Introduction
Gastric cancer (GC) is the third leading cause of
cancer-related mortality (1–3), with ~1,033,701 novel diagnoses and
782,685 mortalities worldwide in 2018 (3). Previous studies have indicated that GC
may be driven by a number of different genetic abnormalities, such
as mutations in cadherin 1 (4) and
catenin α1 (5). Chromosomal
aberrations including Erb-b2 receptor tyrosine kinase 2 (6), adenomatous polyposis coli, tumor
protein p53 and NME/NM23 nucleoside diphosphate kinase 1 (7) have also been frequently identified in
GC. Certain SNPs, such as interleukin 17A (rs2275913) (8), mucin 1 (rs4072037) (9) and prostate stem cell antigen
(rs2976392) (10), have indicated
genetic predispositions towards an increased risk of GC. However,
the aforementioned results are not sufficient to clarify the
complex pathogenesis of GC. Therefore, further research into the
molecular aspects involved in carcinogenesis is required, which
will offer new insights into GC treatment.
The α-enolase (ENO1) gene encodes a
glycolysis-associated enzyme, which contains 434 amino acids and
has a molecular mass of ~57 kDa (11). Previous studies have revealed ENO1 to
be abnormally expressed in a number of cancer types and serves
pivotal roles in tumorigenesis (12–16). For
example, in endometrial cancer, ENO1 silencing significantly
decreased malignant biological behavior; furthermore, the
expression level of ENO1 could affect the prognosis of patients
(14). In breast cancer, ENO1
promoted vascular endothelial cell proliferation, inhibited
apoptosis and accelerated blood vessel formation (15). In non-small cell lung cancer, stably
upregulated ENO1 could activate the focal adhesion
kinase/phosphoinositide 3-kinase (PI3K)/protein kinase B pathway
and its downstream signals, and then activate glycolysis, the cell
cycle and epithelial-mesenchymal transition-associated genes
(13,16). In colorectal cancer tissues, the
expression level of ENO1 was significantly increased, which was
associated with tumorigenesis and metastasis in patients with
colorectal cancer (17). In
addition, an in vitro study suggested that overexpression of
ENO1 promoted proliferation, migration and invasion of the
colorectal cancer cell line HCT116 (17). However, research regarding the role
of ENO1 in GC is insufficient, and further studies are required. To
date, only a few studies have indicated that ENO1 can promote
chemoresistance in GC, and that increased protein levels of ENO1
lead to a poor prognosis for the patient (18). Previous studies indicated that
overexpression of ENO1 can enhance proliferation and migration in
GC cell line AGS (19), and that
ENO1 can be upregulated by a well-known GC-associated protein,
CagA, in AGS cells (20). Combined
with the aforementioned results, we hypothesize that ENO1 serves a
role in the pathogenesis of GC. Microarray is a powerful tool that
can present the whole gene expression profile (21) and, as such, a microarray analysis was
performed on ENO1-silencing GC cells with the aim of gaining
further understanding into the molecular mechanism(s) of ENO1 in
the progression of GC.
Materials and methods
Cell culture and treatment
The human GC cell line MGC-803 (Sun Yat-sen
University Cell Library, Guangdong, China) was cultured as
described previously (22). The
small fragment small interfering RNA (siRNA) against ENO1 and the
scrambled (control) siRNA were synthesized by Beijing Oligobio
(Beijing, China). The siRNA-ENO1 sequences were as follows:
Forward, 5′-GCAUUGGAGCAGAGGUUUATT-3′ and reverse,
5′-UAAACCUCUGCUCCAAUGCTT-3′. The siRNA transfection experiment was
conducted using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific Inc., Waltham, MA, USA), according to the
manufacturer's protocol. The cells were plated onto 6-well plates
at a density of 8.0×104 cells. Following reaching ~50%
confluence, cells were transfected. The cells were assigned to two
groups: NC group, transfected with 50 nM scrambled siRNA; and
ENO1-knockdown group, transfected with 50 nM siRNA against ENO1.
Each group had three parallel samples. Cells were transfected with
Lipofectamine 2000 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) in serum- and antibiotic-free Opti-MEM (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
After 24 h, the cells were treated with TRIzol® (Thermo
Fisher Scientific, Inc.) and the total RNA was extracted.
Microarray analysis
Following extraction of the total RNA from the NC
group and ENO1-knockdown group, the quality was determined using
NanoDrop™ 2,000 (Thermo Fisher Scientific, Inc.), and the 2100
Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The
amplified RNA (aRNA) was prepared using an Affymetrix GeneChip™
3′IVT Express kit (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. The aRNA was
purified, fragmented and hybridized with the chip probes. Following
hybridization, the chip was stained and the final scanned images
and raw data were obtained by the Shanghai GeneChem Co., Ltd.
(Shanghai, China). The raw data were processed using the two-way
semi-linear model, and the genes with fold change (FC) >1.5 and
P<0.05 were regarded as significantly DEGs.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
To determine the interference efficiency of
siRNA-ENO1 and to validate the gene chip results, ENO1 and five
random genes were selected for RT-qPCR analysis and were as
follows: AVL9 cell migration-associated (AVL9), glia maturation
factor β (GFMB), G-protein-coupled receptor 180 (GPR180),
microfibrillar-associated protein 3 (MFAP3)and septin 8 (SEPT8).
The total RNA was extracted from the cells and the quality was
assessed using the aforementioned method. RNA was
reverse-transcribed into cDNA using a reverse transcription kit
(cat. no. 05091284001; Roche Diagnostics, Basel, Switzerland),
according to the manufacturer's protocol. qPCR was carried out in a
volume of 10.0 µl, including 5.0 µl SYBR® Select Master
mix (Roche Diagnostics), 3.4 µl DNase/RNase-free water (Beijing
Solarbio Science and Technology Co., Ltd., Beijing, China), 1.0 µl
cDNA, 0.30 µl forward primer and 0.30 µl reverse primer. β-actin
was selected as the internal reference gene. The Piko Real
detection system (Thermo Fisher Scientific, Inc.) was used for the
amplification according to the manufacturer's protocol. The primers
were synthesized by Generay Biotech Co., Ltd. (Shanghai, China) and
the sequences were as follows: ENO1 forward,
5′-GGGAATCCCACTGTTGAGGT-3′ and reverse, 5′-CGGAGCTCTAGGGCCTCATA-3′;
β-actin forward, 5′-GGGAAATCGTGCGTGACATTAAGG-3′ and reverse,
5′-CAGGAAGGAAGGCTGGAAGAGTG-3′; ALV9 forward,
5′-TTCCATTTCTGGGTGGCAAGT-3′ and reverse,
5′-ACATCGTGGTGGTCGGATTTC-3′; GMFB forward,
5′-CAGCGTTGTTCGTTTCTTTGC-3′ and reverse,
5′-GTCTTTGGTTGTTTGTGATGTTGC-3′; MFAP3 forward,
5′-AATGACATAGATGCCACCTTG-3′ and reverse,
5′-GTGTCCCTCTTCCACCTCTTA-3′; SEPT8 forward,
5′-GGAATAATGTTCACCTTGCTGTCT-3′ and reverse,
5′-TTTGCCTCTACTTCATCACGC-3′. For all RT-qPCR experiments, the
samples were amplified in triplicate, each consisting of three
replicates. The relative levels of target gene mRNA were calculated
and normalized relative to β-actin using the 2−ΔΔCq
method (23).
