Introduction
Breast cancer is one of the most common types of
tumor in women, and has been estimated to account for 29% of all
novel cases of cancer diagnosed among American women in 2016
(1). Breast cancer is an
endocrine-dependent cancer, with ~70% of cases being estrogen
receptor positive (ER+). Estrogen has been reported to
stimulate tumor growth, and endocrine therapy is one of the main
treatment strategies for ER+ breast cancer patients,
decreasing the estrogen levels or inhibiting the function of
estrogen. Effective endocrine therapy is important for the
treatment of breast cancer, and reduces the rate of relapse and
mortality of patients.
Fulvestrant, a selective ER downregulator (SERD), is
specifically used to treat postmenopausal breast cancer patients
following the failure of first-line endocrine therapies (such as
tamoxifen). SERDs function by binding to and blocking the ER, thus
leading to the inhibition of estrogen signaling by targeting the ER
(2). However, drug resistance is a
factor that limits the efficacy of fulvestrant. Certain breast
cancer patients acquire resistance against fulvestrant as a result
of long-term treatment through various mechanisms, including
glycoprotein 88 overexpression (3),
functional interaction of human epidermal growth factor receptor 2
(HER2)/neu with HER3 (4), and
methylation of the ER promoter region (5). It is crucial to understand the
underlying mechanisms involved in fulvestrant resistance, since it
is a major clinical obstacle in breast cancer treatment; however,
these mechanisms remain unknown.
Aberrant expression of microRNAs (miRNAs) is
considered to be one of the numerous mechanisms of drug resistance
(6). miRNAs are a class of small
non-coding RNAs (with a length of ~22 nucleotides) that regulate
gene expression at the post-transcriptional level by binding to the
3′-untranslated region (3′UTR) of target mRNAs to either inhibit
mRNA translation or target the molecule for degradation. miRNAs
participate in a series of important biological processes,
including cell proliferation, differentiation and apoptosis, as
well as signal transduction (7,8). A
number of studies in recent years have reported that aberrant
expression of miRNA is closely associated with drug resistance in
breast cancer, including fulvestrant resistance (9–11).
However, the differential miRNA expression profiles of
fulvestrant-resistant human breast cancer cell lines have rarely
been studied. Thus, three different fulvestrant-resistant cell
lines were established in the current study, which represent a
variety of drug resistance conditions. These cells may exhibit
exogenous or endogenous resistance-associated characteristics, as
reported previously (12–16).
The current study aimed to investigate the
association between differential miRNA expression profiles and
fulvestrant resistance in human breast cancer cells. The known and
predicted miRNAs were detected by next-generation sequencing, which
has a stronger ability to identify genes compared with DNA
microarray (17).
Materials and methods
Cells and reagents
The human breast cancer cell line MCF-7 was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's
medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences) at 37°C in a humidified atmosphere
containing 5% CO2. Fulvestrant was purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
Establishment of three
fulvestrant-resistant cell lines
Establishment of MCF-7-CC cell
line
An MCF-7-CC cell line was established using the
human breast cancer cell line MCF-7, which served as the parental
cell line. Resistance was induced using fulvestrant as the
screening drug by stepwise induction, starting with a low
concentration (12.5 nM) of fulvestrant (13). This method was conducted in two
sequential phases: Adaptation and consolidation. In the adaptation
phase, MCF-7 cells were cultured in complete medium until the
adhesion rate reached 70%; next, 12.5 nM fulvestrant was added to
cells plated in DMEM (containing phenol red) with 5% FBS for 72 h.
Apoptotic cells were then removed and the complete medium was
replaced until the viable cells reached a 70% adherence rate. This
process was repeated twice, and MCF-7 cells were collectively
incubated with 12.5 nM fulvestrant for a total of three times.
Subsequently, the treated cells were incubated with 25, 50, 100,
200, 400, 800 and 1,000 nM fulvestrant, respectively, in the same
manner. Previous studies (14,15)
reported using 100 nM fulvestrant to establish the
fulvestrant-resistant cell line in medium containing
charcoal-stripped FBS. However, medium containing FBS was used in
the current study; thus, it was necessary to increase the
fulvestrant concentration, and 1,000 nM was used as the final dose.
In the consolidation phase, cells incubated with 1,000 nM
fulvestrant in DMEM were then treated with 1,200 nM fulvestrant for
an additional 10 times until normal growth was observed, indicating
that the MCF-7-CC cell line was successfully established.
Establishment of MCF-7-TT cell
line
An MCF-7-TT cell line was established by stepwise
temporal induction, beginning with a high concentration of
fulvestrant (13). The process of
induction was also divided into two stages, including adaptation
and consolidation. Briefly, MCF-7 cells were cultured in complete
medium until the adhesion rate reached 70%. Next, the complete
medium (DMEM with 10% FBS) was discarded, and the screening medium
[DMEM (containing phenol red) with 5% FBS] with 1,000 nM
fulvestrant was used to induce the cells for 1 h. The screening
medium was discarded and replaced with complete culture medium
subsequent to washing three times with PBS (Hyclone; GE Healthcare
Life Sciences). Apoptotic cells were then removed and the complete
medium was replaced until the viable cells reached a 70% adherence
rate. This process was repeated twice, and the MCF-7 cells were
collectively induced with 1,000 nM fulvestrant for 1 h for a total
of three times. Subsequently, 2, 4, 8, 12, 24, 36 and 48 h of
induction were sequentially performed with 1,000 nM fulvestrant
following the aforementioned screening procedure. In the
consolidation phase, the cell line obtained from the adaptation
stage was cultured in 1,000 nM fulvestrant containing medium for 72
h for a total of ten times, following which the MCF-7-TT cell line
was successfully established.
Establishment of MCF-7-21 cell
line
An MCF-7-21 cell line was established using the
human breast cancer cell line MCF-7 as the parental cell line,
which was incubated with a high concentration of fulvestrant for 21
days (16). MCF-7 cells were
cultured in the DMEM (containing phenol red) with 5% FBS, and then
1,000 nM fulvestrant was added to the medium. MCF-7 cells were
cultured for 21 days, following which the MCF-7-21 cell line was
successfully established.
Total cellular RNA extraction
Total RNA of the three fulvestrant-resistant cell
lines was extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. A NanoDrop spectrophotometer (Thermo Fisher Scientific,
Inc.), Qubit 2.0 fluorometer (Thermo Fisher Scientific, Inc.) and
an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa
Clara, CA, USA) were used to detect the purity, concentration and
integrity of RNA samples, respectively.
Small RNA library construction and
sequencing
miRNA high-throughput sequencing was performed by
Beijing Biomarker Biotechnology Co., Ltd. (Beijing, China), and the
small RNA library was constructed and sequenced as follows: A total
of 1.5 µg RNA was used as the initial sample, and the volume was
made up to 6 µl with water. The library was constructed using the
small RNA Sample Prep kit (Illumina, Inc., San Diego, CA, USA)
following the manufacturer's recommendations. As a phosphate group
was present at the 5′ terminus and a hydroxyl group at the 3′
terminus, small RNAs were ligated at the 3′ and 5′ ends using T4
RNA Ligase 1 and T4 RNA Ligase 2 (truncated), respectively. Next,
cDNA was synthesized by reverse transcription (RT) with a RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.),
and the cDNA fragments were subsequently amplified by polymerase
chain reaction (PCR), cloned and sequenced. The target gene
fragment was isolated by a gel separation technique, cut and
collected as a small RNA library. Following the generation of the
library, the concentration of the constructed library was detected
by Qubit 2.0 device, and the library was diluted to a concentration
of 1 ng/µl. The insert size was then tested using the Agilent 2100
bioanalyzer and the working concentration of the library was
determined by a quantitative PCR (qPCR) method to obtain a
high-quality library. High-throughput sequencing of the library was
finally performed on the Illumina HiSeq 2500 sequencing platform
(Illumina, Inc.).
miRNA identification and differential
expression analysis
MiRDeep2 software was used to identify known miRNAs
and predict novel miRNAs (18). The
expression of miRNAs in each sample was analyzed statistically, and
the expression level was normalized using a transcript per million
algorithm (19). The thresholds for
significant miRNA differential expression analysis were set as
follows: |log2 fold change (FC)| of ≥1 and false
discovery rate (FDR) of ≤0.01. The FC represents the ratio of
expression between two samples. The significant P-value obtained
from the hypothesis can be expressed as the probability of miRNAs
expressing no difference between the fulvestrant-resistant cell
line and MCF-7 cell line. As the differential expression analysis
of miRNAs is an independent statistical hypothesis test for the
expression of multiple miRNAs, false-positive results may be
detected. Thus, the Benjamini-Hochberg correction method was used
to test the P-value obtained from the hypothesis. Finally, the FDR
was used as the key indicator of differential expression of miRNAs
during the screening process (20).
