|
1
|
Massimo R, Fassan M and Graham DY:
Epidemiology of gastric cancer. Gastric Cancer. Springer.
(Switzerland). 23–34. 2015.PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global Cancer Statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 Cancers in
185 Countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
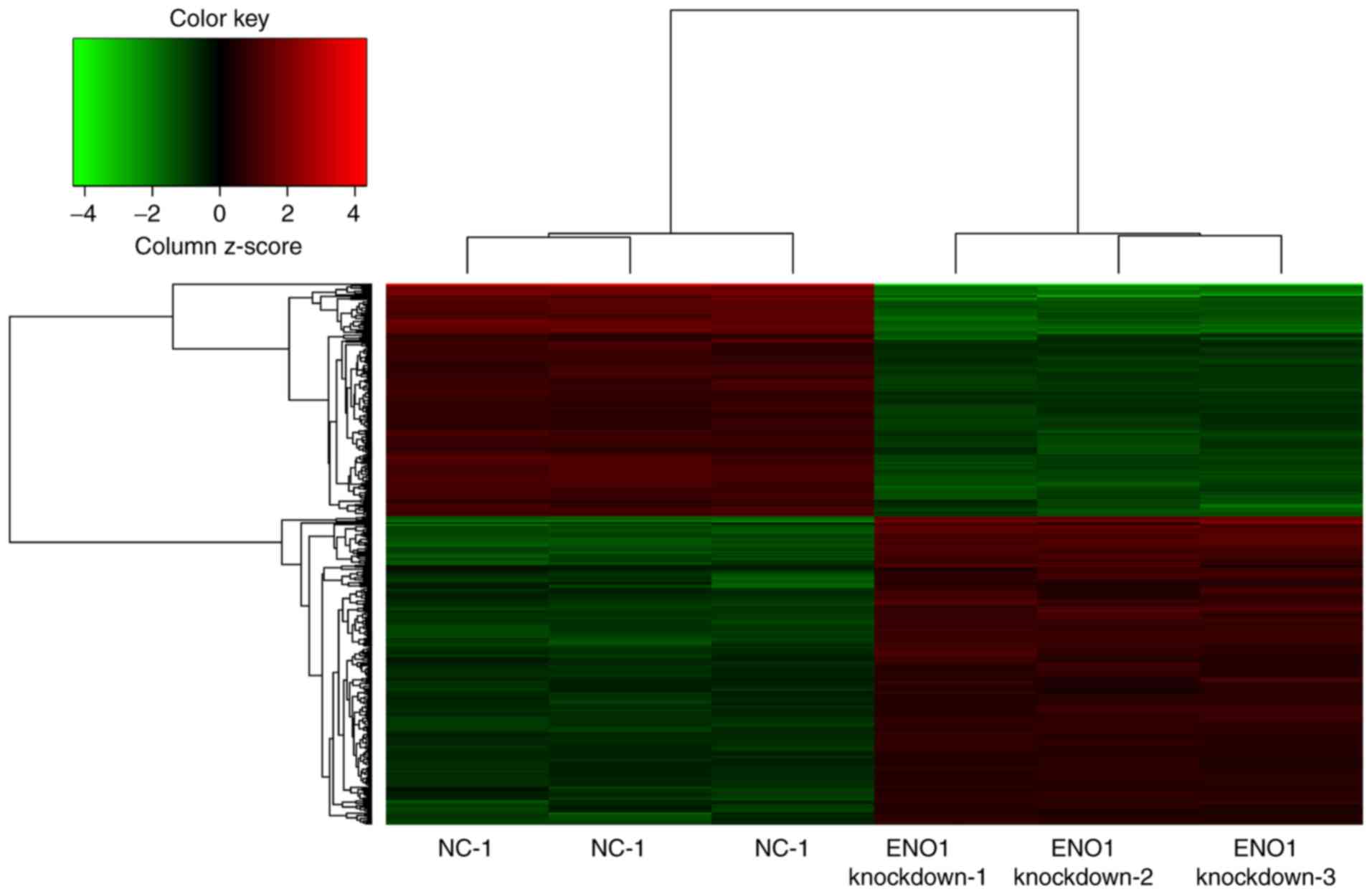
|
|
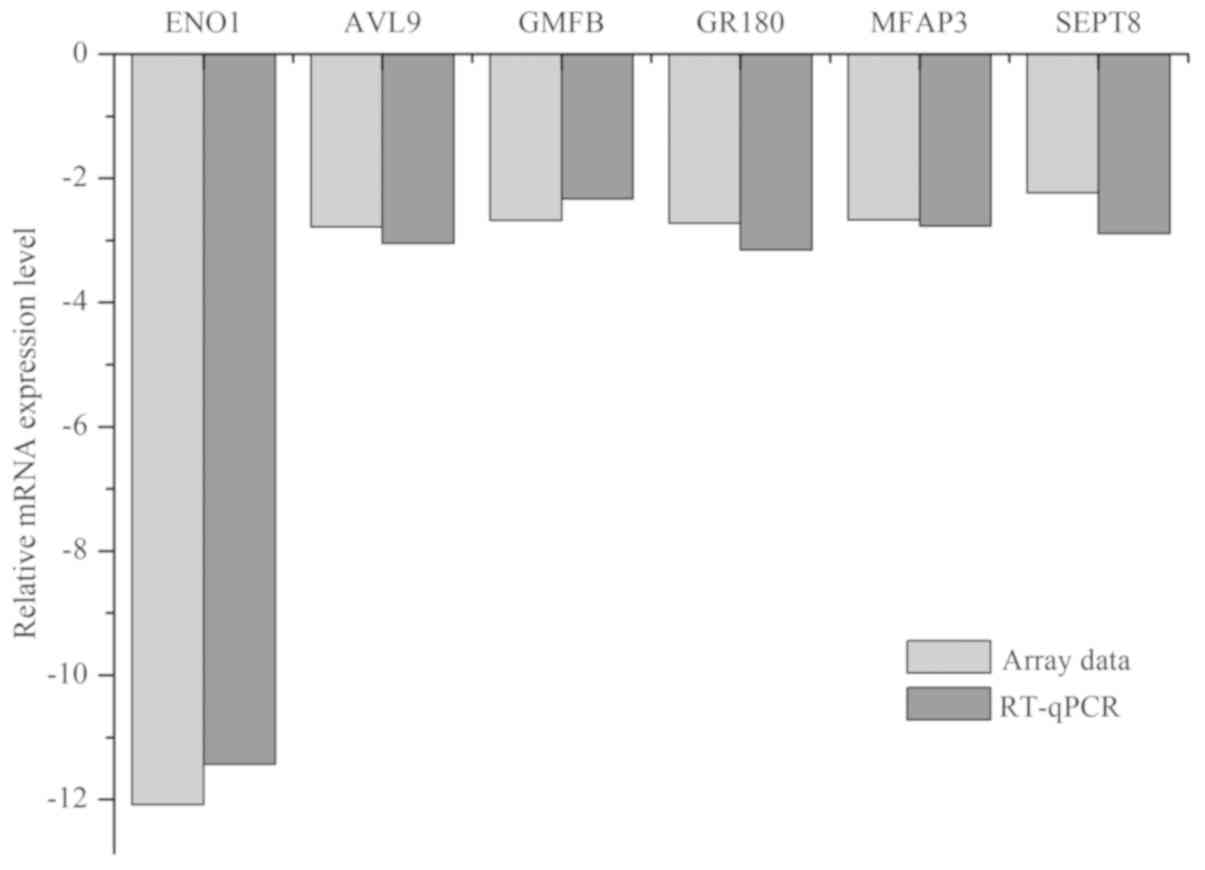
4
|
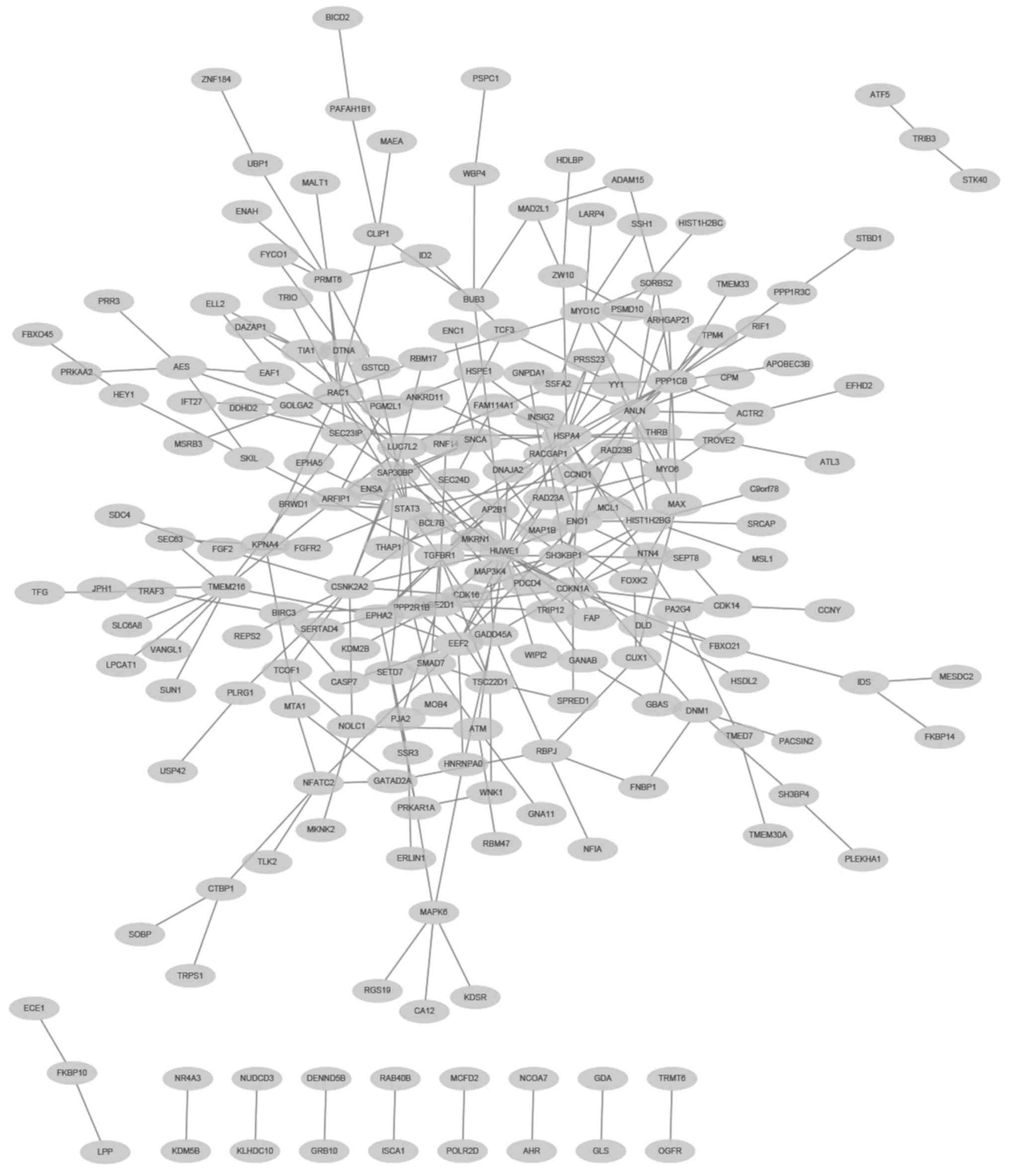
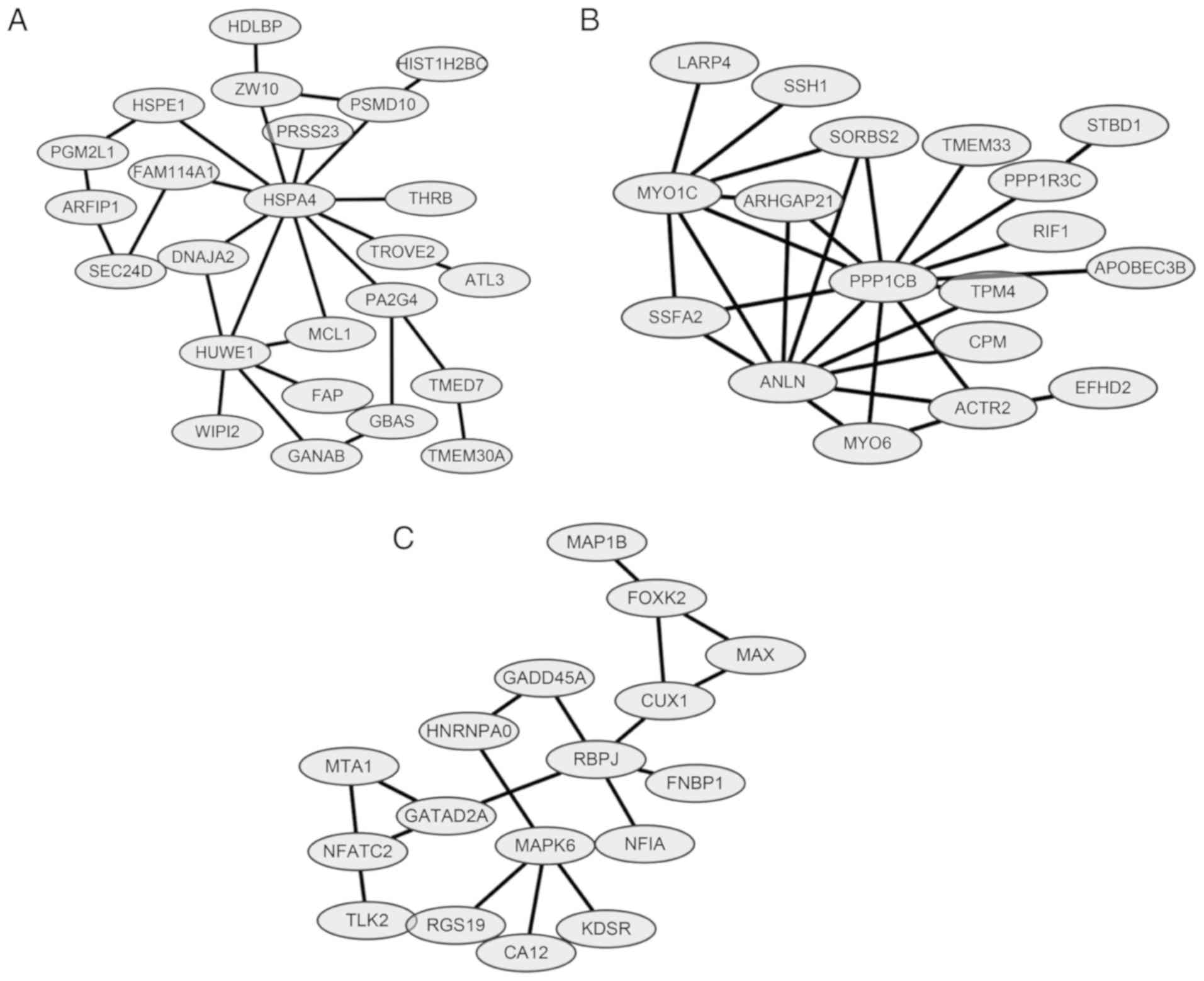
van Roy F: Beyond E-cadherin: Roles of
other cadherin superfamily members in cancer. Nat Rev Cancer.
14:121–134. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Majewski IJ, Kluijt I, Cats A, Scerri TS,