Functional enrichment analysis
The functional enrichment analyses of the DEGs were
performed using DAVID 6.7 (https://david.ncifcrf.gov) (24). Briefly, all the differentially
expressed genes (DEGs) were uploaded in the ‘Functional Annotation’
section of DAVID 6.7, and set E=0.01. The result would indicate the
DEGs mapping to different Gene Ontology (GO) terms. The GO
annotation (www.geneontology.org) includes three parts: Biological
processes (BP), cellular components (CC) and molecular functions
(MF), which provide a descriptive framework and functional
annotation of DEGs. The pathway enrichment analysis was performed
using Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) (25,26).
P<0.05 was considered to indicate statistically significant
functional terms and pathways.
Protein-protein interaction (PPI)
network construction and module selection
A PPI network was constructed based on Biological
General Repository for Interaction Datasets (BioGRID) in WebGestalt
(http://www.webgestalt.org/option.php). The DEGs were
mapped to BioGRID and PPI pairs were acquired. Interactions with a
confidence score >0.4 were retained in the network and were
visualized using Cytoscape (version 3.5.1; http://cytoscape.org). In the PPI network, a node
represents a protein product of a DEG and the degree represents the
number of proteins linked to this node. The nodes with a high
degree (>10) were considered to be important and named ‘hub
genes’ in the present study. The PPI modules were screened using
the ClusterONE plugin (version 1.0; http://www.paccanarolab.org/clusterone) in Cytoscape
(27). Results were considered
statistically significant when P<0.0005.
Statistical analysis
All the data were analyzed using SPSS software
(version 15.0; SPSS, Inc., Chicago, IL, USA). The measurement data
were expressed as the mean ± standard deviation. Comparison between
two groups was performed using an independent sample t-test.
P<0.05 was considered to indicate a statistically significant
result.
Results
Successful knockdown of the ENO1 gene
in MGC-803 cells
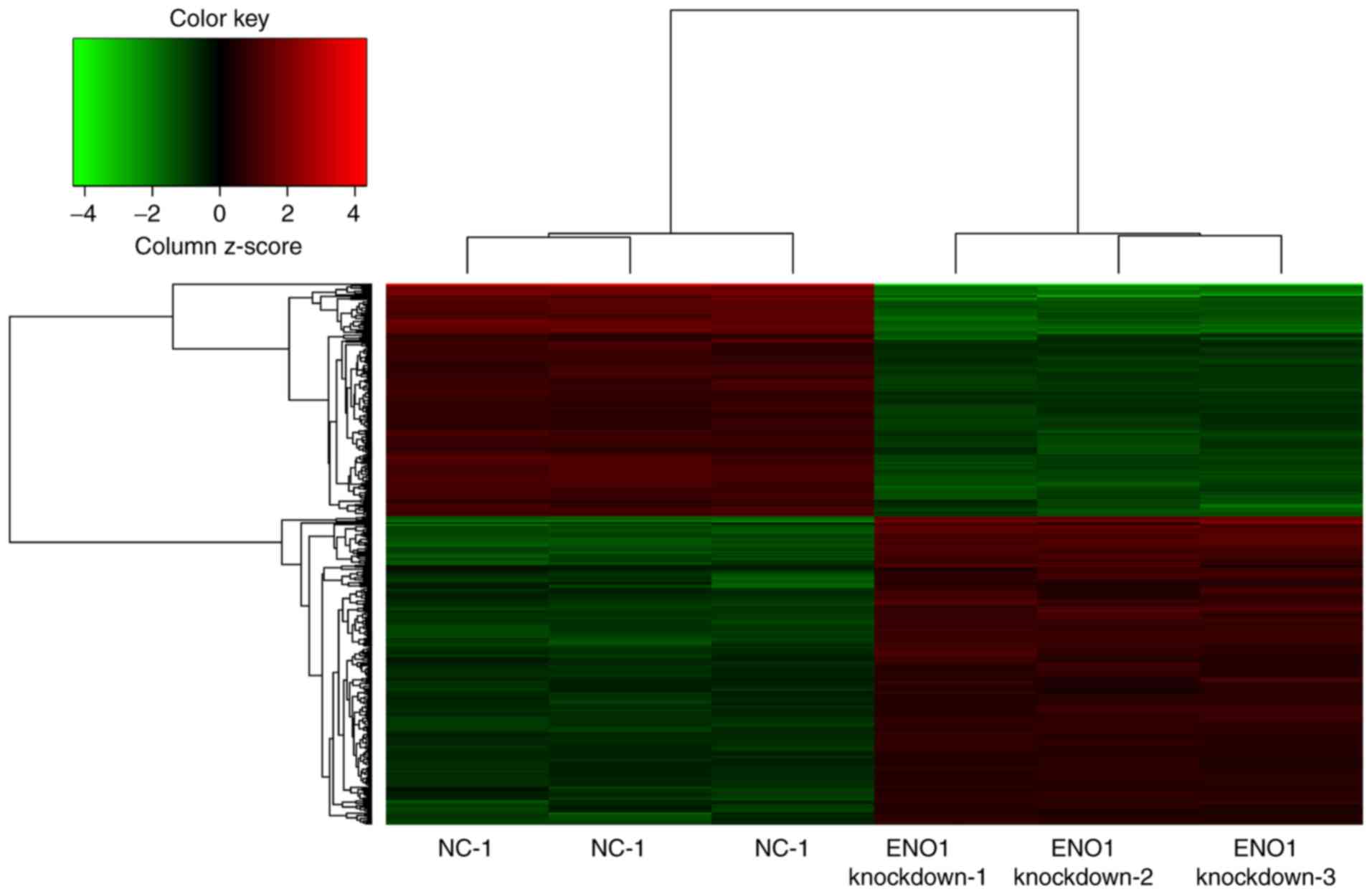
The mRNA expression levels of ENO1 were
downregulated 12.08-fold (array data; Fig. 1) and 11.43±0.39-fold (RT-qPCR data;
Fig. 2) in the ENO1-knockdown group
compared with in the NC group. The results indicated that the siRNA
fragments targeting the ENO1 gene were successful and that
silencing was efficient.
