Introduction
Glioma is one of the most common malignant tumors
observed in China with a high degree of malignancy (1). Gliomas account for between 35 and 50%
of all intracranial brain tumors in adult patients (2). Silent information regulator 1 (SIRT1)
is a conserved class III deacetylase that deacetylates the lysine
residues of nucleoproteins to influence their stability and
transcriptional activities (3,4). In
addition, SIRT1 regulates cellular stress responses and lifespan
(5). Despite great progresses in
surgical treatment options for patients with gliomas, including
chemotherapy and radiotherapy, the prognosis remains poor (6). Further investigation is therefore
crucial to develop novel gene therapies for the treatment of
glioma, and to elucidate the underlying molecular mechanisms of
glioma development and progression.
SIRT1 is associated with cell viability and
inhibition of apoptosis (7,8), and is involved in the growth of certain
tumors (9,10). SIRT1 overexpression promotes tumor
progression by regulating tumor growth-associated signaling
pathways, including Wnt signaling (11,12). A
previous study reported that SIRT1 serves numerous roles in cancer
biology (13). On the one hand,
SIRT1 is upregulated in tumors, while cancer cells can
downregulates the expression of tumor suppressor genes (14). On the other hand, SIRT1 can be
proapoptotic (15) and
anti-proliferative (16), and
consequently has been proposed to behave as a tumor suppressor
in vivo.
The epithelial-mesenchymal transition (EMT) is a
biological process in which epithelial cells lose their polarity
and adhesion properties, and differentiate into mesenchymal cells
that possess migratory and invasive properties (17). EMT is a crucial biological process
involved in the migration and invasion of certain tumor cells
(18), and serves an important role
in cancer progression and fibrosis (19). EMT is associated with tumorigenesis
and metastasis. Previous studies identified that SIRT1 promotes
prostate cancer growth and migration through EMT (20), and stimulates EMT and metastasis in
colorectal cancer (21). Conversely,
other studies revealed that SIRT1 suppresses EMT in cancer
metastasis, organ fibrosis and nasal polypogenesis (22,23).
SIRT1 promotes the viability and inhibits apoptosis of human glioma
cells (24). SIRT1 is also vital for
neural stem cells maintenance and oncogenic transformation
(25).
Numerous studies have identified various
associations of SIRT1 with certain processes involved in cancer;
however, its effects on invasion and EMT in human glioma remain
unclear. Since controversies exist regarding the role of SIRT1 in
tumors, as SIRT1 can downregulate the expression of tumor
suppressor genes (14), SIRT1 also
has been proposed to behave as a tumor suppressor in vivo
(16), therefore, further
investigation is required. To the best of our knowledge, the
present study was the first to examine the effect of SIRT1
silencing on EMT in glioma. To do so, the expression levels of
SIRT1 were analyzed in human glioma tissue samples together with
the effects of SIRT1 on human glioma cell invasion. Previous
studies reported that matrix metalloproteinase-9 (MMP-9) (26), Twist family basic helix-loop-helix
transcription factor 1 (Twist1) and Snail family transcriptional
repressor 1 (Snail1) serve important roles in tumor invasion
(27). Therefore, these protein
expression levels were also detected. The results indicated that
SIRT1 was highly expressed in human glioma tissue samples compared
with in adjacent tissues, and that SIRT1 silencing inhibited human
glioma U87 and U251 cell line viability and invasion. In addition,
SIRT1 silencing suppressed EMT in U87 and U251 cell lines, which
suggested that SIRT1 may serve a role in EMT. In conclusion, the
results of the present study provide an important foundation for
further investigation of the underlying molecular mechanism of
SIRT1 in glioma growth.
Materials and methods
Tissue specimen collection
A total of 20 glioma tissues and adjacent brain
tissues were collected at The Second Affiliated Hospital of Kunming
Medical University (Kunming, China) between April 2016 and April
2017. Tissues were collected following surgical resection. Tissue
histomorphology was confirmed by pathologists. The present study
was approved by the Ethics Committee of The Second Affiliated
Hospital of Kunming Medical University and patients provided
written informed consent.
Immunohistochemistry
Tissues are fixed in 4% paraformaldehyde for 24 h at
room temperature. Fixed tissues were dehydrated with various
concentrations of xylene and ethanol (50% ethanol for 4 h; 75%
ethanol for 4 h; 85% ethanol for 3 h; 95% ethanol for 2 h; 100%
ethanol for 1 h; 100% ethanol for 1 h; 1:1 ethanol-xylene for 1 h;
xylene for 1 h; xylene for 30 min at room temperature), embedded in
paraffin. Sections (4 µm thickness) were cut from a paraffin block.
Sections were dewaxed with various concentrations of xylene and
ethanol (xylene for 10 min; xylene for 5 min; 100% ethanol for 5
min; 95% ethanol for 2 min; 80% ethanol for 2 min; 70% ethanol for
2 min). Antigen repair was performed on the sections with 0.01 M
citric acid buffer (pH 6.0) at 100°C high temperature and 80 kpa
pressure. Sections were blocked by incubation with 5% goat serum
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China) in PBS for 15 min at room temperature. Sections were
incubated with anti-SIRT1 rabbit antibody (1:100; cat. no.
13161-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) overnight at
4°C and with a HRP Goat Anti-Rabbit IgG antibody (1:200; cat. no.
AS014, ABclonal Biotech Co., Ltd., Wuhan, China) for 2 h at room
temperature. The reactions were visualized using a
3,3′-diaminobenzidine visualization kit (Fuzhou Maixin Biotech Co.,
Ltd., Fuzhou, China). Sections were counterstained with hematoxylin
to visualize nuclei, for 5–10 min at room temperature. Sections
were examined under a light microscope, (×400, magnification).
Brown staining indicated immunoreactive positive cells, and blue
staining indicated the nuclei.
Cell culture
The human glioblastoma cell line U251 and the
glioblastoma cell line U87 of unknown origin were purchased from
The Kunming Cell Bank of the Chinese Academy of Sciences (Kunming,
China). Cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) and
antibiotics (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and placed at 37°C in a humidified incubator containing 5%
CO2.