de Jong D, Kluin RJ, Hansford S, Hogervorst FB, Bosma AJ, Hofland
I, et al: An alpha-E-catenin (CTNNA1) mutation in hereditary
diffuse gastric cancer. J Pathol. 229:621–629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pazo Cid RA and Anton A: Advanced
HER2-positive gastric cancer: Current and future targeted
therapies. Crit Rev Oncol Hematol. 85:350–362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gazvoda B, Juvan R, Zupanic-Pajnic I,
Repse S, Ferlan-Marolt K, Balazic J and Komel R: Genetic changes in
Slovenian patients with gastric adenocarcinoma evaluated in terms
of microsatellite DNA. Eur J Gastroenterol Hepatol. 19:1082–1089.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Zheng L, Sun Y and Zhang X:
Analysis of the association of interleukin-17 gene polymorphisms
with gastric cancer risk and interaction with Helicobacter pylori
infection in a Chinese population. Tumour Biol. 35:1575–1580. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saeki N, Saito A, Choi IJ, Matsuo K,
Ohnami S, Totsuka H, Chiku S, Kuchiba A, Lee YS, Yoon KA, et al: A
functional single nucleotide polymorphism in mucin 1, at chromosome
1q22, determines susceptibility to diffuse-type gastric cancer.
Gastroenterology. 140:892–902. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garcia-Gonzalez MA, Bujanda L, Quintero E,
Santolaria S, Benito R, Strunk M, Sopeña F, Thomson C, Pérez-Aisa
A, Nicolás-Pérez D, et al: Association of PSCA rs2294008 gene
variants with poor prognosis and increased susceptibility to
gastric cancer and decreased risk of duodenal ulcer disease. Int J
Cancer. 137:1362–1373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zakrzewicz D, Didiasova M, Zakrzewicz A,
Hocke AC, Uhle F, Markart P, Preissner KT and Wygrecka M: The
interaction of enolase-1 with caveolae-associated proteins
regulates its subcellular localization. Biochem J. 460:295–307.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee SY, Jin CC, Choi JE, Hong MJ, Jung DK,
Do SK, Baek SA, Kang HJ, Kang HG, Choi SH, et al: Genetic
polymorphisms in glycolytic pathway are associated with the
prognosis of patients with early stage non-small cell lung cancer.