Gene expression profile analysis and
hierarchical clustering
The microarray included two groups with six samples,
and the heat map results are presented in Fig. 1. As a result, there were 448 DEGs
with a FC value >1.5 and P<0.05, among which, 183 (40.85%)
were downregulated and 265 (59.15%) were upregulated. The top ten
DEGs with high FC were tropomyosin 4 (TPM4), fibroblast growth
factor 2 (FGF2), inhibitor of DNA binding 2, mitochondrial
ribosomal protein S33, small integral membrane protein 13, cyclin
J, AVL9 cell migration-associated (AVL9), serum/glucocorticoid
regulated kinase family member 3 (SGK3), G protein-coupled receptor
180 (GPR180) and mesoderm development LRP chaperone.
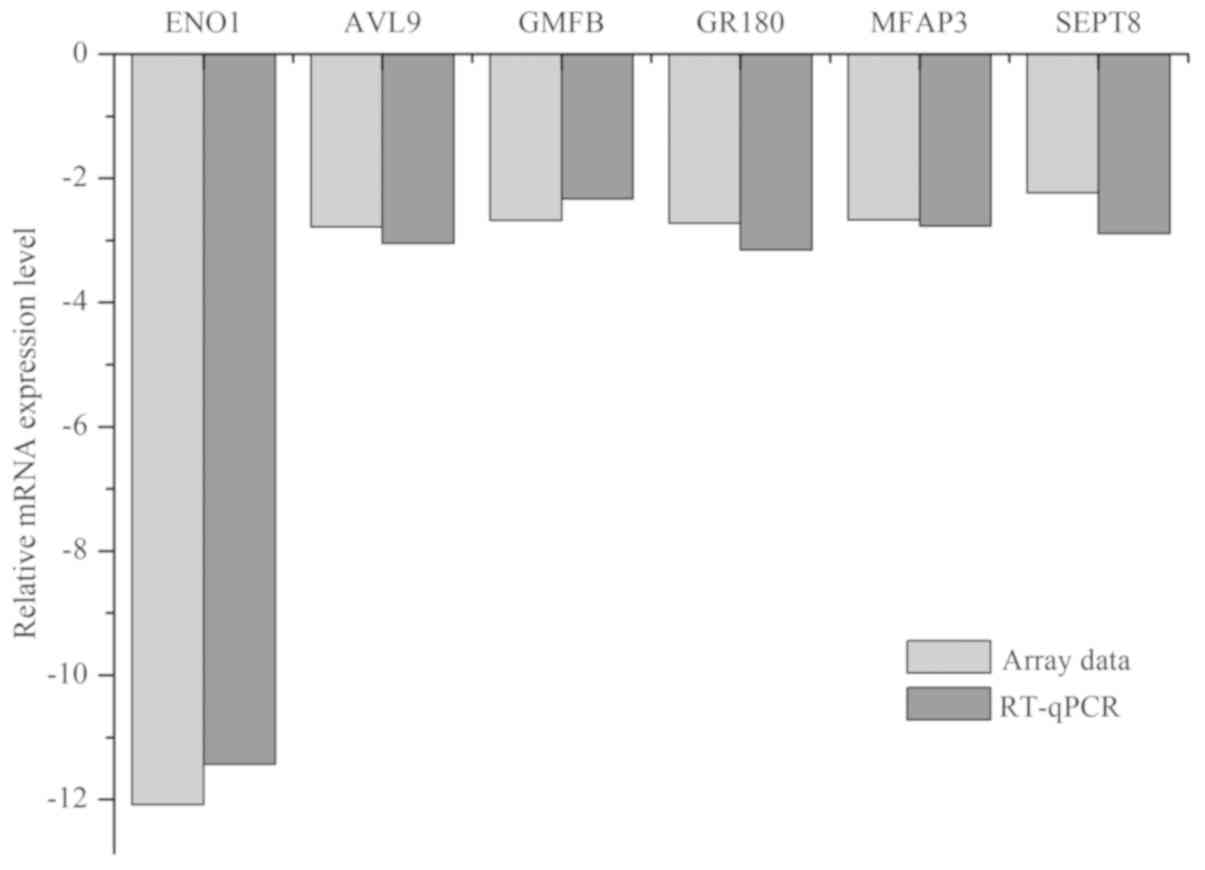
Verification of the array data using
RT-qPCR analysis
Five DEGs (AVL9, GMFB, GPR180, MFAP3 and SEPT8) were
selected for qPCR analysis. The results (Fig. 2) indicated that the mRNA levels of
AVL9, GMFB, GPR180, MFAP3 and SEPT8 were downregulated 2.78-,
2.68-, 2.72-, 2.67- and 2.23-fold, respectively. In the RT-qPCR
experiment, these genes were downregulated 3.05±0.07-, 2.33±0.12-,
3.15±0.06-, 2.77±0.08- and 2.89±0.12-fold, respectively. The array
data were in concordance with the RT-qPCR results.
Functional annotation analysis of the
DEGs
The gene annotation analysis was performed using
DAVID and the detailed results (Table
I) identify the number of significant functional
classifications for BP, CC and MF as 26, 7 and 10, respectively.
The DEGs were mainly enriched in transcription, blood vessel
morphogenesis and cell cycle for BP. CC enrichment was detected for
genes associated with the nuclear lumen, organelle lumen and
nucleoplasm, and MF enrichment was identified for genes associated
with transcription factor activity, transcription regulator
activity and cytoskeletal protein binding.
 | Table I.GO analysis of the DEGs regulated by
ENO1 silencing. |
Table I.
GO analysis of the DEGs regulated by
ENO1 silencing.
