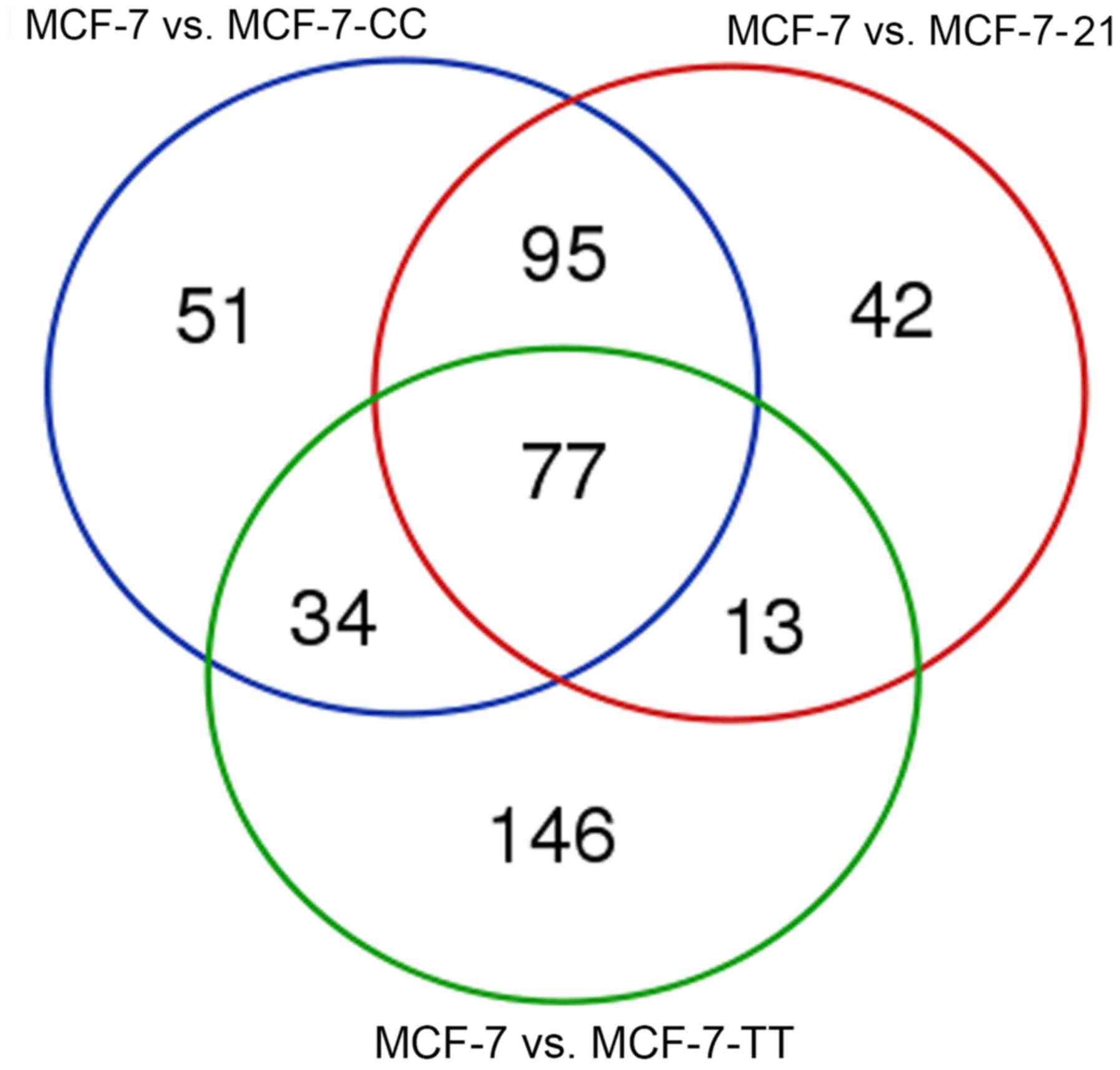
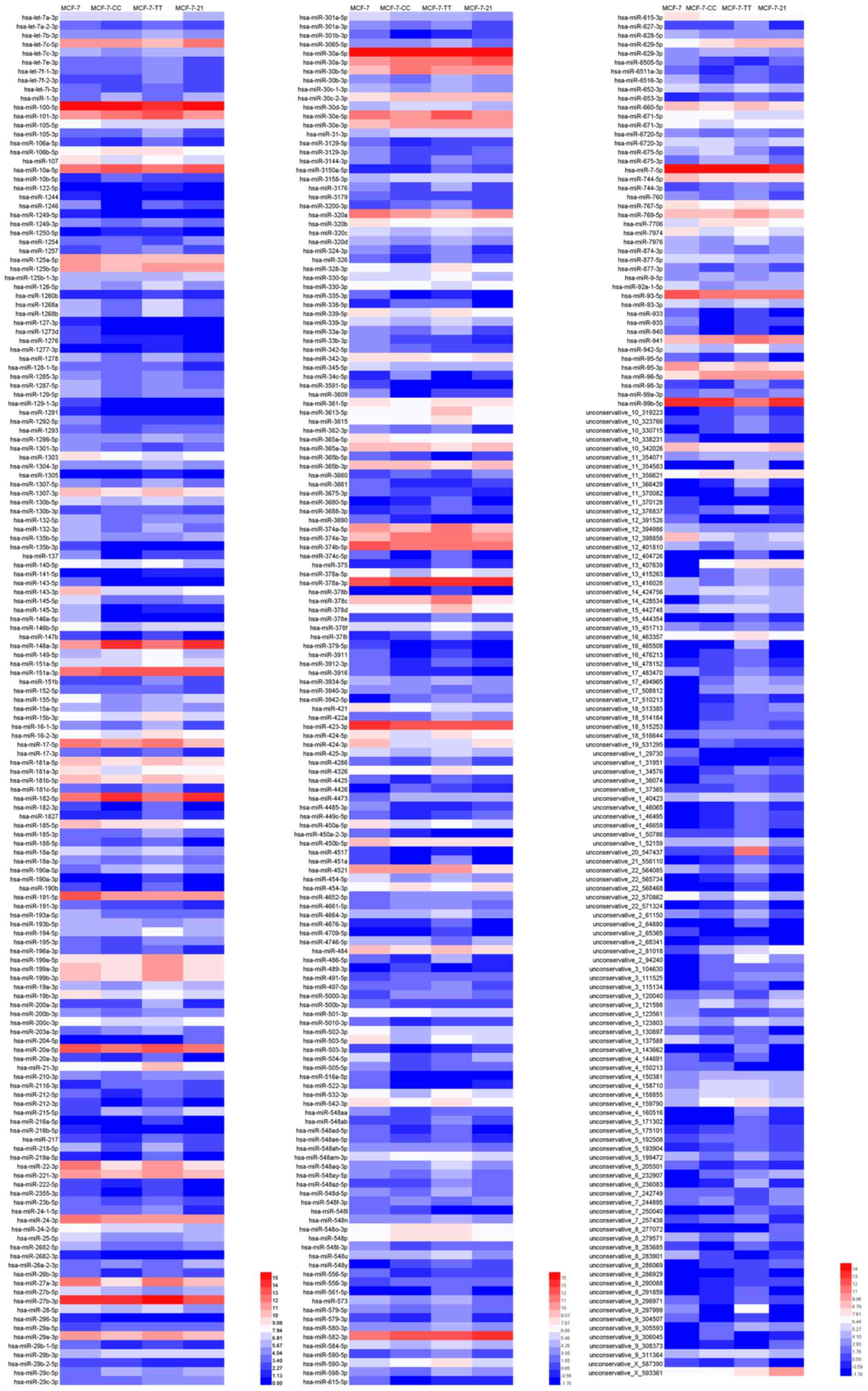
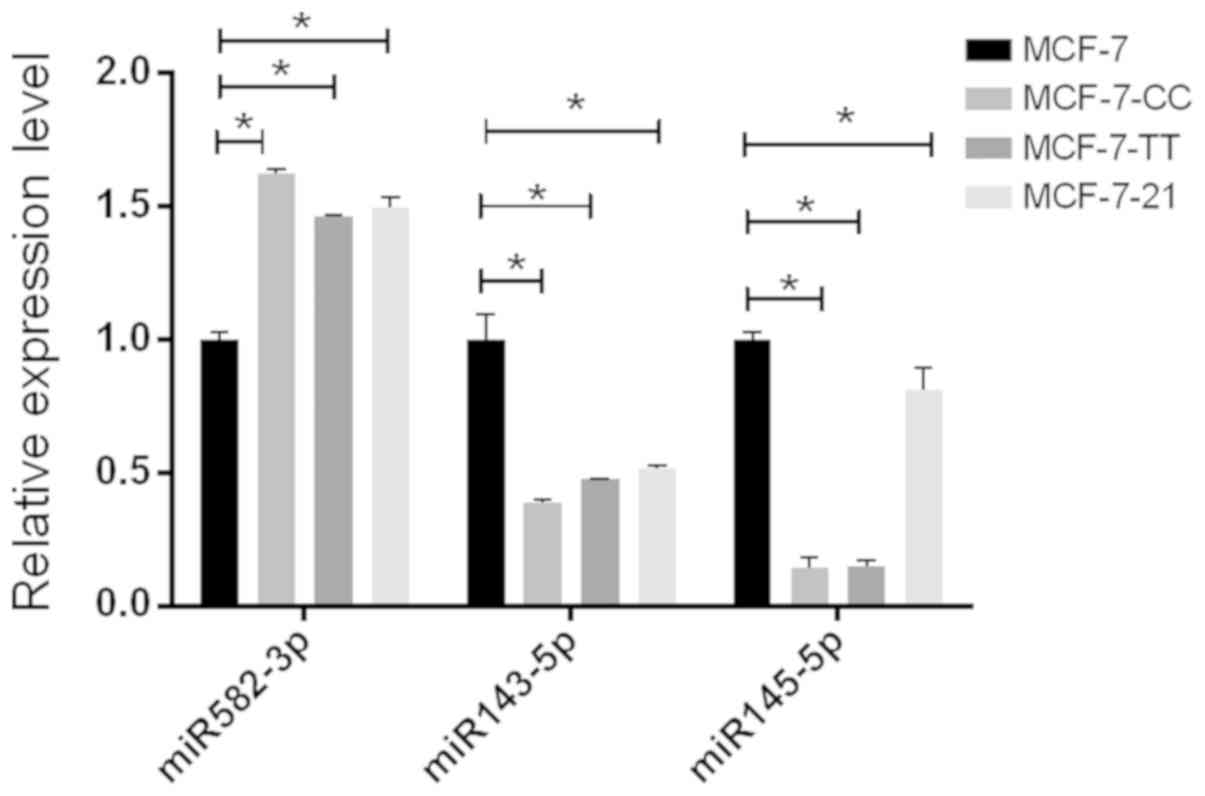
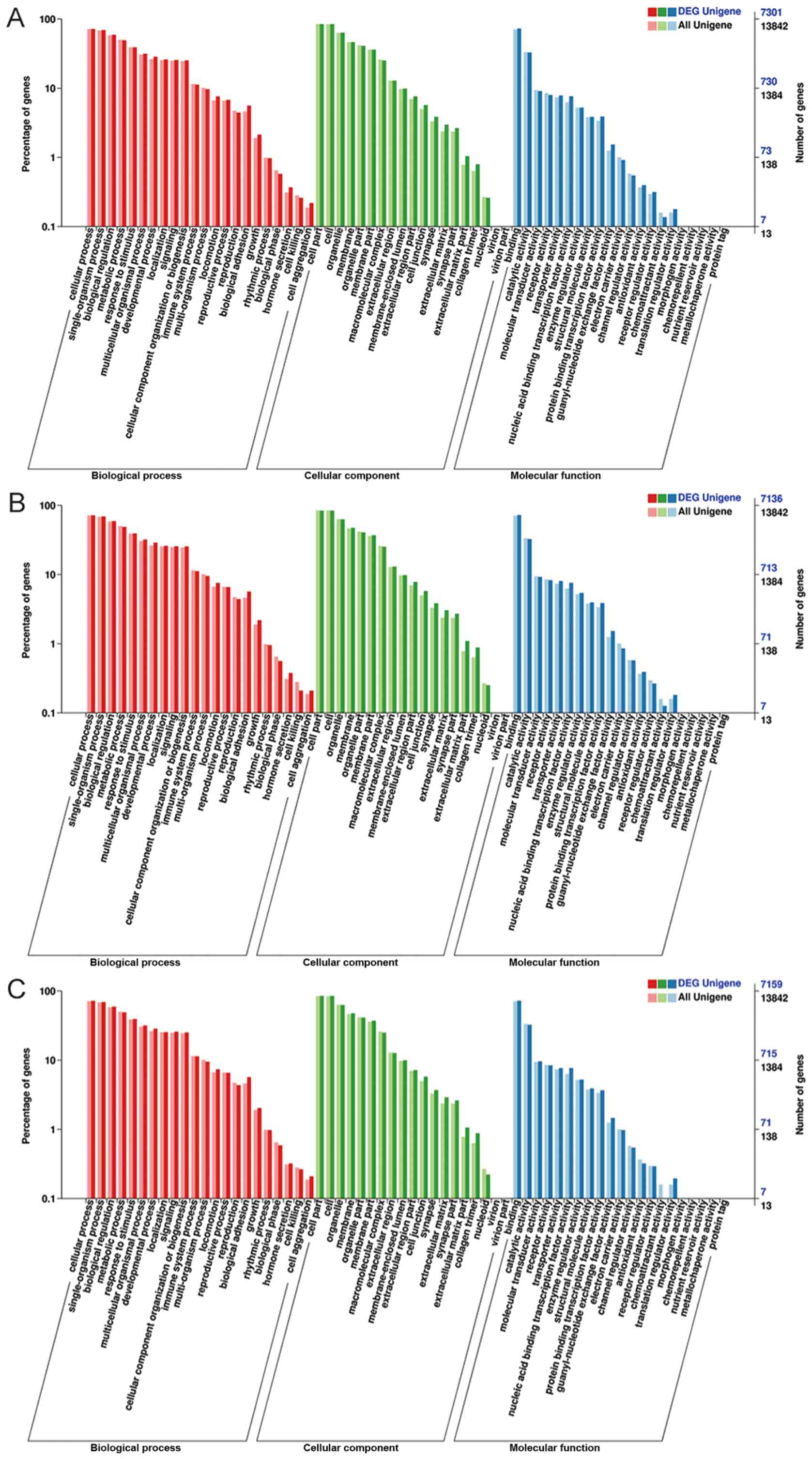
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Osborne CK, Wakeling A and Nicholson RI:
Fulvestrant: An oestrogen receptor antagonist with a novel
mechanism of action. Br J Cancer. 90 Suppl 1:S2–S6. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tangkeangsirisin W and Serrero G: GP88
(Progranulin) confers fulvestrant (Faslodex, ICI 182,780)
resistance to human breast cancer cells. Adv Breast Cancer Res.
03:68–78. 2014. View Article : Google Scholar
|
|
4
|
Osipo C, Meeke K, Cheng D, Weichel A,
Bertucci A, Liu H and Jordan VC: Role for HER2/neu and HER3 in
fulvestrant-resistant breast cancer. Int J Oncol. 30:509–520.
2007.PubMed/NCBI
|
|
5
|
Tsuboi K, Kaneko Y, Nagatomo T, Fujii R,
Hanamura T, Gohno T, Yamaguchi Y, Niwa T and Hayashi SI: Different
epigenetic mechanisms of ERalpha implicated in the fate of
fulvestrant-resistant breast cancer. J Steroid Biochem Mol Biol.