Sci Rep. 6:356032016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai L, Qu Y, Li J, Wang X, Wang K, Wang P,
Jiang BH and Zhang J: Serological proteome analysis approach-based
identification of ENO1 as a tumor-associated antigen and its
autoantibody could enhance the sensitivity of CEA and CYFRA 21-1 in
the detection of non-small cell lung cancer. Oncotarget.
8:36664–36673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao M, Fang W, Wang Y, Guo S, Shu L, Wang
L, Chen Y, Fu Q, Liu Y, Hua S, et al: Enolase-1 is a therapeutic
target in endometrial carcinoma. Oncotarget. 6:15610–15627.
2015.PubMed/NCBI
|
|
15
|
Gao J, Zhao R, Xue Y, Niu Z, Cui K, Yu F,
Zhang B and Li S: Role of enolase-1 in response to hypoxia in
breast cancer: Exploring the mechanisms of action. Oncol Rep.
29:1322–1332. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong
SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al: Alpha-enolase promotes
cell glycolysis, growth, migration, and invasion in non-small cell
lung cancer through FAK-mediated PI3K/AKT pathway. J Hematol Oncol.
8:222015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan P, Zhao S, Yan H, Yin C, Xiao Y, Wang
Y, Ni R, Chen W, Wei G and Zhang P: α-enolase promotes
tumorigenesis and metastasis via regulating AMPK/mTOR pathway in
colorectal cancer. Mol Carcinog. 56:1427–1437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qian X, Xu W, Xu J, Shi Q, Li J, Weng Y,
Jiang Z, Feng L, Wang X, Zhou J and Jin H: Enolase 1 stimulates
glycolysis to promote chemoresistance in gastric cancer.
Oncotarget. 8:47691–47708. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu YQ, Huang ZG, Li GN, Du JL, Ou YP,
Zhang XN, Chen TT and Liang QL: Effects of α-enolase (ENO1)
over-expression on malignant biological behaviors of AGS cells. Int
J Clin Exp Med. 8:231–239. 2015.PubMed/NCBI
|
|
20
|
Chen S, Duan G, Zhang R and Fan Q:
Helicobacter pylori cytotoxin-associated gene A protein upregulates
α-enolase expression via Src/MEK/ERK pathway: implication for
progression of gastric cancer. Int J Oncol. 45:764–770. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin Y and Tian YP: A microarray gene
analysis of peripheral whole blood in normal adult male rats after
long-term GH gene therapy. Cell Mol Biol Lett. 15:177–195. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ouyang P, Lin B, Du J, Pan H, Yu H, He R
and Huang Z: Global gene expression analysis of knockdown
Triosephosphate isomerase (TPI) gene in human gastric cancer cell
line MGC-803. Gene. 647:61–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Q: Identification of biomarkers for
metastatic osteosarcoma based on DNA microarray data. Neoplasma.
62:365–371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li HY, Jin N, Han YP and Jin XF: Pathway
crosstalk analysis in prostate cancer based on protein-protein
network data. Neoplasma. 64:22–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Jiang T, Li Z, Lu L, Zhang R,
Zhang D, Wang X and Tan J: Analysis of differentially co-expressed
genes based on microarray data of hepatocellular carcinoma.