| Identifier | Functional
term | Count | P-value |
|---|
| Biological
process |
|
GO:0006350 | Transcription | 86 |
6.43×10−7 |
|
GO:0006355 | Regulation of
transcription, DNA-dependent | 73 |
5.37×10−6 |
|
GO:0045449 | Regulation of
transcription | 96 |
1.08×10−5 |
|
GO:0051252 | Regulation of RNA
metabolic process | 73 |
1.18×10−5 |
|
GO:0010629 | Negative regulation
of gene expression | 28 |
1.00×10−4 |
|
GO:0016481 | Negative regulation
of transcription | 26 |
1.40×10−4 |
|
GO:0006357 | Regulation of
transcription from RNA polymerase II promoter | 35 |
1.81×10−4 |
|
GO:0048514 | Blood vessel
morphogenesis | 16 |
1.98×10−4 |
|
GO:0010605 | Negative regulation
of macromolecule metabolic process | 35 |
2.17×10−4 |
|
GO:0045934 | Negative regulation
of nucleobase, nucleoside, nucleotide and nucleic acid metabolic
process | 27 |
3.13×10−4 |
|
GO:0010558 | Negative regulation
of macromolecule biosynthetic process | 28 |
3.74×10−4 |
|
GO:0051172 | Negative regulation
of nitrogen compound metabolic process | 27 |
3.82×10−4 |
|
GO:0045892 | Negative regulation
of transcription, DNA-dependent | 21 |
4.39×10−4 |
|
GO:0051253 | Negative regulation
of RNA metabolic process | 21 |
5.34×10−4 |
|
GO:0031327 | Negative regulation
of cellular biosynthetic process | 28 |
5.50×10−4 |
|
GO:0009890 | Negative regulation
of biosynthetic process | 28 |
7.64×10−4 |
|
GO:0001568 | Blood vessel
development | 16 |
9.55×10−4 |
|
GO:0001525 | Angiogenesis | 12 |
9.60×10−4 |
|
GO:0007049 | Cell cycle | 34 |
1.189962×10−3 |
|
GO:0001944 | Vasculature
development | 16 |
1.215671×10−3 |
|
GO:0031099 | Regeneration | 8 |
1.374438×10−3 |
|
GO:0051726 | Regulation of cell
cycle | 18 |
3.03174×10−3 |
|
GO:0031100 | Organ
regeneration | 5 |
3.326962×10−3 |
|
GO:0007507 | Heart
development | 13 |
6.280962×10−3 |
|
GO:0030593 | Neutrophil
chemotaxis | 4 |
8.795164×10−3 |
|
GO:0007167 | Enzyme-linked
receptor protein signaling pathway | 17 |
9.52888×10−3 |
| Cell component |
|
GO:0031981 | Nuclear lumen | 52 |
2.58×10−4 |
|
GO:0070013 | Intracellular
organelle lumen | 60 |
3.75×10−4 |
|
GO:0043233 | Organelle
lumen | 60 |
6.74×10−4 |
|
GO:0031974 | Membrane-enclosed
lumen | 60 |
1.097428×10−3 |
|
GO:0005654 | Nucleoplasm | 34 |
1.312471×10−3 |
|
GO:0005783 | Endoplasmic
reticulum | 34 |
4.951755×10−3 |
|
GO:0044451 | Nucleoplasm
part | 22 |
8.735324×10−3 |
| Molecular
function |
|
GO:0003700 | Transcription
factor activity | 43 |
2.18×10−4 |
|
GO:0030528 | Transcription
regulator activity | 58 |
5.22×10−4 |
|
GO:0008092 | Cytoskeletal
protein binding | 26 |
6.25×10−4 |
|
GO:0016564 | Transcription
repressor activity | 18 |
1.995073×10−3 |
|
GO:0043014 | α-tubulin
binding | 4 |
2.056678×10−3 |
|
GO:0003677 | DNA binding | 77 |
3.813995×10−3 |
|
GO:0046914 | Transition metal
ion binding | 88 |
6.180415×10−3 |
|
GO:0003779 | Actin binding | 17 |
6.453361×10−3 |
|
GO:0008270 | Zinc ion
binding | 75 |
6.826083×10−3 |
|
GO:0004672 | Protein kinase
activity | 26 |