167:115–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hong L, Yang Z, Ma J and Fan D: Function
of miRNA in controlling drug resistance of human cancers. Curr Drug
Targets. 14:1118–1127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
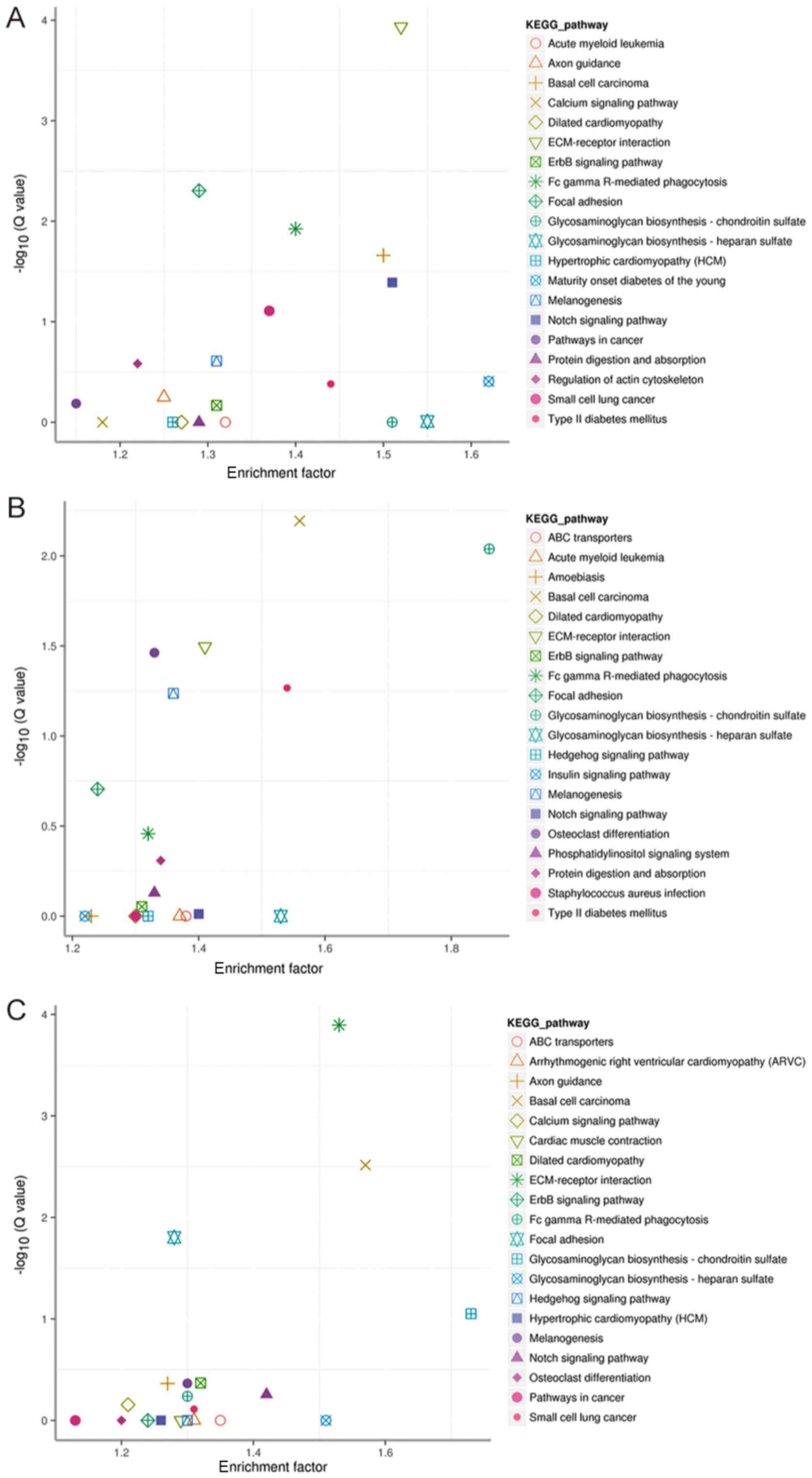
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ji S, Shao G, Lv X, Liu Y, Fan Y, Wu A and
Hu H: Downregulation of miRNA-128 sensitises breast cancer cell to
chemodrugs by targeting Bax. Cell Biol Int. 37:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH and Tang JH: Down-regulation
of miRNA-452 is associated with adriamycin-resistance in breast
cancer cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan P, Yue W, Wang JP, Aiyar S, Li Y, Kim
TH and Santen RJ: Mechanisms of resistance to structurally diverse
antiestrogens differ under premenopausal and postmenopausal
conditions: Evidence from in vitro breast cancer cell models.
