTCs (12,13).
Current technical challenges include methods of increasing CTC
purity and isolating single CTCs for molecular analysis.
Researchers universally acknowledge that no single marker is able
to reliably identify CTCs. Mainstream techniques continue to focus
on utilizing common epithelial cell-specific markers, including
EpCAM and CKs, which were introduced as a basic principle by the
CellSearch® system. A key issue in conducting the
present study was isolating high-quality CTCs. Therefore, the aim
was to isolate CTCs from blood cells based on epithelial
cell-specific markers. The system would need to be simple and
automated for widespread clinical use; however, manual CTC
retrieval under the microscope was an essential step to ensure the
capture of single or pure CTCs. The CTC isolation technique was
tested using 4.5 ml of blood in spiking experiments and exhibited a
recovery rate between 62.5–83.8%, more efficient than that of
previous reports on CTC detection using 7.5 ml blood (8). This increase in capture efficiency may
be attributed to double staining for EpCAM and CKs, as EpCAM is not
expressed by all carcinomas, and as many as 30% express low levels
of EpCAM. Moreover, the expression levels of CKs vary among
different histological subtypes (14,15).
Simultaneous staining with multiple epithelial markers and
confirmation of negative staining for leukocyte markers such as
CD45, has become the standard for reliably identifying CTCs
(16). Additionally, CTCs with low
EpCAM expression have also been detected using a similar capture
system (17). Among patients with
lung cancer, the CTC count significantly increased in advanced
tumor, such as SCLC and tumor progression, particularly with the
development of distant metastasis. These findings are consistent
with previous reports (18,19).
The CellSearch® system has indicates
promise for reliable and reproducible CTC enumeration in the
clinic, and it has been approved for the evaluation of lung cancer.
Patients without clinically detectable distant metastases will
likely exhibit low CTC detection rates (8). Therefore, 91 patients with stage III
and IV diseases were enrolled to evaluate the efficacy of a CTC
detection protocol. In general, the results were similar to
previously reported findings; the CTC count significantly increased
with tumor progression, with a detection rate of 69.3% among
patients with advanced stage IV NSCLC (18,19), and
CTCs were most abundant in patients with SCLC, as CTCs were
detected in 77.8% of patients with this type of cancer (20). A clear trend towards an increased CTC
detection rate with increased tumor size was also observed,
although the association between higher CTC numbers and greater
tumor size was not statistically significant. A detailed analysis
of additional samples may be required to verify the association
between CTC numbers and the actual tumor burden, rather than
primary tumor size (volume). In contrast to a microfluidic
device-confirmed CTC cluster, no CTC clusters were isolated from
the 91 patients using our established protocol; this result was
consistent with those from other reports using the
CellSearch® system. It was concluded that in the present
study, a simplified protocol using double staining resulted in a
higher CTC capture rate and is suitable for clinical use, including
single-cell detection. Moreover, previous studies have confirmed
the prognostic significance of CTCs in patients with lung cancer,
indicating that the CTC number is an independent prognostic factor
for SCLC at baseline and following chemotherapy (19,20).
Notably, ctDNA has been identified as a specific and
sensitive biomarker for the detection of EGFR mutations (21–23).
ctDNA is a part of the total circulating-free DNA, comprising small
fragments of between 70 and 200 base pairs, and is released from
dead cancer cells via apoptosis, necrosis and other mechanisms that
remain to be clarified (24). The
detection of ctDNA is also a characteristic of tumors with high
cellular turnover. Cell-free ctDNA is frequently evaluated in
non-invasive, early diagnostic screens to monitor responses and
disease prognosis and to monitor resistance to treatment (25). In this regard, the biology of CTCs
will hopefully be better understood with the availability of more
comprehensive molecular characterization data, and information on
their clinical implications (26).
In the clinic, decisions regarding therapy rely
heavily on local tumor analyses. Biopsies of primary lesions are
invasive procedures; therefore, an analysis of CTCs as
representative surrogates for primary and metastatic lesions may be
a potential alternative that may also enable the real-time
assessment of resistance to therapy (27). Deep-sequencing genomic analyses of
CTCs from primary tumors and metastases in patients with colorectal
or prostate cancer have revealed that mutations in CTCs resemble
those detected in primary tumors and metastases. These data have
important implications for the use of CTCs as a liquid biopsy
(13,27,28).