The genes obtained from the differential expression analysis were
referred to as differentially expressed genes (DEG). Cluster
analysis for differentially expressed miRNAs was performed using
Heatmap Illustrator software (21).
Validation of three selected
differentially expressed miRNAs by RT-qPCR
RT-qPCR was used to confirm the expression levels of
three miRNAs, namely miR-582-3p, miR-143-5p and miR-145-5p. These
miRNAs were obtained from the next-generation sequencing analysis
and were differentially expressed in all three
fulvestrant-resistant cell lines compared with the parental MCF-7
cell line. Briefly, the extracted total cellular RNA was reverse
transcribed into cDNA using a miScript II Reverse Transcription kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. qPCR was then performed using a LightCycler®
480 II Real-time PCR instrument (Roche Diagnostics, Basel,
Switzerland) with 10-µl PCR reaction mixture, containing 1 µl cDNA,
5 µl 2X LightCycler® 480 SYBR Green I Master (Roche
Diagnostics), 0.2 µl universal primer (Qiagen GmbH), 0.2 µl
miRNA-specific primer and 3.6 µl nuclease-free water. The primer
sequences are presented in Table I.
U6 small nuclear RNA was used as the internal control for the
normalization of miRNA expression. The reaction mixture was
pre-incubated at 95°C for 10 min, followed by 40 cycles of 95°C for
10 sec and 60°C for 30 sec. Each sample was run in triplicate. The
relative expression of the selected differentially expressed miRNAs
was calculated using the 2−ΔΔCq method (22).
 | Table I.Sequences of primers using for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of primers using for
reverse transcription-quantitative polymerase chain reaction.
| Target | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| miR-582-3p |
TAACTGGTTGAACAAC |
GTGCAGGGTCCGAGGT |
| miR-143-5p |
GGTGCAGTGCTGCATC |
GTGCAGGGTCCGAGGT |
| miR-145-5p |
GTCCAGTTTTCCCAGGA |
GTGCAGGGTCCGAGGT |
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses of target
genes
MicroRNA.org, a comprehensive
resource of miRNA target predictions and expression profiles
(23), and the RNAhybrid tool that
is primarily used as a means for miRNA target prediction (24) were used to predict the DEGs.
Wallenius' non-central hypergeometric distribution-based analysis
of GO enrichment of the DEGs was implemented using the GOseq R
packages (25). In addition, KOBAS
software (26) was used to assess
the statistical enrichment of DEGs in KEGG pathways.
Statistical analysis
SPSS version 22 (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. Differences were evaluated using
one-way analysis of variance with a post-hoc Student-Newman-Keuls
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression profile of miRNAs in
fulvestrant-resistant cell lines
A next-generation sequencing instrument (Illumina
HiSeq 2500) was used to detect the expression profile of
differentially expressed miRNAs. The results revealed that 1,536
miRNAs were detected in all samples, including 1,240 known miRNAs
and 296 predicted miRNAs. A total of 257 miRNAs with significant
differences between MCF-7-CC and MCF-7 cells were identified