Endocrinology. 150:2036–2045. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye P, Fang C, Zeng H, Shi Y, Pan Z, An N,
He K, Zhang L and Long X: Differential microRNA expression profiles
in tamoxifen-resistant human breast cancer cell lines induced by
two methods. Oncol Lett. 15:3532–3539. 2018.PubMed/NCBI
|
|
14
|
Fan M, Yan PS, Hartman-Frey C, Chen L,
Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al:
Diverse gene expression and DNA methylation profiles correlate with
differential adaptation of breast cancer cells to the antiestrogens
tamoxifen and fulvestrant. Cancer Res. 66:11954–11966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jensen BL, Skouv J, Lundholt BK and
Lykkesfeldt AE: Differential regulation of specific genes in MCF-7
and the ICI 182780-resistant cell line MCF-7/182R-6. Br J Cancer.
79:386–392. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coser KR, Wittner BS, Rosenthal NF,
Collins SC, Melas A, Smith SL, Mahoney CJ, Shioda K, Isselbacher
KJ, Ramaswamy S and Shioda T: Antiestrogen-resistant subclones of
MCF-7 human breast cancer cells are derived from a common
monoclonal drug-resistant progenitor. Proc Natl Acad Sci USA.
106:14536–14541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stokowy T, Eszlinger M, Świerniak M,
Fujarewicz K, Jarząb B, Paschke R and Krohn K: Analysis options for
high-throughput sequencing in miRNA expression profiling. BMC Res
Notes. 7:1442014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fahlgren N, Howell MD, Kasschau KD,
Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR,
Dangl JL and Carrington JC: High-throughput sequencing of
Arabidopsis microRNAs: Evidence for frequent birth and death of
MIRNA genes. PLoS One. 2:e2192007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Witten DM, Johnstone IM and
Tibshirani R: Normalization, testing, and false discovery rate
estimation for RNA-sequencing data. Biostatistics. 13:523–538.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One. 9:e1119882014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ng EK, Li R, Shin VY, Siu JM, Ma ES and
Kwong A: MicroRNA-143 is downregulated in breast cancer and
regulates DNA methyltransferases 3A in breast cancer cells. Tumour
Biol. 35:2591–2598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
30
|
Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng
L, Zhou H and Zhao RC: miR-145 inhibits breast cancer cell growth
through RTKN. Int J Oncol. 34:1461–1466. 2009.PubMed/NCBI
|
|
31
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye P, Shi Y, An N, Zhou Q, Guo J and Long
X: miR-145 overexpression triggers alteration of the whole
transcriptome and inhibits breast cancer development. Biomed
Pharmacother. 100:72–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gojis O, Rudraraju B, Gudi M, Hogben K,
Sousha S, Coombes RC, Cleator S and Palmieri C: The role of SRC-3
in human breast cancer. Nat Rev Clin Oncol. 7:83–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eedunuri VK, Rajapakshe K, Fiskus W, Geng
C, Chew SA, Foley C, Shah SS, Shou J, Mohamed JS, Coarfa C, et al:
miR-137 targets p160 steroid receptor coactivators SRC1, SRC2, and
SRC3 and inhibits cell proliferation. Mol Endocrinol. 29:1170–1183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Denis H, Van Grembergen O, Delatte B,
Dedeurwaerder S, Putmans P, Calonne E, Rothé F, Sotiriou C, Fuks F
and Deplus R: MicroRNAs regulate KDM5 histone demethylases in
breast cancer cells. Mol Biosyst. 12:404–413. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y
and He F: MiR-137 targets estrogen-related receptor alpha and
impairs the proliferative and migratory capacity of breast cancer
cells. PLoS One. 7:e391022012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han Y, Bi Y, Bi H, Diao C, Zhang G, Cheng
K and Yang Z: miR-137 suppresses the invasion and procedure of EMT
of human breast cancer cell line MCF-7 through targeting CtBP1. Hum
Cell. 29:30–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 restoration sensitizes multidrug-resistant
MCF-7/ADM cells to anticancer agents by targeting YB-1. Acta
Biochim Biophys Sin (Shanghai). 45:80–86. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xin F, Li M, Balch C, Thomson M, Fan M,
Liu Y, Hammond SM, Kim S and Nephew KP: Computational analysis of
microRNA profiles and their target genes suggests significant
involvement in breast cancer antiestrogen resistance.