Using a microfluidic device, the expected activating EGFR mutations
identified in the original tumor biopsy specimens have also been
detected in CTCs from 11 of 12 patients with lung cancer (92%)
(4). More recently, a novel in
vivo device was used to capture circulating lung cancer cells
from one patient, and the same EGFR mutations identified in the
primary tumors were clearly detectable in the collected CTCs
(29). In the present study,
detection of EGFR mutations at the single-cell level was initially
attempted; however, this strategy was less effective, as only ~16%
of cells were positive for mutations in the analysis of 12 single
cells derived from different patients with a known primary
mutation. A subsequent study revealed that a sample with 10 CTCs
exhibited an increased detection rate and proved effective in
detecting the mutation identified in the primary tumor. Another
study reported a similar result of a low percentage of EGFR
mutations in CTCs analyzed using next-generation sequencing
(30). Based on these results, as
little as one CTC is appropriate for detecting EGFR mutations, and
when the CTC number is decreased, sensitivity decreases. Generally,
EGFR mutations were homogeneous. However, we observed one notable
exception, in which a clear rare mutation was detected that
differed from the primary mutation.
Although the present study is limited by the small
sample size, particularly for the EGFR mutation analysis, the
preliminary results support an expanded study using isolated CTCs
to detect EGFR mutations and a potent heterogeneity analysis of
somatic copy number alterations and mutations.
Acknowledgements
Not applicable.
Funding
This study was supported by grants of “Study on the
Application of Clinical Characteristics of Capital” to HTZ (no
Z151100004015104) and to SCZ (no. Z161100000516107) from Beijing
Municipal Science and Technology Project.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conceived and designed the experiments: SCZ, HTZ and
QZ. Performed the experiments: QZ, JYN, ZHY and LY. Analyzed the
data: QZ, JYN, ZDL and JHW. Contributed reagents/materials/analysis
tools: QZ, ZDL and JYN. Wrote the paper: QZ, SCZ and HTZ. Discussed
the results and implications and commented on the manuscript at all
stages: QZ, SCZ and HTZ.
Ethical approval and consent to
participate
The study involved taking peripheral blood from
healthy human, which was approved by Beijing Tuberculosis and
Thoracic Tumor Research Institute Ethics Committee. All
participants provided written informed consent prior to
participation in the study.
Patient consent for publication
Patients consented to the publication of their
data.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CTCs
|
circulating tumor cells
|
|
EGFR
|
epidermal growth factor receptor
|
|
EpCAM
|
epithelial cell adhesion molecule
|
|
CK
|
cytokeratin
|
|
FITC
|
fluorescein isothiocyanate
|
|
CNV
|
copy number variation
|
|
ARMS
|
amplification refractory mutation
system
|
|
WGA
|
whole-genome amplification
|
|
COSMIC
|
The Catalogue of Somatic Mutations in
Cancer
|
|
NSCLC
|
non-small-cell lung cancer
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carmichael J, Wing-san Mak D and O Brien
M: A review of recent advances in the treatment of elderly and poor
performance NSCLC. Cancers (Basel). 10(E236)2018.PubMed/NCBI
|
|
3
|
Nagano T, Tachihara M and Nishimura Y:
Mechanism of resistance to epidermal growth factor
receptor-tyrosine kinase inhibitors and a potential treatment
strategy. Cells. 7(E212)2018.PubMed/NCBI
|
|
4
|
Cooper MR, Alrajhi AM and Durand CR: Role
of immune checkpoint inhibitors in small cell lung cancer. Am J
Ther. 25:e349–e356. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al: Screening for epidermal growth factor receptor mutations in
lung cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu YL, Zhong WZ, Li LY, Zhang XT, Zhang L,
Zhou CC, Liu W, Jiang B, Mu XL, Lin JY, et al: Epidermal growth
factor receptor mutations and their correlation with gefitinib
therapy in patients with non-small cell lung cancer: A
meta-analysis based on updated individual patient data from six
medical centers in mainland China. J Thorac Oncol. 2:430–439. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maheswaran S, Sequist LV, Nagrath S, Ulkus
L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ,
Bell DW, et al: Detection of mutations in EGFR in circulating
lung-cancer cells. N Engl J Med. 359:366–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hou J, Greystoke A, Lancashire L, Cummings
J, Ward T, Board R, Amir E, Hughes S, Krebs M, Hughes A, et al:
Evaluation of circulating tumor cells and serological cell death
biomarkers in small cell lung cancer patients undergoing
chemotherapy. Am J Pathol. 175:808–816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H and Durbin R: Fast and accurate short
read alignment with burrows-wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li H and Durbin R: Fast and accurate
long-read alignment with burrows-wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parkinson DR, Dracopoli N, Petty BG,
Compton C, Cristofanilli M, Deisseroth A, Hayes DF, Kapke G, Kumar
P, Lee JSH, et al: Considerations in the development of circulating
tumor cell technology for clinical use. J Transl Med. 10:1382012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alix-Panabières C and Pantel K: Challenges
in circulating tumour cell research. Nat Rev Cancer. 14:623–631.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Went PT, Lugli A, Meier S, Bundi M,
Mirlacher M, Sauter G and Dirnhofer S: Frequent EpCam protein
expression in human carcinomas. Hum Pathol. 35:122–128. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Broers JL, Ramaekers FC, Rot MK,
Oostendorp T, Huysmans A, van Muijen GN, Wagenaar SS and Vooijs GP:
Cytokeratins in different types of human lung cancer as monitored
by chain-specific monoclonal antibodies. Cancer Res. 48:3221–3229.