(P<0.05), of which 69 miRNAs were upregulated (P<0.05), and
188 were downregulated (P<0.05). There were 270 differentially
expressed miRNAs between MCF-7-TT and MCF-7 cells (P<0.05), of
which 180 miRNAs were upregulated (P<0.05) and 90 were
downregulated (P<0.05). It was also observed that 227 miRNAs
exhibited significantly different expression between MCF-7-21 and
MCF-7 cells (P<0.05), of which 52 miRNAs were upregulated
(P<0.05) and 175 miRNAs were downregulated (P<0.05). These
results are listed in Table II.
 | Table II.Statistical data of differentially
expressed miRNAs in fulvestrant-resistant cell lines as compared
with the MCF-7 parental cell line. |
Table II.
Statistical data of differentially
expressed miRNAs in fulvestrant-resistant cell lines as compared
with the MCF-7 parental cell line.
|
| Differentially
expressed miRNAs |
|---|
|
|
|
|---|
| Cell lines | Total no. | Upregulated | Downregulated |
|---|
| MCF-7 vs.
MCF-7-CC | 257 | 69 | 188 |
| MCF-7 vs.
MCF-7-TT | 270 | 180 | 90 |
| MCF-7 vs.
MCF-7-21 | 227 | 52 | 175 |
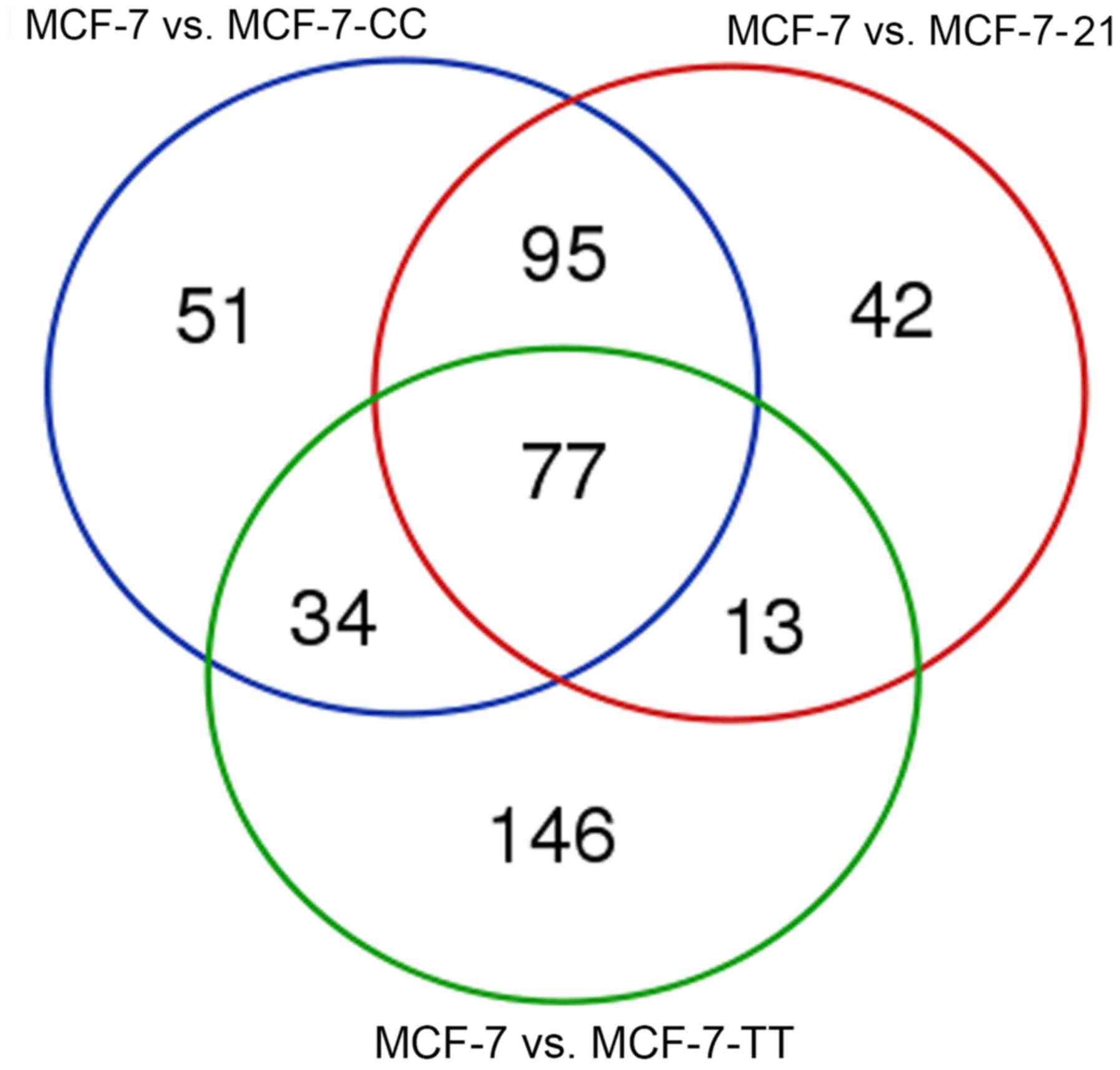
The overlapping results of the differentially
expressed miRNAs in MCF-7-CC vs. MCF-7, MCF-7-TT vs. MCF-7, and
MCF-7-21 vs. MCF-7 cells are displayed in Fig. 1. In total, there were 77 common
differentially expressed miRNAs across the three drug-resistant
cell lines, including miR-1246, miR-143, miR-145, miR-424 and
miR-137. Additionally, 42 miRNAs were differentially expressed only
in MCF-7-21 vs. MCF-7 cells, including miR-188-5p, miR-4326 and
miR-542, while 51 miRNAs were only differentially expressed in
MCF-7-CC vs. MCF-7 cells, including let-7c-3p, miR-199b-3p and
miR-210-3p. Furthermore, 146 miRNAs that were differentially
expressed only in MCF-7-TT vs. MCF-7 cells were reported, including
miR-101-3p, miR-141-5p and miR-15a. Overall, the results revealed
that the differential miRNA expression profiles varied among the
three fulvestrant-resistant cell lines, and that certain miRNAs
were differentially expressed in all three MCF-7-CC, MCF-7-TT and
MCF-7-21 cell lines; however, a number of miRNAs were only reported
in one or two of the cell lines. The significantly upregulated or
downregulated miRNAs in the three drug-resistant breast cancer cell
lines are presented in Table III.
miR-148a, miR-31, miR-21, miR-498 and miR-29b were significantly
upregulated, while miR-143, miR-145, miR-424 and miR-137 were
significantly downregulated in the fulvestrant-resistant cell
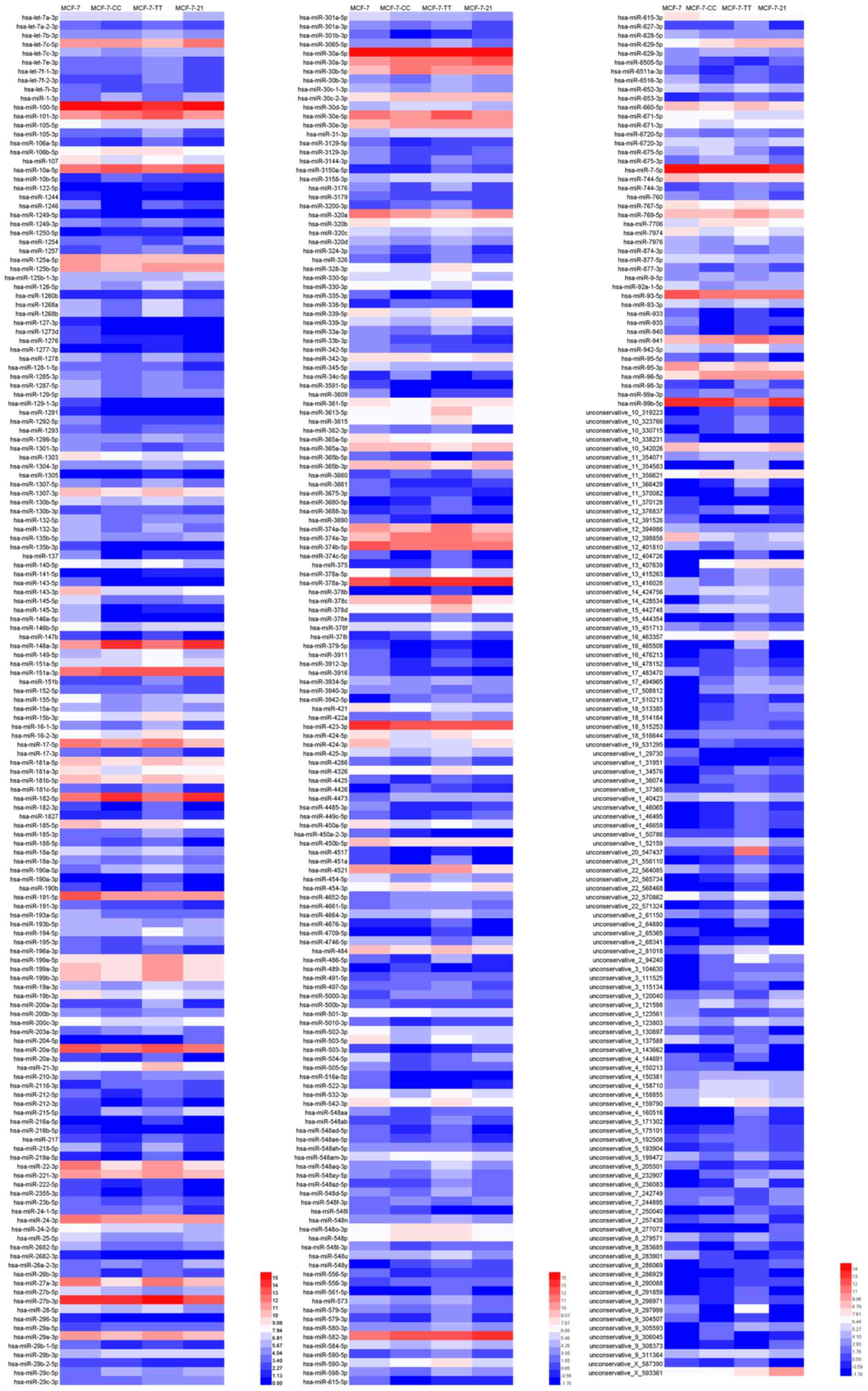
lines. Subsequently, differentially expressed miRNAs with the same
or similar expression profile were clustered by hierarchical
cluster analysis. The clustering results of differently expressed
miRNAs are shown as a heat map in Fig.
2.
 | Table III.Certain of the differentially
expressed miRNA in the three fulvestrant-resistant breast cancer
cell lines. |
Table III.
Certain of the differentially
expressed miRNA in the three fulvestrant-resistant breast cancer
cell lines.
|
| Upregulated
miRNAs |
| Downregulated
miRNAs |
|
|---|
|
|
|
|
|
|
|---|
| Cell lines | miRNA |
log2FC | P-value | miRNA |
log2FC | P-value |
|---|
| MCF-7 vs.