SIRT1 silencing
SIRT1-small interfering RNA (siRNA) was used to
target the SIRT1 gene. The nucleotide sequences were synthesized
from Sangon Biotech Co., Ltd., (Shanghai, China) as follows:
Forward, 5′-ACUUUGCUGUAACCCUGUA-3′ and reverse,
3′-UACAGGGUUACAGCAAAGU-3′ (28). For
comparison, a random nucleotide sequence
(5′-CUAGCUUAUGUGGACCUCG-3′) was used as a negative control. The
transfections were carried out with Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Cells were harvested at 48 h
post-transfection, and mRNA and protein levels were analyzed using
the reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting, respectively.
Cell viability assay
Cell viability was evaluated using the Cell Counting
Kit-8 (CCK-8) (Beyotime Institute of Biotechnology, Haimen, China).
Cells (1×104) were seeded in a 96-well plate. At 48 h
after transfection, cell viability was assessed using the CCK-8
assay, according to the manufacturer's protocol. Briefly, cells
were incubated at 37°C for 1 h, prior to measuring the optical
density (OD) at 450 nm using a multiplate reader.
Cell invasion assays
A cell invasion assay was performed using 8 µm
Transwell chambers (BD Biosciences, San Jose, CA, USA) precoated
with Matrigel. Briefly, 300 µl cell suspension (5×105
cells/ml) was added to the upper chamber, and 500 µl DMEM
containing 10% FBS was added to the lower chamber. Following 48 h
transfection, cells that did not invade the Matrigel were
discarded. Filters were fixed in 90% ethanol for 10 min at room
temperature, stained with 0.1% crystal violet for 5 min at room
temperature, and visualized under a light microscope
(magnification, ×100).
RT-qPCR
Total RNA was isolated from cells and tissues using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
cDNA was generated using a Reverse Transcription (RT) kit (Vazyme
Biotech Co., Ltd, Nanjing, China), according to the manufacturer's
protocol. qPCR was performed using a SYBR qPCR Master Mix kit
(Vazyme Biotech Co., Ltd.) in an ABI 7300 real-time PCR machine
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. qPCRs were performed as follows: 95°C for 5 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The
relative expression of each mRNA of interest were normalized to
endogenous control and analyzed using the 2−ΔΔCq method
(29,30). Epithelial (E-)cadherin, β-catenin,
fibronectin, vimentin and β-actin primers were as follows:
E-cadherin, 5′-ATGCTGAGGATGATTGAGGTGGGT-3′ (forward) and
5′-CAAATGTGTTCAGCTCAGCCAGCA-3′ (reverse); β-catenin,
5′-TGCAGTTCGCCTTCACTATGGACT-3′ (forward) and
5′-GATTTGCGGGACAAAGGGCAAGAT-3′ (reverse); fibronectin,
5′-AAACTTGCATCTGGAGGCAAACCC-3′ (forward) and
5′-AGCTCTGATCAGCATGGACCACTT-3′ (reverse); vimentin,
5′-AGAACCTGCAGGAGGCAGAAGAAT-3′ (forward) and
5′-TTCCATTTCACGCATCTGGCGTTC-3′ (reverse); and β-actin,
5′-TGACGTGGACATCCGCAAAG-3′ (forward) and 5′-CTGGAAGGTGGACAGCGAGG-3′
(reverse).
Western blotting
At 48 h after transfection, total proteins from U87
and U251 cells were extracted using RIPA lysis buffer (Beyotime
Institute of Biotechnology). Protein were quantified by using a
bicinchoninic protein assay kit (Beyotime Institute of
Biotechnology). Proteins (30 µg) were separated by SDS-PAGE (10%
gel) and transferred onto a polyvinylidene fluoride membrane, which
was blocked with 10% skimmed milk in Tris-buffered saline, pH 7.0,
containing 0.1% Tween-20 for 2 h. Following blocking, membranes
were incubated with primary antibodies at 4°C overnight
(ProteinTech Group, Inc.) and followed by HRP Goat Anti-Rabbit IgG
secondary antibodies (dilution 1:1,000; cat. no. AS014; ABclonal,
Wuhan, China) for 2 h at room temperature. The primary antibodies
as follows: SIRT1 antibody (dilution, 1:1,000; cat. no. 13161-1-AP;
ProteinTech), MMP-9 antibody (dilution, 1:1,000; cat. no.
10375-2-AP; ProteinTech), Twist1 antibody (dilution, 1:1,000; cat.
no. 25465-1-AP; ProteinTech), MMP-9 antibody (dilution, 1:1,000;
cat. no. 10375-2-AP; ProteinTech), Snail1 antibody (dilution,
1:1,000; cat. no. 13099-1-AP; ProteinTech), E-cadherin antibody
(dilution, 1:1,000; cat. no. 20874-1-AP; ProteinTech), β-catenin
antibody (dilution, 1:1,000; cat. no. 51067-2-AP; ProteinTech),
Fibronectin antibody (dilution, 1:1,000; cat. no. 15613-1-AP;
ProteinTech), Vimentin antibody (dilution, 1:1,000; cat. no.
10366-1-AP; ProteinTech). Membrane signals were visualized with
enhanced chemiluminescence reagent (Beyotime Institute of
Biotechnology). The OD of the bands was determined using ImageJ 2×
software (National Institutes of Health, Bethesda, MD, USA) and
normalized to the expression of the internal control (β-actin).
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 30 min
at room temperature. Following three washes with 1X PBS, cells were
blocked with normal goat serum for 2 h. The cells were then
incubated overnight at 4°C with anti-E-cadherin (1:100; 20874-1-AP;
ProteinTech Group, Inc.) or anti-fibronectin antibodies (dilution,
1:100; cat. no. 15613-1-AP; ProteinTech Group, Inc.) and with
fluorescein isothiocyanate-conjugated secondary antibody (dilution,
1:100; cat. no. SA00003-2; ProteinTech Group, Inc.) or
CoraLite594-conjugated secondary antibody (dilution, 1:100; cat.
no. SA00013-4; ProteinTech Group, Inc.). Nuclei were stained with
10 µg/ml DAPI in the dark for 5 min at room temperature. Cells were
observed using a fluorescence microscope (magnification, ×100).
Three representative fields of stained cells were analyzed using
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.,
Rockville, MD, USA) to obtain the mean OD, which represents the
staining strength per positive pixel.