7.297114×10−3 |
KEGG pathway enrichment analysis
The DEGs responding to ENO1 silencing were enriched
in six significant pathways: Systemic lupus erythematosus [Homo
sapiens (hsa)05322; P<0.001], viral carcinogenesis
(hsa05203; P=0.00141), alcoholism (hsa05034; P=0.003), forkhead box
O (FoxO) signaling pathway (hsa04068; P=0.0077), miRNAs in cancer
(hsa05206; P=0.0183) and cAMP signaling pathway (hsa04024;
P=0.0415) (Table II).
 | Table II.Detailed information of the Kyoto
Encyclopedia of Genes and Genomes pathway analysis. |
Table II.
Detailed information of the Kyoto
Encyclopedia of Genes and Genomes pathway analysis.
| Identifier | Name | Count | Gene | P-value |
|---|
| hsa05322 | Systemic lupus
erythematosus | 6 | HIST1H2BD,
HIST1H2BG, HIST1H2BF, HIST1H2BE, HIST1H2BI, HIST1H2BC | <0.001 |
| hsa05203 | Viral
carcinogenesis | 10 | CDKN1A, HIST1H2BD,
CCND1, STAT3, TRAF3, HIST1H2BG, HIST1H2BF, HIST1H2BE, HIST1H2BI,
HIST1H2BC |
1.41×10−3 |
| hsa05034 | Alcoholism | 6 | HIST1H2BD,
HIST1H2BG, HIST1H2BF, HIST1H2BE, HIST1H2BI, HIST1H2BC |
3.11×10−3 |
| hsa04068 | FoxO signaling
pathway | 6 | CDKN1A, SGK3,
CCND1, STAT3, TGFBR1, SETD7 |
7.7×10−3 |
| hsa05206 | MicroRNAs in
cancer | 8 | CDKN1A, CYP1B1,
DICER1, PDCD4, MCL1, PLAU, CCND1, STAT3 |
1.83×10−2 |
| hsa04024 | cAMP signaling
pathway | 5 | ADCY9, HTR1D,
PPP1CB, RAC1, CREB3L2 |
4.15×10−2 |
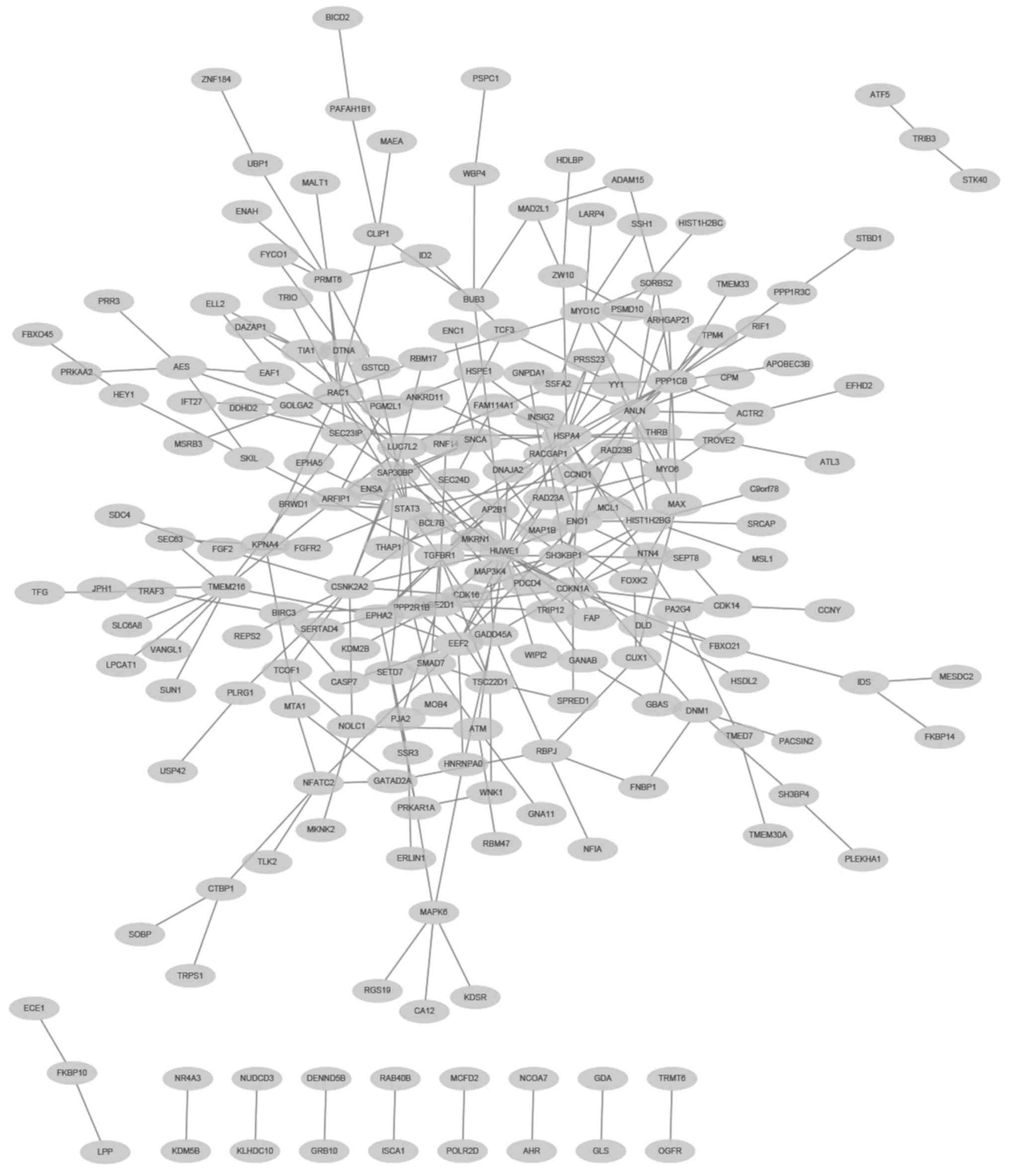
PPI network analysis
The PPIs among the 448 DEGs were predicted using
WebGestalt with information from BioGRID. The constructed network
consisted of 209 proteins (nodes) and 293 interactions (edges)
(Fig. 3). In addition, there were
seven genes that had high degrees with edges ≥10 in the PPI
network. These seven genes were HECT, UBA and WWE domain-containing
1, E3 ubiquitin protein ligase (HUWE1; degree, 16), protein
phosphatase 1 catalytic subunit β (PPP1CB; degree, 16), heat shock
protein family A (Hsp70) member 4 (HSPA4; degree, 16), signal
transducer and activator of transcription 3 (STAT3; degree, 13),
anillin actin-binding protein (degree, 12), Src homology 3
domain-containing kinase-binding protein 1 (degree, 10) and casein
kinase 2α2 (degree, 10), respectively. Among these, HUWE1, PPP1CB
and HSPA4 were the top three nodes with 16 edges.
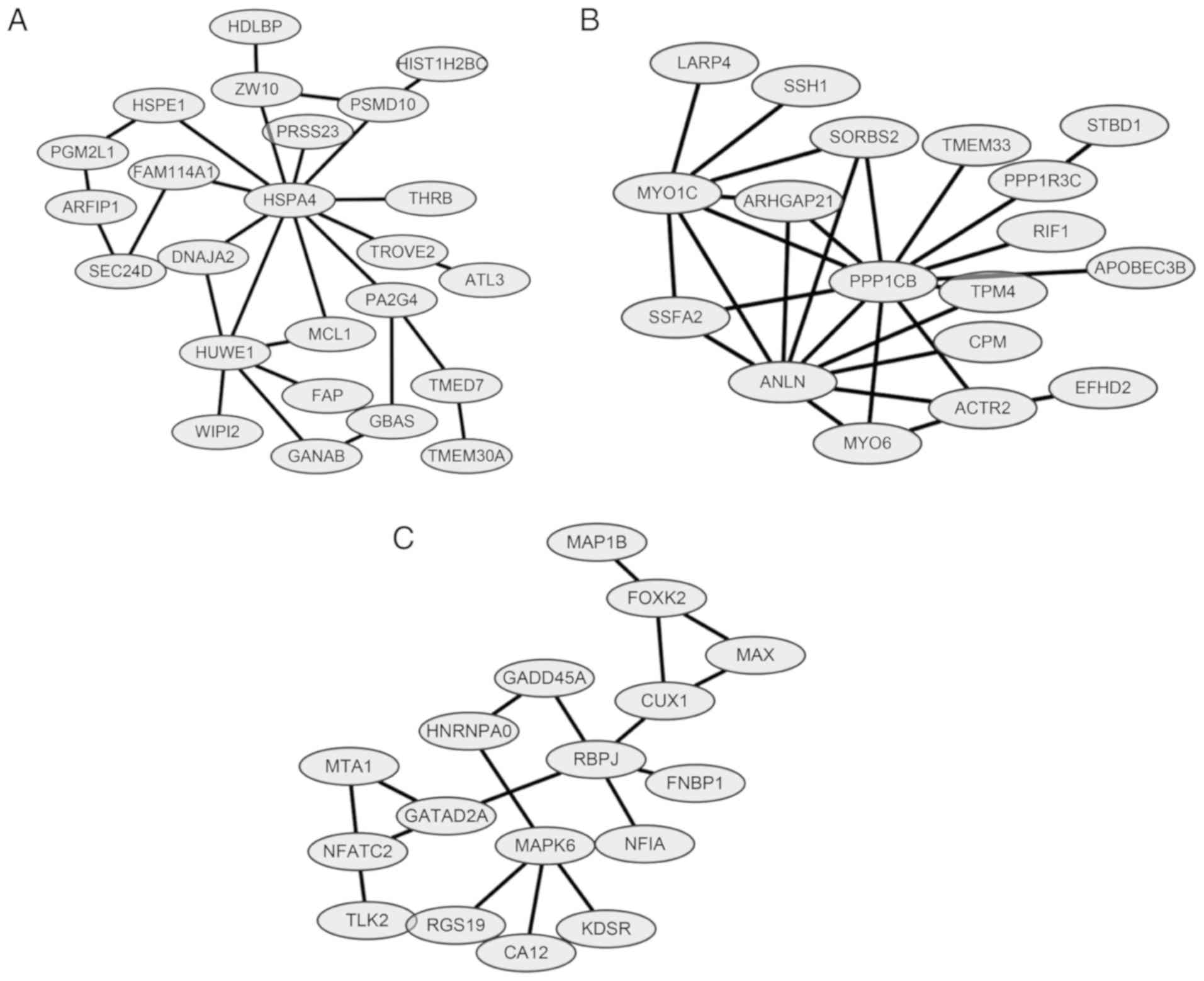
Module analysis and protein domain
analysis
ClusterONE was applied for module analysis to
further predict potential protein complexes. For the network
constructed above, there were three significant modules
(P<0.0005; Fig. 4), as follows:
Module A (nodes, 24; density, 0.101; quality, 0.549; P=0.00000791);
Module B (Nodes, 18; density, 0.183; quality, 0.683; P=0.000136)
and Module C (nodes, 17; density, 0.132; quality, 0.581; P=0.00045)
(Table III). For the protein
domain analysis, one significant domain was found for Module B:
Myosin head, motor domain (IPR001609) (P=0.034). No significantly
enriched protein domains were identified in Modules A and C.