MCF-7-CC | miR-148a-3p | 2.9113 | P<0.01 | miR-137 | −4.2516 | P<0.01 |
|
| miR-31-3p | 1.3064 | P<0.01 | miR-143-5p | −5.4926 | P<0.01 |
|
| miR-215-5p | 3.3344 | P<0.01 | miR-145-5p | −3.4729 | P<0.01 |
|
| miR-489-5p | 24.5083 | P<0.01 | miR-424-3p | −2.807 | P<0.01 |
|
| miR-182-5p | 1.5574 | P<0.01 | miR-335-3p | −26.0166 | P<0.01 |
|
| miR-1-3p | 2.0917 | P<0.01 | miR-221-3p | −1.3765 | P<0.01 |
|
| miR-498-3p | 24.5083 | P<0.01 | miR-222-5p | −1.221 | P<0.01 |
|
| miR-582-3p | 1.4994 | P<0.01 | miR-26b-3p | −2.1457 | P<0.01 |
| MCF-7 vs.
MCF-7-TT | miR-122-5p | 3.9638 | P<0.01 | miR-137 | −2.5543 | P<0.01 |
|
| miR-21-3p | 1.5632 | P<0.01 | miR-143-5p | −3.0583 | P<0.01 |
|
| miR-148a-3p | 1.5609 | P<0.01 | miR-145-5p | −2.6236 | P<0.01 |
|
| miR-422a | 2.3538 | P<0.01 | miR-146a-5p | −5.0017 | P<0.01 |
|
| miR-29a-5p | 1.1446 | P<0.01 | miR-424-3p | −3.4994 | P<0.01 |
|
| miR-31-3p | 1.0498 | P<0.01 | miR-504-5p | −2.5182 | P<0.01 |
|
| miR-629-5p | 2.0143 | P<0.01 | miR-342-3p | −1.9604 | P<0.01 |
|
| miR-582-3p | 1.0648 | P<0.01 | miR-935 | −2.3213 | P<0.01 |
| MCF-7 vs.
MCF-7-21 | miR-148a-3p | 3.3875 | P<0.01 | miR-137 | −2.5542 | P<0.01 |
|
| miR-182-5p | 1.6882 | P<0.01 | miR-143-5p | −3.6957 | P<0.01 |
|
| miR-99a-3p | 1.0275 | P<0.01 | miR-145-5p | −1.8248 | P<0.01 |
|
| miR-100-5p | 1.2957 | P<0.01 | miR-221-3p | −1.2638 | P<0.01 |
|
| miR-29b-3p | 1.4432 | P<0.01 | miR-222-5p | −2.3876 | P<0.01 |
|
| miR-215-5p | 4.251 | P<0.01 | miR-424-3p | −1.072 | P<0.01 |
|
| miR-182-5p | 1.6882 | P<0.01 | miR-127-3p | −25.3999 | P<0.01 |
|
| miR-582-3p | 2.3284 | P<0.01 | miR-1276 | −3.828267 | P<0.01 |
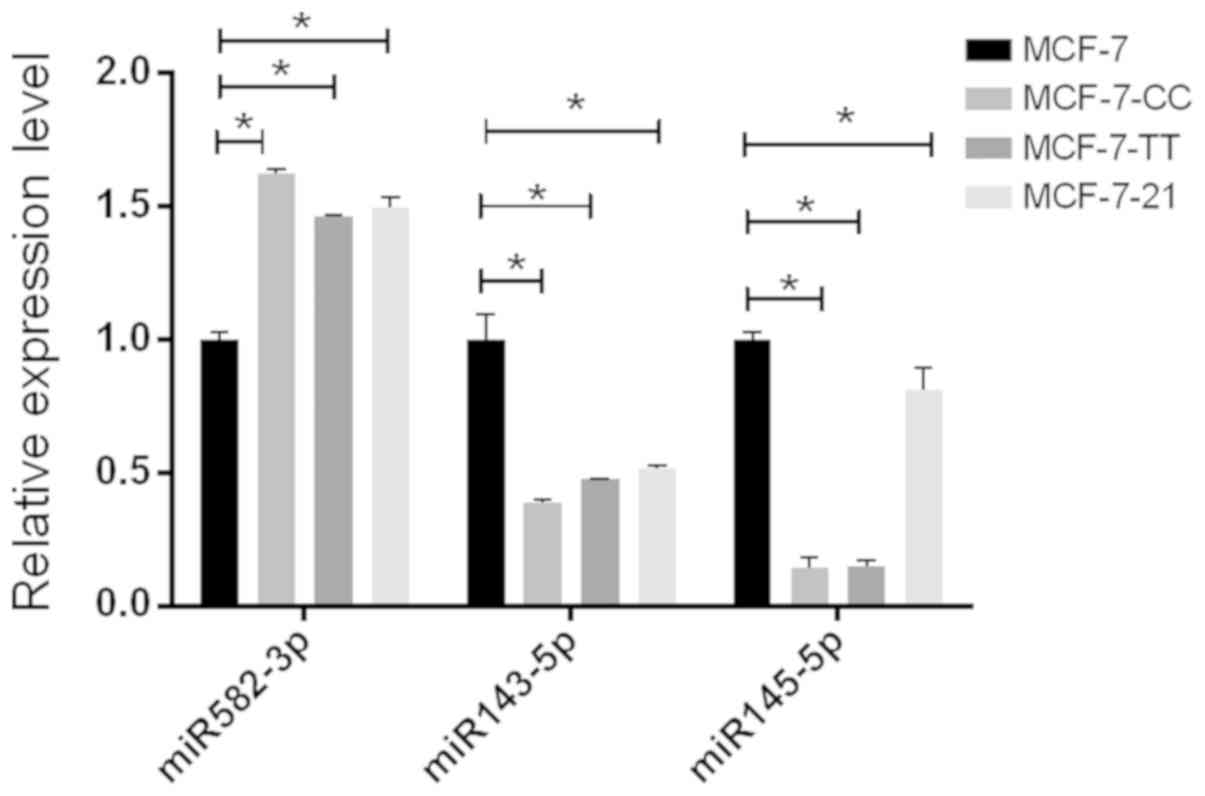
RT-qPCR validation of three selected
differentially expressed miRNAs
RT-qPCR was used to validate the expression of three
selected differentially expressed miRNAs, namely miR-582-3p,
miR-143-5p and miR-145-5p, which were significantly differentially
expressed in the three fulvestrant-resistant cell lines compared
with the parental MCF-7 cell line (Fig.
3). This demonstrated that the expression profiles of these
miRNAs detected by RT-qPCR were consistent with the result of
next-generation sequencing.