Statistical analysis
Differences between two groups were analyzed using
Student's t-tests with GraphPad Prism software (version 5.0a;
GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as
the mean ± standard deviation (n=3). P<0.05 was considered to
indicate a statistically significant difference.
Results
SIRT1 expression in glioma
tissues
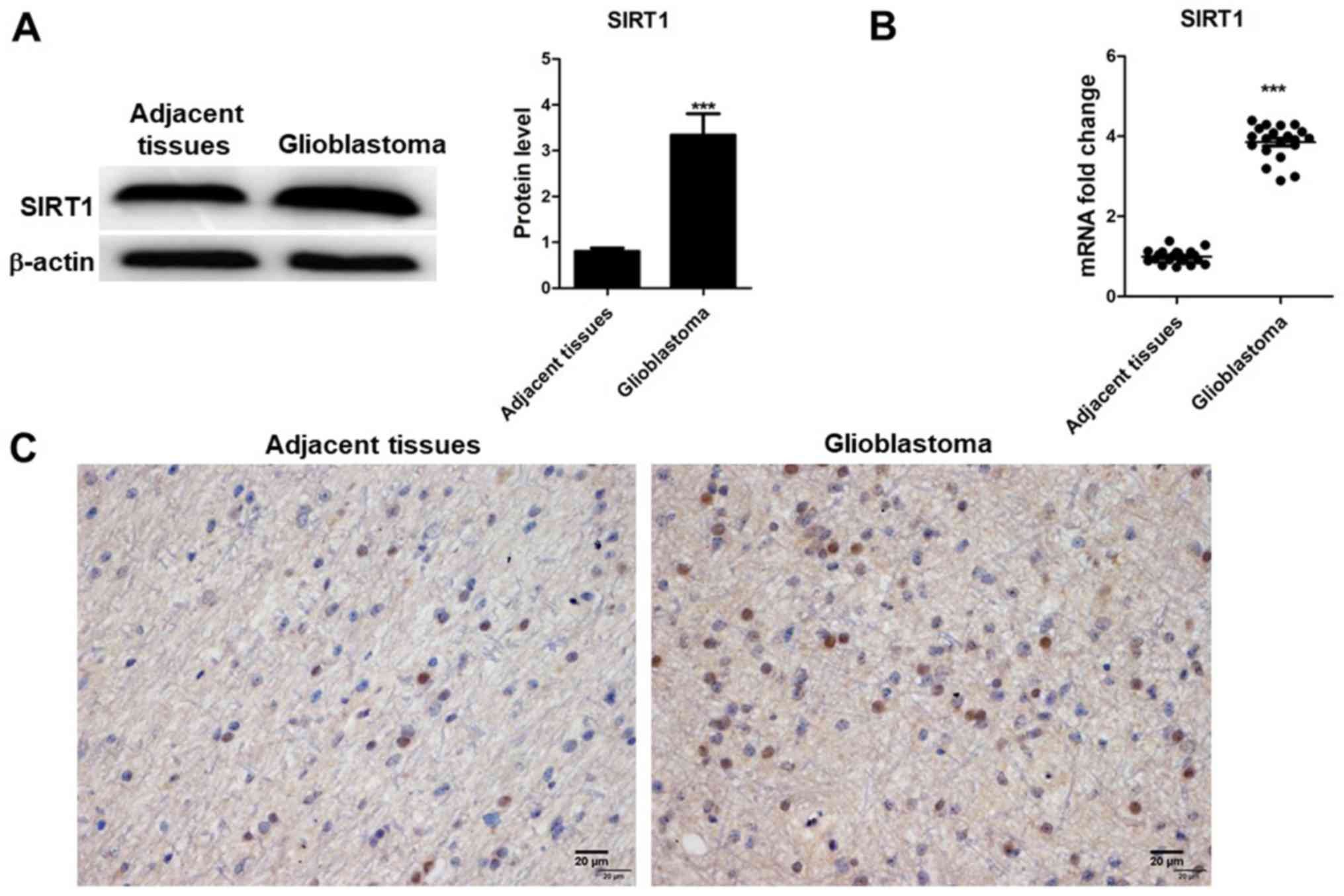
SIRT1 expression was detected in 20 glioblastoma
tissues and their corresponding adjacent non-tumor tissues using
RT-qPCR, western blotting and immunohistochemistry. The results
indicated that SIRT1 mRNA and protein expression levels were
upregulated in tumor tissues compared with in adjacent non-tumor
tissues (Fig. 1A and B), which was
determined to be significant. Results from immunohistochemistry
indicated that SIRT1 was present in the nucleus of glioma tissues
(Fig. 1C). SIRT1 levels were
different in glioma tissues compared with adjacent tissues. These
results indicated that SIRT1 expression was associated with glioma
development.