 | Table III.Detailed information of three modules
screened from the PPI network of all DEGs. |
Table III.
Detailed information of three modules
screened from the PPI network of all DEGs.
| Module | Nodes | Density | Quality | P-value | Members |
|---|
| A | 24 | 0.101 | 0.549 |
7.905×10−6 | HSPE1, PGM2L1,
HSPA4, ARFIP1, TMED7, TMEM30A, HUWE1, GANAB, GBAS, TROVE2, ATL3,
SEC24D, FAM114A1, ZW10, HDLBP, FAP, PA2G4, DNAJA2, WIPI2, MCL1,
PRSS23, THRB, PSMD10, HIST1H2BC |
| B | 18 | 0.183 | 0.683 |
1.356×10−4 | SORBS2, ARHGAP21,
ANLN, PPP1CB, TPM4, SSFA2, MYO1C, LARP4, SSH1, ACTR2, EFHD2, STBD1,
PPP1R3C, MYO6, CPM, RIF1, APOBEC3B, TMEM33 |
| C | 17 | 0.132 | 0.581 |
4.502×10−4 | RGS19, MAPK6, CUX1,
FOXK2, NFATC2, TLK2, MAP1B, GATAD2A, RBPJ, MTA1, HNRNPA0, NFIA,
KDSR, CA12, MAX, GADD45A, FNBP1 |
Discussion
In the present study, a total of 448 DEGs responded
to ENO1 knockdown in the human GC cell line MGC-803. Certain DEGs
that demonstrated significantly decreased expression in the present
study, including TPM4, have been reported to be associated with
clinical progression in patients with colon cancer (28). Another gene, SGK3, which was
identified to be downregulated in the present study has previously
been reported to serve an important role in the development of
breast cancer (29). In addition,
FGF2, which also demonstrated significantly decreased expression,
is a well-known oncogene (30,31).
The DEGs were enriched in six significant pathways:
Systemic lupus erythematosus, viral carcinogenesis, alcoholism,
FoxO signaling pathway, miRNAs in cancer and cAMP signaling pathway
(Table II). Among these, some were
reported to be associated with cancer development, such as the cAMP
signaling pathway and FoxO signaling pathway. cAMP signaling
regulates protein levels by controlling gene transcription via
transcriptional activators that are involved in cancer cell
migration, proliferation, apoptosis, and cytoskeleton remodeling in
bladder cancer (32), breast cancer
(33) and lung cancer (34). Furthermore, an exchange protein
directly activated by cAMP (EPAC1) has been regarded as a
prognostic marker and may be a potential therapy target for GC
(35), which suggests the important
role that the cAMP signaling pathway serves in the pathological
processes of GC. The FoxO signaling pathway has also been reported
to be associated with breast cancer (36), bladder cancer (37), prostate cancer (38) and lung cancer (39). Previous studies have demonstrated
that there may be crosstalk between the cAMP signaling pathway and
the FoxO signaling pathway. On one hand, activation of the cAMP
signaling pathway increased FoxO1 phosphorylation (40). On the other hand, FoxOs supported the
metabolic requirements of normal and tumor cells via the PI3K
signaling pathway, which was reported to interact with the cAMP
signaling pathway in a number of physiological processes (41). In addition, systemic lupus
erythematosus was the most significant pathway response to ENO1
inhibition, and this result was similar to that in our previous
study on TPI silencing (22). As
previously discussed, although no definitive evidence has suggested
the involvement of systemic lupus erythematosus in the pathogenesis
of GC, there may be an association between the two since certain
sporadic patients were affected by GC and systemic lupus
erythematosus simultaneously (22,42,43). The
molecular mechanisms underlying the involvement of systemic lupus
erythematosus in the pathogenesis of GC are not currently fully
understood and further research is required.
The constructed PPI network based on BioGRID
included 209 nodes and 293 edges. There were seven DEGs with a
degree ≥10, among which, the first three were PPP1CB, HUWE1 and
HSPA4, which were regarded as hub genes and may interact with ENO1
in GC progression. These three genes have been reported to be
associated with cancer development. PPP1CB, encoding protein
phosphatase 1 catalytic subunit β isoform, has been reported in
prostate cancer, chronic lymphocytic leukemia (44) and even used as a potential biomarker
for distinguishing malignant melanoma from other melanocytic
lesions (45). HUWE1, as a ubiquitin
ligase, has been regarded as a tumor suppressor and served key
roles in tumorigenesis (46). For
example, compared with normal thyroid tissue, HUWE1 was
downregulated in human thyroid cancer tissues, and overexpression
of HUWE1 in thyroid cancer cells enhanced chemotherapeutic
sensitivity and inhibition of cell proliferation, cell migration
and invasion (47). The third hub
gene HSPA4 encodes a heat shock 70 protein. It has been reported
that upregulation of heat shock proteins (HSPA12A, HSP90B1, HSPA4,
HSPA5 and HSPA6) in tumor tissues is associated with poor outcomes
from hepatitis B virus-associated early-stage hepatocellular
carcinoma (48). Furthermore, HSPA4
has been reported to regulate cell migration and delay gastric
ulcer healing (49). To the best of
our knowledge, the present study is the first to identify the
possible association between PPP1CB, HUWE1, HSPA4 and ENO1. Further
studies are required to confirm these connections and their
functions in the pathogenesis in GC tumorigenesis.
In summary, the results of the present study provide
a comprehensive bioinformatic analysis of the genes associated with
ENO1. The important signaling pathways (such as cAMP signaling
pathway and FoxO signaling pathway) and key genes (such as PPP1CB,
HUWE1 and HSPA4) may help to narrow down the role of ENO1 in the
pathogenesis and treatment of GC.