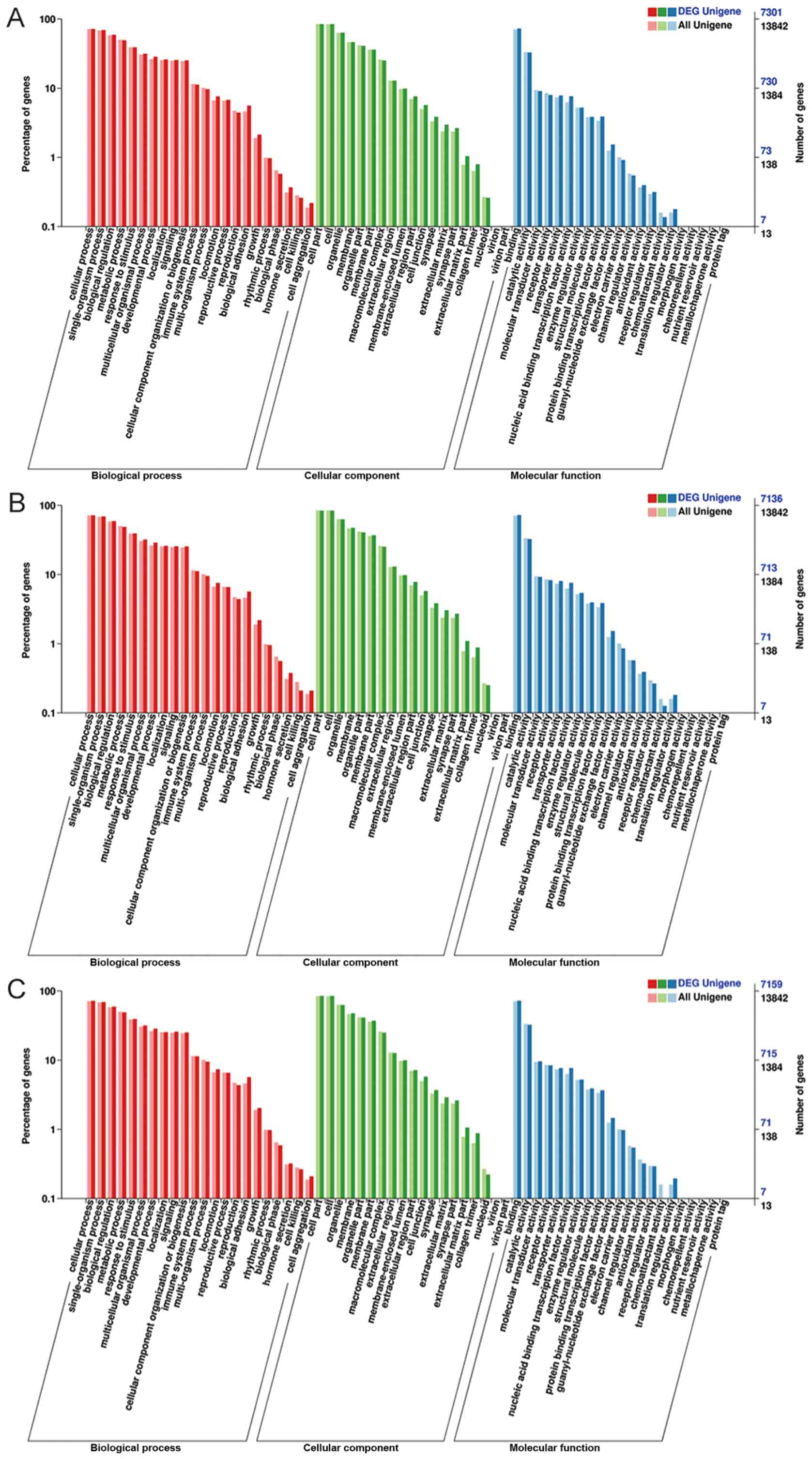
GO and KEGG pathway analyses of
differentially expressed miRNAs
To further understand the biological function of the
predicted targets, GO and KEGG pathway enrichment analyses were
performed. The results of GO enrichment analysis were similar in
the three fulvestrant-resistant cell lines. The analysis of
differential miRNA expression in MCF-7 vs. MCF-7-CC, MCF-7 vs.
MCF-7-TT, and MCF-7 vs. MCF-7-21 cells are shown in Fig. 4A-C, respectively. GO annotation
analysis included three categories, as follows: Biological process,
cellular component and molecular function. The targets of the
differentially expressed miRNAs were associated to the biological
processes of biological regulation, metabolism, response to
stimulus, hormone secretion and cell killing. The enriched cellular
components were primarily associated with cell, organelle, membrane
and cell junction. In addition, the main molecular functions were
reported to be binding, catalytic activity, molecular transducer
activity and chemoattractant activity.
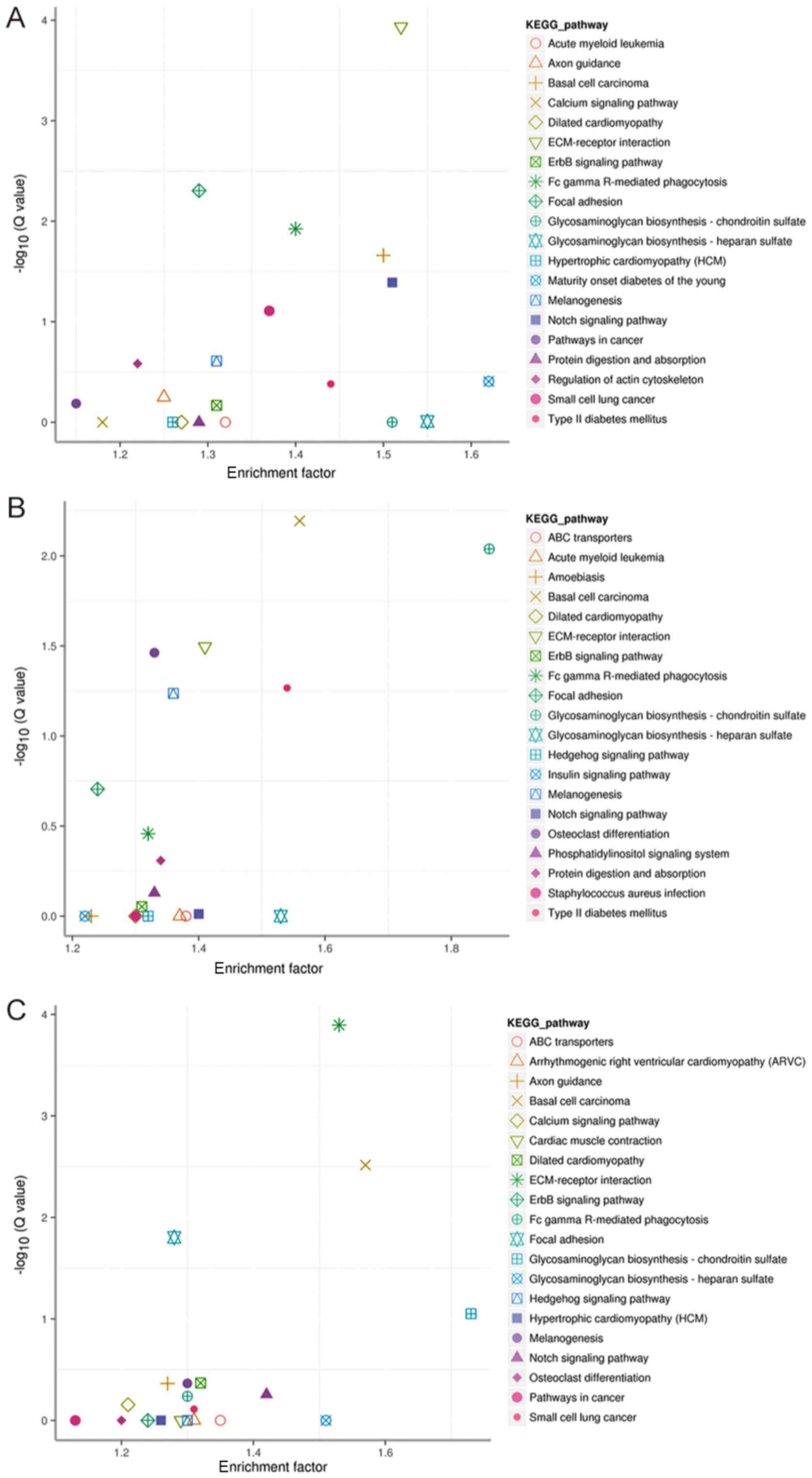
Furthermore, the significant results of KEGG pathway
enrichment analysis for MCF-7 vs. MCF-7-CC, MCF-7 vs. MCF-7-TT, and
MCF-7 vs. MCF-7-21 cells are listed in Fig. 5A-C, respectively. The targets of the
differentially expressed miRNAs in MCF-7 vs. MCF-7-CC cells were
markedly enriched in the extracellular matrix (ECM)-receptor
interaction, Notch signaling pathway, focal adhesion and ErbB
signaling pathways. In MCF-7 vs. MCF-7-TT cells, the markedly
enriched pathways included osteoclast differentiation,
melanogenesis, basal cell carcinoma, glycosaminoglycan
biosynthesis-chondroitin sulfate and ECM-receptor interaction.