SIRT1 silencing inhibits glioma cell
viability
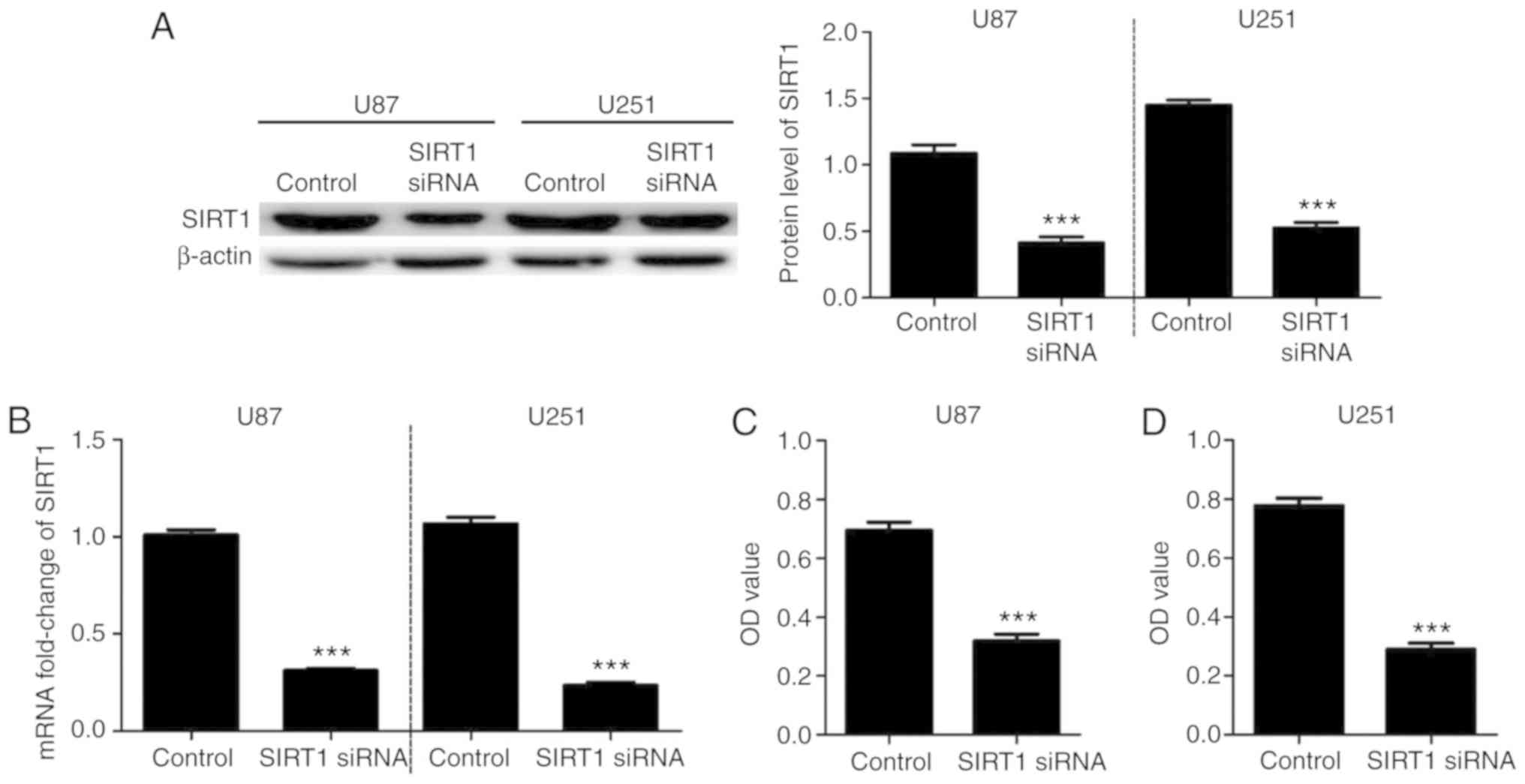
To investigate the effects of SIRT1 on U87 and U251
cell viability, cells were transfected with SIRT1-siRNA. RT-qPCR
and western blotting confirmed the successful SIRT1 silencing in
U87 and U251 cell lines (Fig. 2A and
B). A CCK-8 assay was used to detect the cell viability of U87
and U251 cells at 48 h after transfection with SIRT1-siRNA. Results
indicated that U87 and U251 cell viabilities were significantly
decreased following SIRT1-siRNA transfection compared with control
transfection (Fig. 2C and D). These
data suggested that SIRT1 expression may be involved in U87 and
U251 cell viability.
Silencing SIRT1 inhibits invasion of
glioma cells
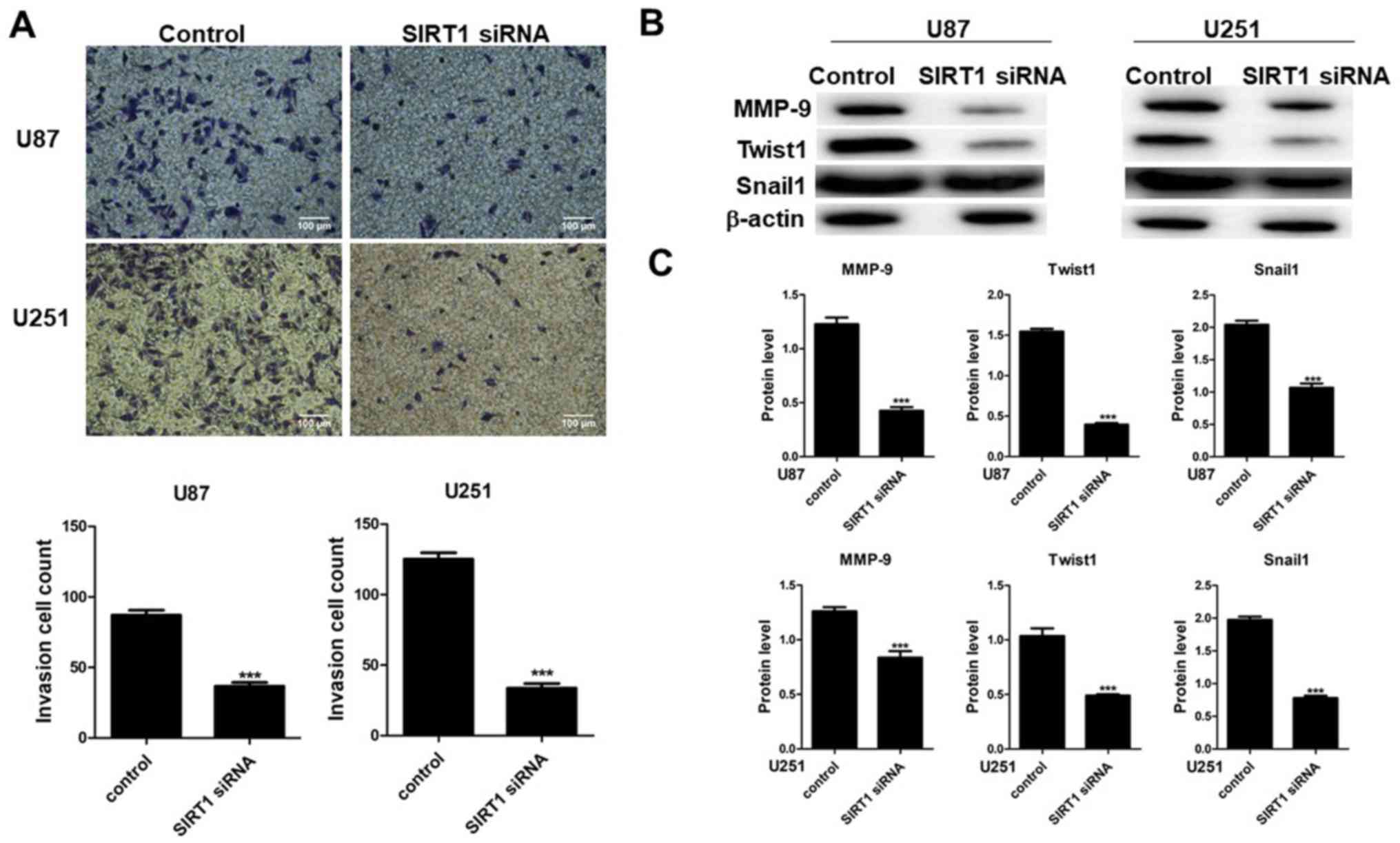
To determine the effect of SIRT1 on cell invasion,
Transwell invasion assays were performed on U87 and U251 cells
transfected with SIRT1-siRNA. Results indicated that SIRT1-siRNA
suppressed U87 and U251 cell compared with cells transfected with
control siRNA (Fig. 3A). Images of
cells present on the lower membranes of the invasion assay chambers
and quantification of crystal violet-positive cells are presented
in Fig. 3A, which indicated a
significant difference for the two cell lines. Results indicated
that SIRT1-siRNA inhibited U87 and U251 cell invasion. The
expression levels of MMP-9, Twist1 and Snail1 were therefore
determined in U87 and U251 cell lines by western blotting. Results
indicated that SIRT1-siRNA decreased the protein levels of MMP-9,
Twist1 and Snail1 in the two cell lines (Fig. 3B and C), which suggested that SIRT1
may have a beneficial effect on glioma cell invasion.