1988.PubMed/NCBI
|
|
16
|
O Flaherty JD, Gray S, Richard D, Fennell
D, O Leary JJ, Blackhall FH and O Byrne KJ: Circulating tumour
cells, their role in metastasis and their clinical utility in lung
cancer. Lung Cancer. 76:19–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Punnoose EA, Atwal SK, Spoerke JM, Savage
H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS and
Lackner MR: Molecular biomarker analyses using circulating tumor
cells. PLoS One. 5:e125172010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krebs MG, Sloane R, Priest L, Lancashire
L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G,
et al: Evaluation and prognostic significance of circulating tumor
cells in patients with non-small-cell lung cancer. J Clin Oncol.
29:1556–1563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka F, Yoneda K, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T,
et al: Circulating tumor cell as a diagnostic marker in primary
lung cancer. Clin Cancer Res. 15:6980–6986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hou J, Krebs MG, Lancashire L, Sloane R,
Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, et
al: Clinical significance and molecular characteristics of
circulating tumor cells and circulating tumor microemboli in
patients with small-cell lung cancer. J Clin Oncol. 30:525–532.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thress KS, Brant R, Carr TH, Dearden S,
Jenkins S, Brown H, Hammett T, Cantarini M and Barrett JC: EGFR
mutation detection in ctDNA from NSCLC patient plasma: A
cross-platform comparison of leading technologies to support the
clinical development of AZD9291. Lung Cancer. 90:509–515. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bordi P, Del Re M, Danesi R and Tiseo M:
Circulating DNA in diagnosis and monitoring EGFR gene mutations in
advanced non-small cell lung cancer. Transl Lung Cancer Res.
4:584–597. 2015.PubMed/NCBI
|
|
23
|
Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang
F, Xu L and Yin R: Circulating tumor DNA is effective for the
detection of EGFR mutation in non-small cell lung cancer: A
meta-analysis. Cancer Epidemiol Biomarkers Prev. 24:206–212. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
25
|
Alix-Panabières C and Pantel K:
Circulating tumor cells: Liquid biopsy of cancer. Clin Chem.
59:110–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Effenberger KE, Schroeder C, Eulenburg C,
Reeh M, Tachezy M, Riethdorf S, Vashist YK, Izbicki JR, Pantel K
and Bockhorn M: Disseminated tumor cells in pancreatic cancer-an
independent prognosticator of disease progression and survival. Int
J Cancer. 131:E475–E483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagrath S, Sequist LV, Maheswaran S, Bell
DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky
A, et al: Isolation of rare circulating tumour cells in cancer
patients by microchip technology. Nature. 450:1235–1239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gorges TM, Penkalla N, Schalk T, Joosse
SA, Riethdorf S, Tucholski J, Lücke K, Wikman H, Jackson S, Brychta
N, et al: Enumeration and molecular characterization of tumor cells
in lung cancer patients using a novel in vivo device for capturing
circulating tumor cells. Clin Cancer Res. 22:2197–2206. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marchetti A, Del Grammastro M, Felicioni
L, Malatesta S, Filice G, Centi I, De Pas T, Santoro A, Chella A,
Brandes AA, et al: Assessment of EGFR mutations in circulating
tumor cell preparations from NSCLC patients by next generation
sequencing: Toward a real-time liquid biopsy for treatment. PLoS
One. 9:e1038832014. View Article : Google Scholar : PubMed/NCBI
|