Bioinformatics. 25:430–434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vilquin P, Donini CF, Villedieu M, Grisard
E, Corbo L, Bachelot T, Vendrell JA and Cohen PA: MicroRNA-125b
upregulation confers aromatase inhibitor resistance and is a novel
marker of poor prognosis in breast cancer. Breast Cancer Res.
17:132015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rodriguez-Barrueco R, Nekritz EA, Bertucci
F, Yu J, Sanchez-Garcia F, Zeleke TZ, Gorbatenko A, Birnbaum D,
Ezhkova E, Cordon-Cardo C, et al: miR-424(322)/503 is a breast
cancer tumor suppressor whose loss promotes resistance to
chemotherapy. Genes Dev. 31:553–566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F,
Wang X, He X and Zhao Y and Zhao Y: Prognostic role of microRNA-21
in various carcinomas: A systematic review and meta-analysis. Eur J
Clin Invest. 41:1245–1253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blower PE, Chung JH, Verducci JS, Lin S,
Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et
al: MicroRNAs modulate the chemosensitivity of tumor cells. Mol
Cancer Ther. 7:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu X, Li R, Shi W, Jiang T, Wang Y, Li C
and Qu X: Silencing of microRNA-21 confers the sensitivity to
tamoxifen and fulvestrant by enhancing autophagic cell death
through inhibition of the PI3K-AKT-mTOR pathway in breast cancer
cells. Biomed Pharmacother. 77:37–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou Q, Zeng H, Ye P, Shi Y, Guo J and
Long X: Differential microRNA profiles between
fulvestrant-resistant and tamoxifen-resistant human breast cancer
cells. Anticancer Drugs. 29:539–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Welshons WV, Wolf MF, Murphy CS and Jordan
VC: Estrogenic activity of phenol red. Mol Cell Endocrinol.
57:169–178. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Di Leva G, Gasparini P, Piovan C, Ngankeu
A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, et
al: MicroRNA cluster 221–222 and estrogen receptor alpha
interactions in breast cancer. J Natl Cancer Inst. 102:706–721.
2010. View Article : Google Scholar
|
|
51
|
Yeh MH, Tzeng YJ, Fu TY, You JJ, Chang HT,
Ger LP and Tsai KW: Extracellular matrix-receptor interaction
signaling genes associated with inferior breast cancer survival.
Anticancer Res. 38:4593–4605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dontu G, Jackson KW, McNicholas E,
Kawamura MJ, Abdallah WM and Wicha MS: Role of Notch signaling in
cell-fate determination of human mammary stem/progenitor cells.
Breast Cancer Res. 6:R605–R615. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li L, Tang P, Li S, Qin X, Yang H, Wu C
and Liu Y: Notch signaling pathway networks in cancer metastasis: A
new target for cancer therapy. Med Oncol. 34:1802017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alketbi A and Attoub S: Notch signaling in
cancer: Rationale and strategies for targeting. Curr Cancer Drug
Targets. 15:364–374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hoskin V, Szeto A, Ghaffari A, Greer PA,
Cote GP and Elliott BE: Ezrin regulates focal adhesion and
invadopodia dynamics by altering calpain activity to promote breast
cancer cell invasion. Mol Biol Cell. 26:3464–3479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Negoro K, Yamano Y, Fushimi K, Saito K,
Nakatani K, Shiiba M, Yokoe H, Bukawa H, Uzawa K, Wada T, et al:
Establishment and characterization of a cisplatin-resistant cell
line, KB-R, derived from oral carcinoma cell line, KB. Int J Oncol.
30:1325–1332. 2007.PubMed/NCBI
|
|
58
|
Watson MB, Lind MJ and Cawkwell L:
Establishment of in-vitro models of chemotherapy resistance.
Anticancer Drugs. 18:749–754. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang LY and Trujillo JM: Biological
characterization of multidrug-resistant human colon carcinoma
sublines induced/selected by two methods. Cancer Res. 50:3218–3225.
1990.PubMed/NCBI
|