SIRT1 silencing inactivates EMT
pathway in glioma cells
The aforementioned results suggested that SIRT1 may
promote glioma cell viability invasion. EMT is a crucial biological
process for the invasion of certain types of malignant tumor cells.
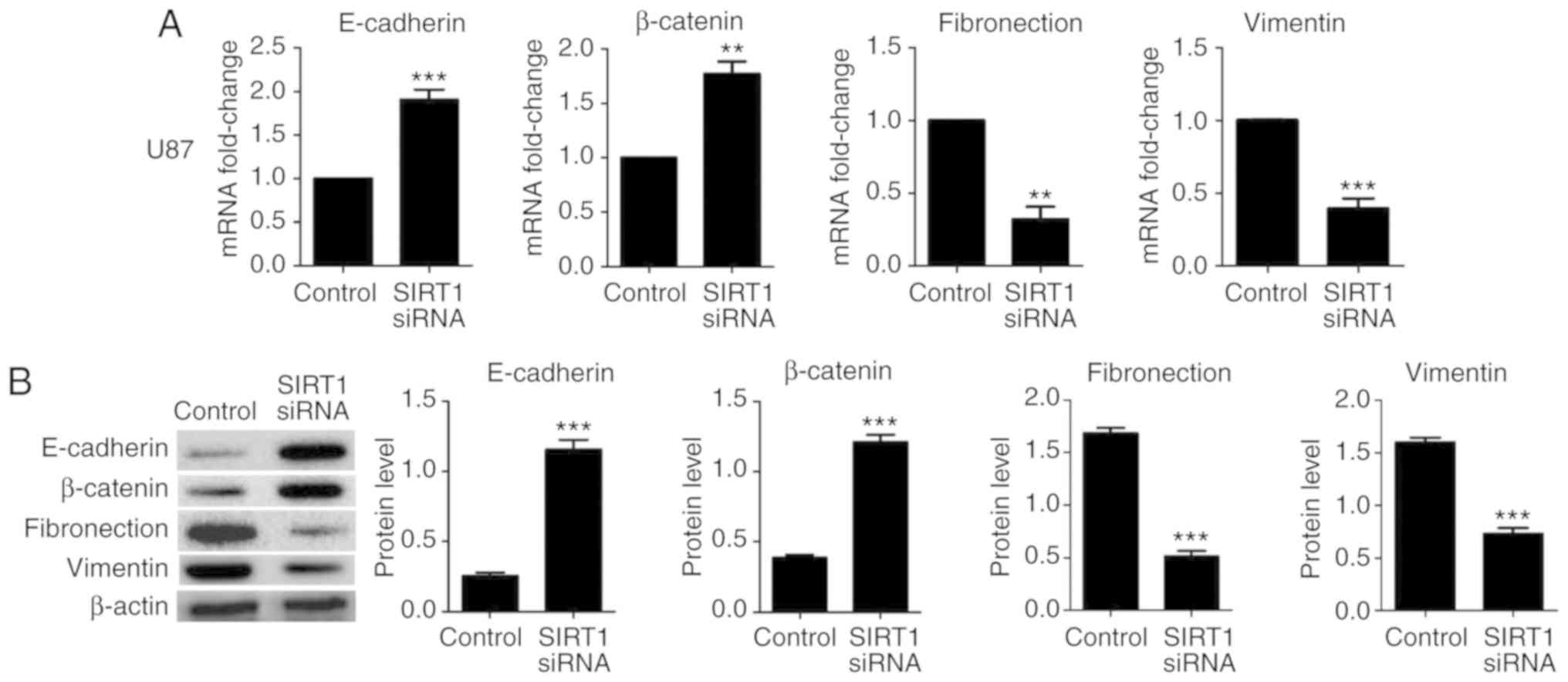
To identify the effect of SIRT1 on EMT in U87 and U251 cells,
RT-qPCR and western blotting were used to detect the expression
levels of the epithelial markers E-cadherin and β-catenin, and the
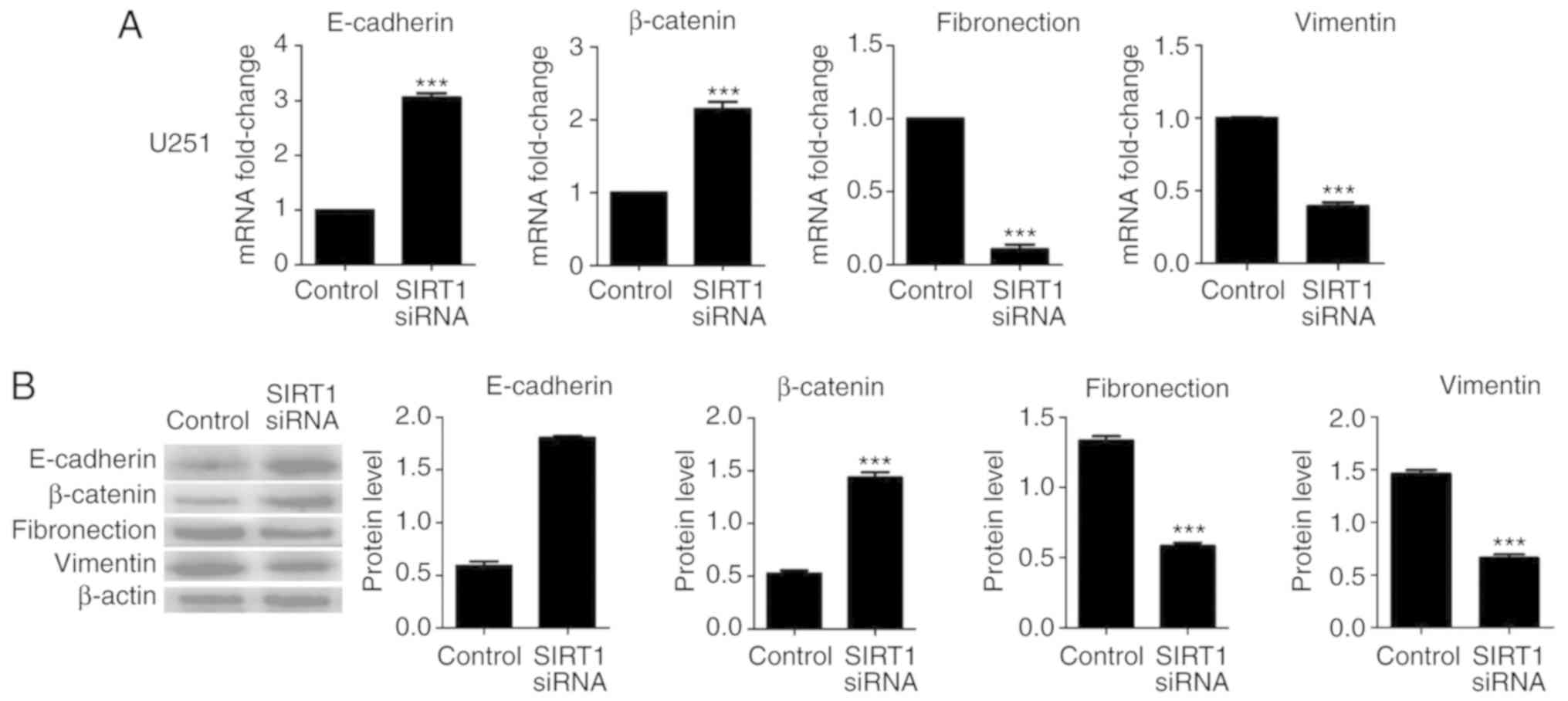
mesenchymal markers fibronectin and vimentin. Results indicated