Acknowledgements
The authors would like to thank, Dr Tao Li, Dr
Yuqing He, Dr Zunnan Huang, Dr Jindong Ni and Dr Huanwen Tang of
Guangdong Medical University, for their advice on the present
study.
Funding
The present study was supported by grants from the
Natural Science Fund Project of Guangdong Province, China (no.
2016A030313683), the Social Science and Technology Development
Project of Dongguan, Guangdong Province, China (nos. 2014108101051
and 2016108101039) and the National Natural Science Foundation of
China (no. 81200082).
Availability of data and materials
The datasets generated and/or analyzed during the
study are available from the corresponding author upon request.
Authors' contributions
ZH and PO designed the experiments, performed the
bioinformatic analysis and wrote the paper; BL and HP identified
the correct siRNA fragment and performed the array; JD carried out
the statistical analysis; RH performed the RT-qPCR experiment; SZ
cultured the cells.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AVL9
|
AVL9 cell migration-associated
|
|
DEGs
|
differentially expressed genes
|
|
ENO1
|
α-enolase
|
|
FGF2
|
fibroblast growth factor 2
|
|
GC
|
gastric cancer
|
|
GMFB
|
glia maturation factor β
|
|
GPR180
|
G-protein-coupled receptor 180
|
|
HSPA4
|
heat shock protein family A, Hsp70
member 4
|
|
HUWE1
|
HECT, UBA and WWE domain-containing 1,
E3 ubiquitin protein ligase
|
|
MFAP3
|
microfibrillar-associated protein
3
|
|
PPP1CB
|
protein phosphatase 1 catalytic
subunit β
|
|
SEPT8
|
septin 8
|
|
SGK3
|
serum/glucocorticoid-regulated kinase
family member 3
|
|
TPM4
|
tropomyosin 4
|
References
|
1
|
Massimo R, Fassan M and Graham DY:
Epidemiology of gastric cancer. Gastric Cancer. Springer.
(Switzerland). 23–34. 2015.PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global Cancer Statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 Cancers in
185 Countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Roy F: Beyond E-cadherin: Roles of
other cadherin superfamily members in cancer. Nat Rev Cancer.
14:121–134. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Majewski IJ, Kluijt I, Cats A, Scerri TS,
de Jong D, Kluin RJ, Hansford S, Hogervorst FB, Bosma AJ, Hofland
I, et al: An alpha-E-catenin (CTNNA1) mutation in hereditary
diffuse gastric cancer. J Pathol. 229:621–629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pazo Cid RA and Anton A: Advanced
HER2-positive gastric cancer: Current and future targeted
therapies. Crit Rev Oncol Hematol. 85:350–362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gazvoda B, Juvan R, Zupanic-Pajnic I,
Repse S, Ferlan-Marolt K, Balazic J and Komel R: Genetic changes in
Slovenian patients with gastric adenocarcinoma evaluated in terms
of microsatellite DNA. Eur J Gastroenterol Hepatol. 19:1082–1089.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Zheng L, Sun Y and Zhang X:
Analysis of the association of interleukin-17 gene polymorphisms
with gastric cancer risk and interaction with Helicobacter pylori
infection in a Chinese population. Tumour Biol. 35:1575–1580. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saeki N, Saito A, Choi IJ, Matsuo K,
Ohnami S, Totsuka H, Chiku S, Kuchiba A, Lee YS, Yoon KA, et al: A
functional single nucleotide polymorphism in mucin 1, at chromosome
1q22, determines susceptibility to diffuse-type gastric cancer.
Gastroenterology. 140:892–902. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garcia-Gonzalez MA, Bujanda L, Quintero E,
Santolaria S, Benito R, Strunk M, Sopeña F, Thomson C, Pérez-Aisa
A, Nicolás-Pérez D, et al: Association of PSCA rs2294008 gene
variants with poor prognosis and increased susceptibility to
gastric cancer and decreased risk of duodenal ulcer disease. Int J
Cancer. 137:1362–1373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zakrzewicz D, Didiasova M, Zakrzewicz A,
Hocke AC, Uhle F, Markart P, Preissner KT and Wygrecka M: The
interaction of enolase-1 with caveolae-associated proteins
regulates its subcellular localization. Biochem J. 460:295–307.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee SY, Jin CC, Choi JE, Hong MJ, Jung DK,
Do SK, Baek SA, Kang HJ, Kang HG, Choi SH, et al: Genetic
polymorphisms in glycolytic pathway are associated with the
prognosis of patients with early stage non-small cell lung cancer.
Sci Rep. 6:356032016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai L, Qu Y, Li J, Wang X, Wang K, Wang P,
Jiang BH and Zhang J: Serological proteome analysis approach-based
identification of ENO1 as a tumor-associated antigen and its
autoantibody could enhance the sensitivity of CEA and CYFRA 21-1 in
the detection of non-small cell lung cancer. Oncotarget.
8:36664–36673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao M, Fang W, Wang Y, Guo S, Shu L, Wang
L, Chen Y, Fu Q, Liu Y, Hua S, et al: Enolase-1 is a therapeutic
target in endometrial carcinoma. Oncotarget. 6:15610–15627.
2015.PubMed/NCBI
|
|
15
|
Gao J, Zhao R, Xue Y, Niu Z, Cui K, Yu F,
Zhang B and Li S: Role of enolase-1 in response to hypoxia in
breast cancer: Exploring the mechanisms of action. Oncol Rep.
29:1322–1332. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong
SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al: Alpha-enolase promotes
cell glycolysis, growth, migration, and invasion in non-small cell
lung cancer through FAK-mediated PI3K/AKT pathway. J Hematol Oncol.
8:222015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan P, Zhao S, Yan H, Yin C, Xiao Y, Wang
Y, Ni R, Chen W, Wei G and Zhang P: α-enolase promotes
tumorigenesis and metastasis via regulating AMPK/mTOR pathway in
colorectal cancer. Mol Carcinog. 56:1427–1437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qian X, Xu W, Xu J, Shi Q, Li J, Weng Y,
Jiang Z, Feng L, Wang X, Zhou J and Jin H: Enolase 1 stimulates
glycolysis to promote chemoresistance in gastric cancer.