Finally, in MCF-7 vs. MCF-7-21 cells, the markedly enriched
pathways included the ECM-receptor interaction, focal adhesion,
glycosaminoglycan biosynthesis-chondroitin sulfate and Notch
signaling.
Discussion
miRNAs regulate gene expression at the
post-transcriptional level. Evidence suggests that miRNAs are often
dysregulated in human tumors, and can function as oncogenes or
tumor suppressors (27), serving
important roles in the tumor progression. It has been reported that
the aberrant expression of miRNA is associated with drug resistance
(6). Understanding the targets of
miRNAs and the underlying mechanism of drug resistance can
contribute to new developments in the treatment of drug resistance
in breast cancer.
In the present study, the differential expression of
miRNAs in MCF-7 cells and three fulvestrant-resistant breast cancer
cell lines was detected by next-generation sequencing. It was
observed that the differential miRNA expression profiles were
different among the three fulvestrant-resistant cell lines as
compared with the parental MCF-7 cells, and certain miRNAs were
only differentially expressed in only one or two of the cell lines.
RT-qPCR was then performed to further validate the expression
levels of miR-582-3p, miR-143-5p and miR-145-5p in MCF-7 and
fulvestrant-resistant breast cancer cell lines, and the results
were consistent with those of next-generation sequencing. According
to the study findings, miR-148a, miR-31, miR-21, miR-498 and
miR-29b were significantly upregulated, while miR-143, miR-145,
miR-424 and miR-137 were significantly downregulated in
fulvestrant-resistant cell lines.
Several previous studies have demonstrated that
miR-143 and miR-145 are downregulated in breast cancer. For
instance, miR-143 has been reported to function as a tumor
suppressor in breast cancer by downregulating DNA methyltransferase
3A (DNMT3A) expression. The downregulation of miR-143 in breast
cancer can lead to the overexpression of DNMT3A, which induces the
hypermethylation and silencing of tumor suppressor genes, while
contributing to tumorigenesis (28).
A number of studies reported that the overexpression of miR-145
inhibited breast cancer cell invasion and metastasis by regulating
the expression of tumor metastasis-associated suppressor genes,
such as mucin 1, matrix metalloproteinase-11 and ADAM
metallopeptidase domain 17, while this miRNA was also able to
inhibit cell growth by targeting rhotekin and c-Myc (29–31). Our
previous study also demonstrated that miR-145 overexpression
induced alterations in the whole transcriptome and inhibited breast
cancer development (32). Tumor
suppressor miRNAs, such as miR-143 and miR-145, inhibit ERBB3
expression, and consequently suppress the proliferation and
invasion of breast cancer cells (33). In the current study, the results of
next-generation sequencing revealed that miR-143 and miR-145 were
significantly downregulated in the three fulvestrant-resistant cell
lines, which may be associated to drug resistance. Thus, it can be
speculated that the interaction between miR-143/145 and their
targets may serve an important role in fulvestrant resistance;
however, this field requires further study.
Steroid receptor coactivator 3 (SRC3) is a SRC
member of the p160 family, which is frequently amplified in breast
cancer (34). miR-137 downregulates
the SRC3 gene by targeting its 3′UTR, and inhibits cell
proliferation and drug resistance (35). Overexpression of miR-137 in breast
cancer cells can also reduce proliferation and migration by
downregulating lysine demethylase 5B, estrogen-related receptor α
and C-terminal-binding protein 1 (36–38). Zhu
et al (39) reported that
miR-137 was downregulated in multiple drug-resistant cell lines,
including MCF-7/ADM. The overexpression of miR-137 was able to
enhance the sensitivity of breast cancer cells to chemotherapeutic
agents by modulating the expression of P-glycoprotein by targeting
Y box binding protein 1. In the present study, miR-137 was
downregulated in fulvestrant-resistant cell lines, suggesting that
drug sensitivity was reduced and drug resistance was enhanced.
Therefore, based on these findings, it is speculated that the
expression of miR-137 may alter the sensitivity of breast cancer
cells to fulvestrant, and thus miR-137 may serve as a predictor of
endocrine therapy.
The present study results also indicated that
miR-424 expression was significantly downregulated in
fulvestrant-resistant cell lines, which is consistent with the
findings of a recent study (40). It
has been reported that miR-424 was associated with drug resistance
in breast cancer. Silencing of miR-424 was reported to activate the
phosphoinositide 3-kinase/protein kinase B/mammalian target of
rapamycin (PI3K/AKT/mTOR) signaling pathway and lead to the
resistance of MCF-7 cells to letrozole (41). Rodriguez-Barrueco et al
(42) demonstrated that a loss of
the miRNA cluster miR-424(322)/503 may induce resistance to
chemotherapeutic drugs by modulating the expression levels of
insulin-like growth factor 1 receptor and B-cell lymphoma 2.
According to the aforementioned observations, it can be speculated
that miR-424 may affect its corresponding targets and alter the
signaling pathway to induce fulvestrant resistance. Thus, miR-424
may be used as a potential biomarker in the diagnosis and prognosis
of fulvestrant-resistant breast cancer.
As an oncomiRNA, miR-21 has been reported to be
overexpressed in numerous types of tumor, including head and neck
squamous cell carcinoma, and colon and breast cancer (43). miR-21 may promote breast cancer
invasion and metastasis by regulating the metastasis-associated
tumor suppressor gene, tropomyosin 1 (44). Blower et al (45) reported that miR-21 was associated
with the potency of anticancer agents and chemoresistance,
suggesting that miR-21 may be involved in breast cancer drug
resistance. It has also been demonstrated that the silencing of
miR-21 confers sensitivity to tamoxifen and fulvestrant by
enhancing autophagic cell death through inhibition of the
PI3K/AKT/mTOR signaling pathway in breast cancer cells (46). The results of the present study
indicated that miR-21 expression was increased in the MCF-7-TT cell
line; thus, it is proposed that the overexpression of miR-21 may
reduce the sensitivity of cells to fulvestrant and enhance drug
resistance. Zhou et al (47)
also reported that miR-21 may be associated with fulvestrant
resistance. Estradiol was able to repress the expression of miR-21
by activating the ER in MCF-7 cells, which may explain how
fulvestrant, as a pure antiestrogen agent, can increase the
expression of miR-21 (48).
Therefore, the interaction between Estradiol and miR-21 may be
closely associated with fulvestrant resistance.
In the present study, miR-221-3p and miR-222-5p were
downregulated in MCF-7-CC and MCF-7-21 cell lines, which was
inconsistent with previous studies. Rao et al (9) and Xin et al (40) reported that miR-221 and miR-222 were
upregulated in a fulvestrant-resistant cell line. In these studies,
hormone-free medium with 10% charcoal-stripped FBS was used without
phenol red to culture a fulvestrant-resistant cell line, as
reported previously (14). However,
the present study used DMEM (originally containing phenol red) with
5% FBS, which has estrogenic activity (49). Thus, the different results may be due
to the effects of estrogenic activity in the media used. It has
been reported that medium containing charcoal-stripped serum mimics
postmenopausal conditions, while unstripped serum mimics
premenopausal conditions (12),
suggesting that the fulvestrant-resistant cell lines obtained in
the current study may exhibit cell behaviors associated with
premenopausal conditions. Furthermore, ER expression levels were
indicated to be markedly reduced in fulvestrant-resistant cells
under premenopausal conditions, whereas almost no expression of ER
was detected in fulvestrant-resistant cells under postmenopausal
conditions (12). The mechanisms of
resistance to estrogen antagonists differ under premenopausal and
postmenopausal conditions, which may account for the
inconsistencies between the present study results and those of
previous studies (9,40). In addition, the ER directly
suppresses miR-221 and miR-222 expression in breast cancer by
recruiting the corepressors nuclear receptor corepressor and
silencing mediator of retinoic acid and thyroid hormone receptor
(50). Further studies are required
to determine the various mechanisms underlying fulvestrant
resistance in premenopausal and postmenopausal conditions, and to
elucidate the roles of miR-221 and miR-222 in fulvestrant
resistance.