that SIRT1 silencing significantly increased the mRNA and protein
levels of E-cadherin and β-catenin in U87 and U251 cells;
conversely, SIRT1-siRNA significantly decreased the mRNA and
protein levels of fibronectin and vimentin in U87 and U251 cells
(Figs. 4 and 5, respectively). In addition,
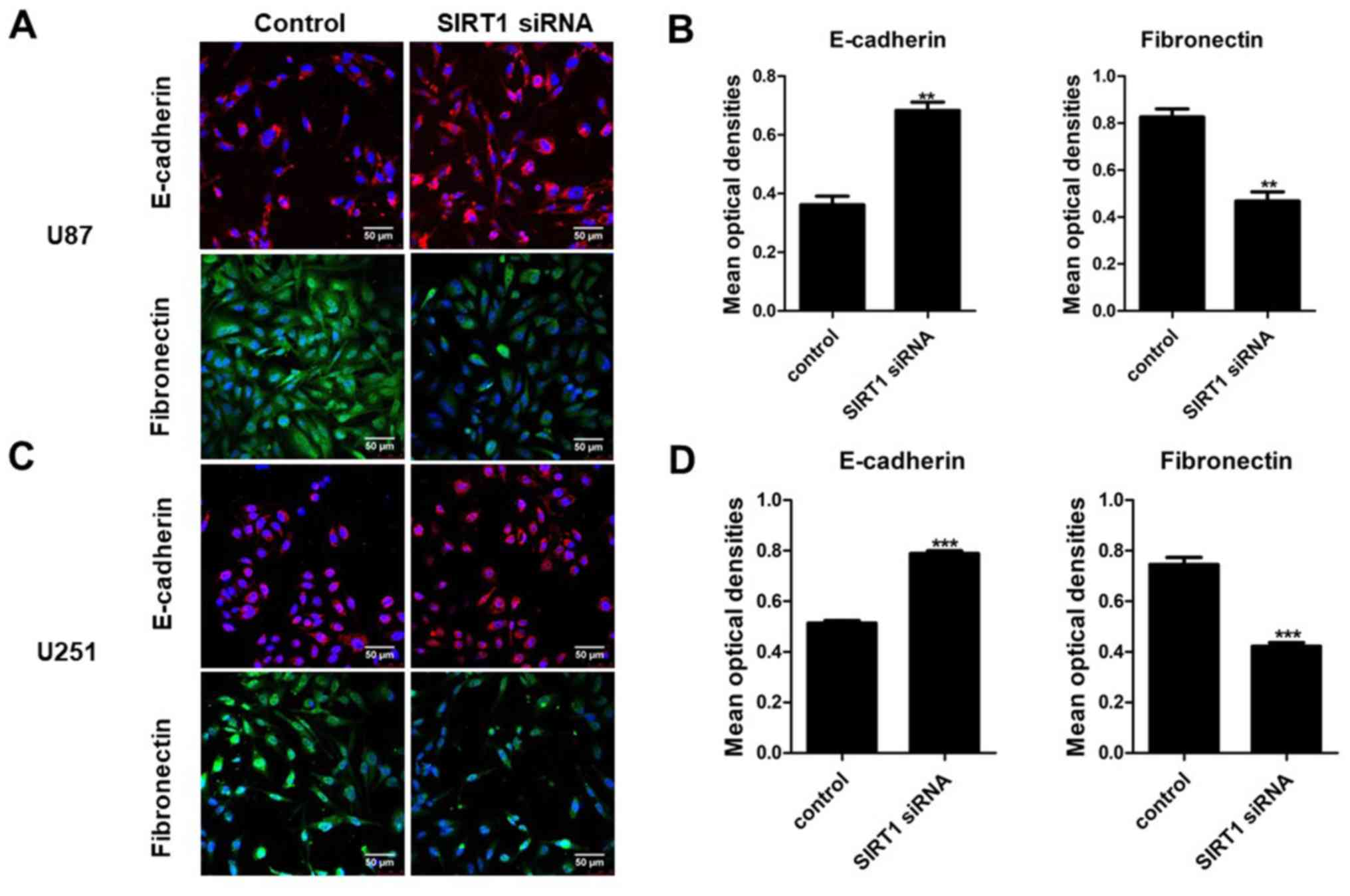
immunofluorescence analysis for E-cadherin and fibronectin revealed
a similar significant fluorescence decrease following SIRT1
silencing (Fig. 6). These results
indicate that SIRT1 may promote EMT in glioma cells.