Oncotarget. 8:47691–47708. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu YQ, Huang ZG, Li GN, Du JL, Ou YP,
Zhang XN, Chen TT and Liang QL: Effects of α-enolase (ENO1)
over-expression on malignant biological behaviors of AGS cells. Int
J Clin Exp Med. 8:231–239. 2015.PubMed/NCBI
|
|
20
|
Chen S, Duan G, Zhang R and Fan Q:
Helicobacter pylori cytotoxin-associated gene A protein upregulates
α-enolase expression via Src/MEK/ERK pathway: implication for
progression of gastric cancer. Int J Oncol. 45:764–770. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin Y and Tian YP: A microarray gene
analysis of peripheral whole blood in normal adult male rats after
long-term GH gene therapy. Cell Mol Biol Lett. 15:177–195. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ouyang P, Lin B, Du J, Pan H, Yu H, He R
and Huang Z: Global gene expression analysis of knockdown
Triosephosphate isomerase (TPI) gene in human gastric cancer cell
line MGC-803. Gene. 647:61–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Q: Identification of biomarkers for
metastatic osteosarcoma based on DNA microarray data. Neoplasma.
62:365–371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li HY, Jin N, Han YP and Jin XF: Pathway
crosstalk analysis in prostate cancer based on protein-protein
network data. Neoplasma. 64:22–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Jiang T, Li Z, Lu L, Zhang R,
Zhang D, Wang X and Tan J: Analysis of differentially co-expressed
genes based on microarray data of hepatocellular carcinoma.
Neoplasma. 64:216–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Li D, Tang Y, Wu F and Wang J:
CytoCluster: A cytoscape plugin for cluster analysis and
visualization of biological networks. Int J Mol Sci. 18(pii):
E18802017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang R, Zheng G, Ren D, Chen C, Zeng C, Lu
W and Li H: The clinical significance and biological function of
tropomyosin 4 in colon cancer. Biomed Pharmacother. 101:1–7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun X, Liu X, Liu BO, Li S, Zhang D and
Guo H: Serum- and glucocorticoid-regulated protein kinase 3
overexpression promotes tumor development and aggression in breast
cancer cells. Oncol Lett. 12:437–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Coleman SJ, Chioni AM, Ghallab M, Anderson
RK, Lemoine NR, Kocher HM and Grose RP: Nuclear translocation of
FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic
cancer cell invasion. EMBO Mol Med. 6:467–481. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu M, Gu M, Zhang K, Zhou J, Wang Z and Da
J: miR-203 inhibition of renal cancer cell proliferation, migration
and invasion by targeting of FGF2. Diagn Pathol. 10:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ichikawa H, Itsumi M, Kajioka S, Maki T,
Lee K, Tomita M and Yamaoka S: Overexpression of exchange protein
directly activated by cAMP-1 (EPAC1) attenuates bladder cancer cell
migration. Biochem Biophys Res Commun. 495:64–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumar N, Gupta S, Dabral S, Singh S and
Sehrawat S: Role of exchange protein directly activated by cAMP
(EPAC1) in breast cancer cell migration and apoptosis. Mol Cell
Biochem. 430:115–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park JY and Juhnn YS: cAMP signaling
increases histone deacetylase 8 expression via the Epac2-Rap1A-Akt
pathway in H1299 lung cancer cells. Exp Mol Med. 49:e2972017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun DP, Fang CL, Chen HK, Wen KS, Hseu YC,
Hung ST, Uen YH and Lin KY: EPAC1 overexpression is a prognostic
marker and its inhibition shows promising therapeutic potential for
gastric cancer. Oncol Rep. 37:1953–1960. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bullock M: FOXO factors and breast cancer:
Outfoxing endocrine resistance. Endocr Relat Cancer. 23:R113–130.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang M, Zhao Y, Liu N, Chen E, Quan Z, Wu
X and Luo C: Overexpression of HepaCAM inhibits bladder cancer cell
proliferation and viability through the AKT/FoxO pathway. J Cancer
Res Clin Oncol. 143:793–805. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Y, Blee AM, Wang D, An J, Pan Y, Yan
Y, Ma T, He Y, Dugdale J, Hou X, et al: Loss of FOXO1 cooperates
with TMPRSS2-ERG overexpression to promote prostate tumorigenesis
and cell invasion. Cancer Res. 77:6524–6537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu Z, Ju Y and Liu H: Antilung cancer
effect of glucosamine by suppressing the phosphorylation of FOXO.
Mol Med Rep. 16:3395–3400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Machado J, Manfredi LH, Silveira WA,
Gonçalves DAP, Lustrino D, Zanon NM, Kettelhut IC and Navegantes
LC: Calcitonin gene-related peptide inhibits autophagic-lysosomal
proteolysis through cAMP/PKA signaling in rat skeletal muscles. Int
J Biochem Cell Bio. 72:40–50. 2016. View Article : Google Scholar
|
|
41
|
Formosa R and Vassallo J: cAMP signalling
in the normal and tumorigenic pituitary gland. Mol Cell Endocrinol.
392:37–50. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Haroon N, Aggarwal A, Garg N, Krishnani N,
Kumari N and Agarwal V: An unusual case of systemic lupus
erythematosus mimic: disseminated gastric signet ring cell
carcinoma. Indian J Med Sci. 60:520–522. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shimoda T, Matsutani T, Yoshida H, Hosone
M and Uchida E: A case of gastric cancer associated with systemic
lupus erythematosus and nephrotic syndrome. Nihon Shokakibyo Gakkai
Zasshi. 110:1797–1803. 2013.(In Japanese). PubMed/NCBI
|
|
44
|
Velusamy T, Palanisamy N, Kalyana-Sundaram
S, Sahasrabuddhe AA, Maher CA, Robinson DR, Bahler DW, Cornell TT,
Wilson TE, Lim MS, et al: Recurrent reciprocal RNA chimera
involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proc
Natl Acad Sci USA. 110:3035–3040. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun D, Zhou M, Kowolik CM, Trisal V, Huang
Q, Kernstine KH, Lian F and Shen B: Differential expression
patterns of capping protein, protein phosphatase 1, and casein
kinase 1 may serve as diagnostic markers for malignant melanoma.
Melanoma Res. 21:335–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sander B, Xu W, Eilers M, Popov N and
Lorenz S: A conformational switch regulates the ubiquitin ligase
HUWE1. Elife. 6:e210362017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma W, Zhao P, Zang L, Zhang K, Liao H and
Hu Z: Tumour suppressive function of HUWE1 in thyroid cancer. J
Biosci. 41:395–405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang Z, Zhuang L, Szatmary P, Wen L, Sun
H, Lu Y, Xu Q and Chen X: Upregulation of heat shock proteins
(HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is
associated with poor outcomes from HBV-related early-stage
hepatocellular carcinoma. Int J Med Sci. 12:256–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Park JM, Kim JW and Hahm KB: HSPA4, the
‘Evil Chaperone’ of the HSP family, delays gastric ulcer healing.
Dig Dis Sci. 60:824–826. 2015. View Article : Google Scholar : PubMed/NCBI
|