GO and KEGG pathway analyses were performed in the
current study to further understand the biological function of the
predicted targets and their potential roles in the development of
fulvestrant resistance. Although there were a few differences in
the KEGG enriched pathways among the three resistant cell lines,
the results indicated that the targets of the differentially
expressed miRNAs were primarily involved in ECM-receptor
interaction, the Notch signaling pathway and focal adhesion.
Previous studies (51–56) have demonstrated that these pathways
were involved in the progression and metastasis of cancer. It has
also been reported that abnormal Notch signaling may contribute to
mammary carcinogenesis by deregulating the self-renewal ability of
normal mammary stem cells (52).
Further studies are needed to verify whether these pathways are
associated with fulvestrant resistance.
The establishment of a drug-resistant cell line
model in vitro may aid future investigation into the
mechanism underlying drug resistance. Additionally, different
methods to induce resistance are likely to result in various
mechanisms of resistance. DMEM with 5% FBS and phenol red was used
to culture the fulvestrant-resistant cell lines in the present
study, which may be considered to represent the patient condition
in vitro (12). In the
present study, three fulvestrant-resistant cell lines were
established by three different methods, which may represent
different drug resistance conditions. These cell lines may have
exogenous or endogenous resistance characteristics, similar to the
conditions of patients. Thus, these different fulvestrant-resistant
cell lines were used to investigate the association between
differential miRNA expression profiles and fulvestrant resistance
of human breast cancer cell lines. The MCF-7-CC cell line was
established by stepwise induction, starting with low concentrations
of fulvestrant. Following treatment with the corresponding
concentration of fulvestrant screening medium, replacement of the
drug-free complete medium, removal of apoptotic cells and
proliferation of viable cells, tumor cells were allowed an
appropriate buffer time to activate certain physiological pathways
and adapt to stimulation with the drug in order to acquire
resistance. It has been reported the induction of a drug-resistant
cell line in a stepwise manner resulted in alterations in the
physiology, genetics and remodeling, and cells acquired resistance
during the process of screening and culturing (57,58).
Therefore, it can be speculated that the MCF-7-CC cell line
established in the current study may be an acquired drug-resistant
cell line. For the drug-resistant cell line MCF-7-TT, which was
established in a stepwise temporal manner starting with a high
concentration of fulvestrant, the induction time increased
gradually from 1 to 48 h, and then to 72 h. During the induction,
apoptotic cells were removed and viable cells proliferated, which
provided a buffer time for repair of slightly damaged cells and
eliminate relatively drug sensitive and severely damaged cells;
however, cells with inherent relatively resistant properties were
maintained. Yang and Trujillo (59)
reported that discontinuous screening and cultivating resistant
cells with high concentrations is mainly conducted by inducing the
death of sensitive cells with high concentrations of drugs and
maintaining those with inherently drug-resistant properties.
Therefore, it is speculated that the MCF-7-TT cell line obtained in
the current study may arise from a combination of exogenous and
endogenous resistance-associated factors. Finally, for the
drug-resistant cell line MCF-7-21, drug resistance was established
by incubating the cells with a high concentration of fulvestrant
for 21 days; apoptotic cells were not removed, while viable cells
did not proliferate during the induction. As a period of time for
cells to adapt to treatment with fulvestrant was not provided, it
is speculated that MCF-7-21 may be an endogenous resistant cell
line. However, it was difficult to distinguish between the
endogenous and acquired forms of resistance. The differential miRNA
expression profiles varied among the three fulvestrant-resistant
cell lines, suggesting that the drug-resistant cell lines may
undertake different molecular mechanisms of fulvestrant resistance.
Therefore, further study is required to elucidate the association
among these mechanisms.
In conclusion, the current study investigated the
association between differential miRNA expression profiles and
fulvestrant resistance in human breast cancer cells. A number of
differentially expressed miRNAs were identified in the present
study. The current results suggest that miR-143, miR-145, miR-424,
miR-137 and miR-21 may serve important roles in fulvestrant
resistance in breast cancer and may be potential targets for breast
cancer treatment. As there are few studies regarding the mechanisms
of miRNAs in fulvestrant resistance, the present study may provide
the basis for future research into fulvestrant resistance and
development of new strategies for the treatment of patients with
fulvestrant-resistant breast cancer. The functional analysis of
miRNAs in fulvestrant resistance will also serve an important role
in the treatment of drug-resistant breast cancer. According to the
results of this preliminary study, one or several differentially
expressed miRNAs of interest will be selected in our further
studies, which will investigate the association between miRNAs and
fulvestrant resistance.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (nos. 81272372 and
30873044) and the Zhongnan Hospital of Wuhan University Science,
Technology and Innovation Seed Fund (grant no. znpy2016033). The
project was also sponsored by the Scientific Research Foundation
for Returned Overseas Chinese Scholars, State Education
Ministry.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, KH and HZ designed the study. KH and HZ
performed the experiments; JG, KH, HZ, YS, PY, QZ and ZP collected
and analyzed the data. XL, JG, KH and HZ wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Osborne CK, Wakeling A and Nicholson RI:
Fulvestrant: An oestrogen receptor antagonist with a novel
mechanism of action. Br J Cancer. 90 Suppl 1:S2–S6. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tangkeangsirisin W and Serrero G: GP88
(Progranulin) confers fulvestrant (Faslodex, ICI 182,780)
resistance to human breast cancer cells. Adv Breast Cancer Res.
03:68–78. 2014. View Article : Google Scholar
|
|
4
|
Osipo C, Meeke K, Cheng D, Weichel A,
Bertucci A, Liu H and Jordan VC: Role for HER2/neu and HER3 in
fulvestrant-resistant breast cancer. Int J Oncol. 30:509–520.
2007.PubMed/NCBI
|
|
5
|
Tsuboi K, Kaneko Y, Nagatomo T, Fujii R,
Hanamura T, Gohno T, Yamaguchi Y, Niwa T and Hayashi SI: Different
epigenetic mechanisms of ERalpha implicated in the fate of
fulvestrant-resistant breast cancer. J Steroid Biochem Mol Biol.
167:115–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hong L, Yang Z, Ma J and Fan D: Function
of miRNA in controlling drug resistance of human cancers. Curr Drug
Targets. 14:1118–1127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ji S, Shao G, Lv X, Liu Y, Fan Y, Wu A and
Hu H: Downregulation of miRNA-128 sensitises breast cancer cell to
chemodrugs by targeting Bax. Cell Biol Int. 37:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH and Tang JH: Down-regulation
of miRNA-452 is associated with adriamycin-resistance in breast
cancer cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan P, Yue W, Wang JP, Aiyar S, Li Y, Kim
TH and Santen RJ: Mechanisms of resistance to structurally diverse
antiestrogens differ under premenopausal and postmenopausal
conditions: Evidence from in vitro breast cancer cell models.
Endocrinology. 150:2036–2045. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye P, Fang C, Zeng H, Shi Y, Pan Z, An N,
He K, Zhang L and Long X: Differential microRNA expression profiles
in tamoxifen-resistant human breast cancer cell lines induced by
two methods. Oncol Lett. 15:3532–3539. 2018.PubMed/NCBI
|
|
14
|
Fan M, Yan PS, Hartman-Frey C, Chen L,
Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al:
Diverse gene expression and DNA methylation profiles correlate with
differential adaptation of breast cancer cells to the antiestrogens
tamoxifen and fulvestrant. Cancer Res. 66:11954–11966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jensen BL, Skouv J, Lundholt BK and
Lykkesfeldt AE: Differential regulation of specific genes in MCF-7
and the ICI 182780-resistant cell line MCF-7/182R-6. Br J Cancer.