Discussion
Glioma is one of the most common malignancies in
China. In addition, it is the most frequent type of intracranial
tumor and the most primary malignant brain tumor (31,32). The
World Health Organization classifies glioma according to its
malignancy with grades ranging from I to IV (33). Although microsurgical tumor
resection, adjuvant chemotherapy and radiotherapy are available,
75% of patients diagnosed with glioma succumb within 18 months of
diagnosis (34). Further
investigation aiming to develop novel therapeutic targets to
prevent glioma metastasis is therefore crucial. To the best of our
knowledge, the effect of SIRT1 on glioma remains poorly documented
and the role of SIRT1 in EMT in glioma remains unclear.
SIRT1 is a class III histone deacetylase that
belongs to the sirtuin family. The sirtuins have been reported to
serve crucial roles in genome stability, stress responses and
tumorigenesis (35). However, they
serve a dual role in cancer. SIRT1 is a bifunctional sirtuin that
is involved in tumor suppression and other oncogenic factors
(36); however, the mechanisms
underlying these contradictory functions remain unclear. SIRT1 is
upregulated in lung adenocarcinoma, colorectal, hepatocellular and
prostate cancer (10,36,37).
SIRT1 inhibition can suppress tumor growth (38). In addition, the tumor suppressor p53
downregulates SIRT1 expression in glioma tumor cells (39). SIRT1 also induces p53 inactivation
and inhibits p53-dependent apoptosis (7). Furthermore, SIRT1 promotes viability
and inhibits apoptosis of glioma tumor cell lines (24). However, the effects of SIRT1 on
invasion and metastasis and its underlying molecular mechanisms in
glioma remain unknown. In the present study, the expression levels
of SIRT1 in glioma tissues and adjacent brain tissue were
determined using RT-qPCR, western blotting and
immunohistochemistry. Results revealed that SIRT1 was overexpressed
in glioma tissues, compared with adjacent brain tissues that only
exhibited weak signal. In addition, SIRT1 silencing was used to
investigate SIRT1 effects on invasion and metastasis of glioma U87
cells.
EMT is an important process involved in tumor
metastasis (40,41). Previous studies have identified that
increased migration and invasion are positively associated with EMT
(42,43). In addition, decreased expression of
E-cadherin and β-catenin, and increased expression of fibronectin
and vimentin serve crucial roles in tumor progression (44). Furthermore, it was reported
previously that EMT serves a crucial role in glioma (45,46) and
that SIRT1 induces EMT in various types of tumor (47). However, it was identified previously
that SIRT1 suppresses EMT in cancer metastasis (22). In the present study, SIRT1 silencing
suppressed viability and invasion of glioma U87 and U251 cells,
increased E-cadherin and β-catenin expression, and decreased
fibronectin and vimentin expression.
To the best of our knowledge, the present study was
the first to assess the mRNA and protein expression levels of SIRT1
in human glioma and adjacent brain tissues. To do so, the effects
of SIRT1 on viability and invasion of glioma U87 and U251 cells
were investigated. Results indicated that SIRT1 expression was
higher in glioma tissues compared with in adjacent brain tissues,
and that SIRT1 silencing inhibited cell viability, invasion and
metastasis in glioma U87 and U251 cells. Furthermore, SIRT1
silencing significantly decreased EMT in U87 and U251 cells, which
suggested that EMT may contribute to the inhibition of viability,
invasion and metastasis of glioma cells.
The preliminary results of the present study
highlight the potential association of SIRT1 with EMT during human
glioma progression; however, the underlying molecular mechanisms
remain unclear and further investigation is required. Future work
will investigate how SIRT1 influences the EMT associated pathway in
human glioma.
In conclusion, the results of the present study may
serve to further understand SIRT1 functions and mechanisms in
glioma cells. These results suggested that SIRT1 may be considered
as a potential novel therapeutic target for glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and are freely available to any
researchers.
Authors' contributions
YL, XC and YC guided the experiments; XW and SC
designed the experiments; XC, YC, QW and YL performed the
experiments and analyzed the data; and YL wrote the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All human tissue samples were collected by The
Second Affiliated Hospital of Kunming Medical University and
written informed consent was obtained from all patients. All
methods were approved by the Research Medical Ethics Committee of
Kunming Medical University and were performed in accordance with
the approved guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Penas-Prado M, Armstrong S and Gilbert MR:
Glioblastoma. Handb Clin Neurol. 105:485–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith JS, Brachmann CB, Celic I, Kenna MA,
Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer
C, Wolberger C and Boeke JD: A phylogenetically conserved
NAD+-dependent protein deacetylase activity in the Sir2 protein
family. Proc Natl Acad Sci USA. 97:6658–6663. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu T, Liu PY and Marshall GM: The
critical role of the class III histone deacetylase SIRT1 in cancer.