Neoplasma. 64:216–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Li D, Tang Y, Wu F and Wang J:
CytoCluster: A cytoscape plugin for cluster analysis and
visualization of biological networks. Int J Mol Sci. 18(pii):
E18802017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang R, Zheng G, Ren D, Chen C, Zeng C, Lu
W and Li H: The clinical significance and biological function of
tropomyosin 4 in colon cancer. Biomed Pharmacother. 101:1–7. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun X, Liu X, Liu BO, Li S, Zhang D and
Guo H: Serum- and glucocorticoid-regulated protein kinase 3
overexpression promotes tumor development and aggression in breast
cancer cells. Oncol Lett. 12:437–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Coleman SJ, Chioni AM, Ghallab M, Anderson
RK, Lemoine NR, Kocher HM and Grose RP: Nuclear translocation of
FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic
cancer cell invasion. EMBO Mol Med. 6:467–481. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu M, Gu M, Zhang K, Zhou J, Wang Z and Da
J: miR-203 inhibition of renal cancer cell proliferation, migration
and invasion by targeting of FGF2. Diagn Pathol. 10:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ichikawa H, Itsumi M, Kajioka S, Maki T,
Lee K, Tomita M and Yamaoka S: Overexpression of exchange protein
directly activated by cAMP-1 (EPAC1) attenuates bladder cancer cell
migration. Biochem Biophys Res Commun. 495:64–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumar N, Gupta S, Dabral S, Singh S and
Sehrawat S: Role of exchange protein directly activated by cAMP
(EPAC1) in breast cancer cell migration and apoptosis. Mol Cell
Biochem. 430:115–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park JY and Juhnn YS: cAMP signaling
increases histone deacetylase 8 expression via the Epac2-Rap1A-Akt
pathway in H1299 lung cancer cells. Exp Mol Med. 49:e2972017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun DP, Fang CL, Chen HK, Wen KS, Hseu YC,
Hung ST, Uen YH and Lin KY: EPAC1 overexpression is a prognostic
marker and its inhibition shows promising therapeutic potential for
gastric cancer. Oncol Rep. 37:1953–1960. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bullock M: FOXO factors and breast cancer:
Outfoxing endocrine resistance. Endocr Relat Cancer. 23:R113–130.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang M, Zhao Y, Liu N, Chen E, Quan Z, Wu
X and Luo C: Overexpression of HepaCAM inhibits bladder cancer cell
proliferation and viability through the AKT/FoxO pathway. J Cancer
Res Clin Oncol. 143:793–805. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Y, Blee AM, Wang D, An J, Pan Y, Yan
Y, Ma T, He Y, Dugdale J, Hou X, et al: Loss of FOXO1 cooperates
with TMPRSS2-ERG overexpression to promote prostate tumorigenesis
and cell invasion. Cancer Res. 77:6524–6537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu Z, Ju Y and Liu H: Antilung cancer
effect of glucosamine by suppressing the phosphorylation of FOXO.
Mol Med Rep. 16:3395–3400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Machado J, Manfredi LH, Silveira WA,
Gonçalves DAP, Lustrino D, Zanon NM, Kettelhut IC and Navegantes
LC: Calcitonin gene-related peptide inhibits autophagic-lysosomal
proteolysis through cAMP/PKA signaling in rat skeletal muscles. Int
J Biochem Cell Bio. 72:40–50. 2016. View Article : Google Scholar
|
|
41
|
Formosa R and Vassallo J: cAMP signalling
in the normal and tumorigenic pituitary gland. Mol Cell Endocrinol.
392:37–50. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Haroon N, Aggarwal A, Garg N, Krishnani N,
Kumari N and Agarwal V: An unusual case of systemic lupus
erythematosus mimic: disseminated gastric signet ring cell
carcinoma. Indian J Med Sci. 60:520–522. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shimoda T, Matsutani T, Yoshida H, Hosone
M and Uchida E: A case of gastric cancer associated with systemic
lupus erythematosus and nephrotic syndrome. Nihon Shokakibyo Gakkai
Zasshi. 110:1797–1803. 2013.(In Japanese). PubMed/NCBI
|
|
44
|
Velusamy T, Palanisamy N, Kalyana-Sundaram
S, Sahasrabuddhe AA, Maher CA, Robinson DR, Bahler DW, Cornell TT,
Wilson TE, Lim MS, et al: Recurrent reciprocal RNA chimera
involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proc
Natl Acad Sci USA. 110:3035–3040. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun D, Zhou M, Kowolik CM, Trisal V, Huang
Q, Kernstine KH, Lian F and Shen B: Differential expression
patterns of capping protein, protein phosphatase 1, and casein
kinase 1 may serve as diagnostic markers for malignant melanoma.
Melanoma Res. 21:335–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sander B, Xu W, Eilers M, Popov N and
Lorenz S: A conformational switch regulates the ubiquitin ligase
HUWE1. Elife. 6:e210362017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma W, Zhao P, Zang L, Zhang K, Liao H and
Hu Z: Tumour suppressive function of HUWE1 in thyroid cancer. J
Biosci. 41:395–405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang Z, Zhuang L, Szatmary P, Wen L, Sun
H, Lu Y, Xu Q and Chen X: Upregulation of heat shock proteins
(HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is
associated with poor outcomes from HBV-related early-stage
hepatocellular carcinoma. Int J Med Sci. 12:256–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Park JM, Kim JW and Hahm KB: HSPA4, the
‘Evil Chaperone’ of the HSP family, delays gastric ulcer healing.
Dig Dis Sci. 60:824–826. 2015. View Article : Google Scholar : PubMed/NCBI
|