79:386–392. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coser KR, Wittner BS, Rosenthal NF,
Collins SC, Melas A, Smith SL, Mahoney CJ, Shioda K, Isselbacher
KJ, Ramaswamy S and Shioda T: Antiestrogen-resistant subclones of
MCF-7 human breast cancer cells are derived from a common
monoclonal drug-resistant progenitor. Proc Natl Acad Sci USA.
106:14536–14541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stokowy T, Eszlinger M, Świerniak M,
Fujarewicz K, Jarząb B, Paschke R and Krohn K: Analysis options for
high-throughput sequencing in miRNA expression profiling. BMC Res
Notes. 7:1442014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fahlgren N, Howell MD, Kasschau KD,
Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR,
Dangl JL and Carrington JC: High-throughput sequencing of
Arabidopsis microRNAs: Evidence for frequent birth and death of
MIRNA genes. PLoS One. 2:e2192007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Witten DM, Johnstone IM and
Tibshirani R: Normalization, testing, and false discovery rate
estimation for RNA-sequencing data. Biostatistics. 13:523–538.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One. 9:e1119882014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ng EK, Li R, Shin VY, Siu JM, Ma ES and
Kwong A: MicroRNA-143 is downregulated in breast cancer and
regulates DNA methyltransferases 3A in breast cancer cells. Tumour
Biol. 35:2591–2598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
30
|
Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng
L, Zhou H and Zhao RC: miR-145 inhibits breast cancer cell growth
through RTKN. Int J Oncol. 34:1461–1466. 2009.PubMed/NCBI
|
|
31
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye P, Shi Y, An N, Zhou Q, Guo J and Long
X: miR-145 overexpression triggers alteration of the whole
transcriptome and inhibits breast cancer development. Biomed
Pharmacother. 100:72–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gojis O, Rudraraju B, Gudi M, Hogben K,
Sousha S, Coombes RC, Cleator S and Palmieri C: The role of SRC-3
in human breast cancer. Nat Rev Clin Oncol. 7:83–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eedunuri VK, Rajapakshe K, Fiskus W, Geng
C, Chew SA, Foley C, Shah SS, Shou J, Mohamed JS, Coarfa C, et al:
miR-137 targets p160 steroid receptor coactivators SRC1, SRC2, and
SRC3 and inhibits cell proliferation. Mol Endocrinol. 29:1170–1183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Denis H, Van Grembergen O, Delatte B,
Dedeurwaerder S, Putmans P, Calonne E, Rothé F, Sotiriou C, Fuks F
and Deplus R: MicroRNAs regulate KDM5 histone demethylases in
breast cancer cells. Mol Biosyst. 12:404–413. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y
and He F: MiR-137 targets estrogen-related receptor alpha and
impairs the proliferative and migratory capacity of breast cancer
cells. PLoS One. 7:e391022012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han Y, Bi Y, Bi H, Diao C, Zhang G, Cheng
K and Yang Z: miR-137 suppresses the invasion and procedure of EMT
of human breast cancer cell line MCF-7 through targeting CtBP1. Hum
Cell. 29:30–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 restoration sensitizes multidrug-resistant
MCF-7/ADM cells to anticancer agents by targeting YB-1. Acta
Biochim Biophys Sin (Shanghai). 45:80–86. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xin F, Li M, Balch C, Thomson M, Fan M,
Liu Y, Hammond SM, Kim S and Nephew KP: Computational analysis of
microRNA profiles and their target genes suggests significant
involvement in breast cancer antiestrogen resistance.
Bioinformatics. 25:430–434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vilquin P, Donini CF, Villedieu M, Grisard
E, Corbo L, Bachelot T, Vendrell JA and Cohen PA: MicroRNA-125b
upregulation confers aromatase inhibitor resistance and is a novel
marker of poor prognosis in breast cancer. Breast Cancer Res.
17:132015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rodriguez-Barrueco R, Nekritz EA, Bertucci
F, Yu J, Sanchez-Garcia F, Zeleke TZ, Gorbatenko A, Birnbaum D,
Ezhkova E, Cordon-Cardo C, et al: miR-424(322)/503 is a breast
cancer tumor suppressor whose loss promotes resistance to
chemotherapy. Genes Dev. 31:553–566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F,
Wang X, He X and Zhao Y and Zhao Y: Prognostic role of microRNA-21
in various carcinomas: A systematic review and meta-analysis. Eur J
Clin Invest. 41:1245–1253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blower PE, Chung JH, Verducci JS, Lin S,
Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et
al: MicroRNAs modulate the chemosensitivity of tumor cells. Mol
Cancer Ther. 7:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu X, Li R, Shi W, Jiang T, Wang Y, Li C
and Qu X: Silencing of microRNA-21 confers the sensitivity to
tamoxifen and fulvestrant by enhancing autophagic cell death
through inhibition of the PI3K-AKT-mTOR pathway in breast cancer
cells. Biomed Pharmacother. 77:37–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou Q, Zeng H, Ye P, Shi Y, Guo J and
Long X: Differential microRNA profiles between
fulvestrant-resistant and tamoxifen-resistant human breast cancer
cells. Anticancer Drugs. 29:539–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Welshons WV, Wolf MF, Murphy CS and Jordan
VC: Estrogenic activity of phenol red. Mol Cell Endocrinol.
57:169–178. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Di Leva G, Gasparini P, Piovan C, Ngankeu
A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, et
al: MicroRNA cluster 221–222 and estrogen receptor alpha
interactions in breast cancer. J Natl Cancer Inst. 102:706–721.
2010. View Article : Google Scholar
|
|
51
|
Yeh MH, Tzeng YJ, Fu TY, You JJ, Chang HT,
Ger LP and Tsai KW: Extracellular matrix-receptor interaction
signaling genes associated with inferior breast cancer survival.
Anticancer Res. 38:4593–4605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dontu G, Jackson KW, McNicholas E,
Kawamura MJ, Abdallah WM and Wicha MS: Role of Notch signaling in
cell-fate determination of human mammary stem/progenitor cells.
Breast Cancer Res. 6:R605–R615. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li L, Tang P, Li S, Qin X, Yang H, Wu C
and Liu Y: Notch signaling pathway networks in cancer metastasis: A
new target for cancer therapy. Med Oncol. 34:1802017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alketbi A and Attoub S: Notch signaling in
cancer: Rationale and strategies for targeting. Curr Cancer Drug
Targets. 15:364–374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hoskin V, Szeto A, Ghaffari A, Greer PA,
Cote GP and Elliott BE: Ezrin regulates focal adhesion and
invadopodia dynamics by altering calpain activity to promote breast
cancer cell invasion. Mol Biol Cell. 26:3464–3479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Negoro K, Yamano Y, Fushimi K, Saito K,
Nakatani K, Shiiba M, Yokoe H, Bukawa H, Uzawa K, Wada T, et al:
Establishment and characterization of a cisplatin-resistant cell
line, KB-R, derived from oral carcinoma cell line, KB. Int J Oncol.
30:1325–1332. 2007.PubMed/NCBI
|
|
58
|
Watson MB, Lind MJ and Cawkwell L:
Establishment of in-vitro models of chemotherapy resistance.
Anticancer Drugs. 18:749–754. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang LY and Trujillo JM: Biological
characterization of multidrug-resistant human colon carcinoma
sublines induced/selected by two methods. Cancer Res. 50:3218–3225.
1990.PubMed/NCBI
|