Cancer Res. 69:1702–1705. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin SJ, Defossez PA and Guarente L:
Requirement of NAD and SIR2 for life-span extension by calorie
restriction in Saccharomyces cerevisiae. Science. 289:2126–2128.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marumoto T and Saya H: Molecular biology
of glioma. Adv Exp Med Biol. 746:2–11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo J, Nikolaev AY, Imai S, Chen D, Su F,
Shiloh A, Guarente L and Gu W: Negative control of p53 by Sir2alpha
promotes cell survival under stress. Cell. 107:137–148. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2(SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lim CS: Human SIRT1: A potential biomarker
for tumorigenesis? Cell Biol Int. 31:636–637. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi HN, Bae JS, Jamiyandorj U, Noh SJ,
Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG and Moon WS: Expression
and role of SIRT1 in hepatocellular carcinoma. Oncol Rep.
26:503–510. 2011.PubMed/NCBI
|
|
11
|
Holloway KR, Calhoun TN, Saxena M, Metoyer
CF, Kandler EF, Rivera CA and Pruitt K: SIRT1 regulates Dishevelled
proteins and promotes transient and constitutive Wnt signaling.
Proc Natl Acad Sci USA. 107:9216–9221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Zhang M, Dong H, Yong S, Li X,
Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, et al: Deacetylation
of cortactin by SIRT1 promotes cell migration. Oncogene.
28:445–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pruitt K, Zinn RL, Ohm JE, McGarvey KM,
Kang SH, Watkins DN, Herman JG and Baylin SB: Inhibition of SIRT1
reactivates silenced cancer genes without loss of promoter DNA
hypermethylation. PLoS Genet. 2:e402006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeung F, Hoberg JE, Ramsey CS, Keller MD,
Jones DR, Frye RA and Mayo MW: Modulation of NF-kappaB-dependent
transcription and cell survival by the SIRT1 deacetylase. EMBO J.
23:2369–2380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chua KF, Mostoslavsky R, Lombard DB, Pang
WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N,
et al: Mammalian SIRT1 limits replicative life span in response to
chronic genotoxic stress. Cell Metab. 2:67–76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zavadil J, Haley J, Kalluri R, Muthuswamy
SK and Thompson E: Epithelial-mesenchymal transition. Cancer Res.
68:9574–9577. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyazono K: Transforming growth factor-β
signaling and cancer: The 28th sapporo cancer seminar, 25–27 June
2008. Cancer Sci. 100:363–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Y, Li J, Zheng F, Ouyang Y, Chen X,
Zhang L, Chen Y, Wang L, Mu S and Zhang H: Effect of SIRT1 Gene on
epithelial-mesenchymal transition of human prostate cancer PC-3
Cells. Med Sci Monit. 22:380–386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng F, Su L, Yao C, Liu L, Shen J, Liu
C, Chen X, Luo Y, Jiang L, Shan J, et al: SIRT1 promotes
epithelial-mesenchymal transition and metastasis in colorectal
cancer by regulating Fra-1 expression. Cancer Lett. 375:274–283.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Simic P, Williams EO, Bell EL, Gong JJ,
Bonkowski M and Guarente L: SIRT1 suppresses the
epithelial-to-mesenchymal transition in cancer metastasis and organ
fibrosis. Cell Rep. 3:1175–1186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee M, Kim DW, Yoon H, So D, Khalmuratova
R, Rhee CS, Park JW and Shin HW: Sirtuin 1 attenuates nasal
polypogenesis by suppressing epithelial-to-mesenchymal transition.
J Allergy Clin Immunol. 137:87–98.e7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu Y, Zhang J, Wu S, Li B, Liu S and Cheng
J: SIRT1 promotes proliferation and inhibits apoptosis of human
malignant glioma cell lines. Neurosci Lett. 525:168–172. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JS, Park JR, Kwon OS, Lee TH, Nakano
I, Miyoshi H, Chun KH, Park MJ, Lee HJ, Kim SU and Cha HJ: SIRT1 is
required for oncogenic transformation of neural stem cells and for
the survival of ‘cancer cells with neural stemness’ in a
p53-dependent manner. Neuro Oncol. 17:95–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu WB, Wang W, Du YH, Li H, Xia SJ and Liu
HT: MicroRNA-3713 regulates bladder cell invasion via MMP9. Sci
Rep. 6:323742016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun T, Fu J, Shen T, Lin X, Liao L, Feng
XH and Xu J: The Small C-terminal domain phosphatase 1 inhibits
cancer cell migration and invasion by dephosphorylating
Ser(P)68-Twist1 to accelerate Twist1 protein degradation. J Biol
Chem. 291:11518–11528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ford J, Jiang M and Milner J:
Cancer-specific functions of SIRT1 enable human epithelial cancer
cell growth and survival. Cancer Res. 65:10457–10463. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Z, Long X, Chao C, Yan C, Wu Q, Hua S,
Zhang Y, Wu A and Fang W: Knocking down CDK4 mediates the elevation
of let-7c suppressing cell growth in nasopharyngeal carcinoma. BMC
Cancer. 14:2742014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Qu Q, Qu J, Luo WM, Wang SY, He YZ,
Luo QS, Xu YX and Wang YF: Association between XRCC1 polymorphisms
and glioma risk among Chinese population. Med Oncol. 31:1862014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sai K, Zhong MG, Wang J, Chen YS, Mou YG,
Ke C, Zhang XH, Yang QY, Lin FH, Guo CC, et al: Safety evaluation
of high-dose BCNU-loaded biodegradable implants in Chinese patients
with recurrent malignant gliomas. J Neurol Sci. 343:60–65. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Q: WHO classification of tumors of
central nervous system (2007): An introduction. Zhonghua Bing Li
Xue Za Zhi. 37:5–7. 2008.(In Chinese). PubMed/NCBI
|
|
34
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bosch-Presegué L and Vaquero A: The dual
role of sirtuins in cancer. Genes Cancer. 2:648–662. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Hokka D, Maniwa Y, Ohbayashi C,
Itoh T and Hayashi Y: Sirt1 is a tumor promoter in lung
adenocarcinoma. Oncol Lett. 8:387–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu DF, Jiang SJ, Pan ZP, Cheng WD, Zhang
WJ, Yao XK, Li YC and Lun YZ: Expression and clinical significance
of Sirt1 in colorectal cancer. Oncol Lett. 11:1167–1172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oon CE, Strell C, Yeong KY, Östman A and
Prakash J: SIRT1 inhibition in pancreatic cancer models:
Contrasting effects in vitro and in vivo. Eur J Pharmacol.
757:59–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yuan F, Liu L, Lei Y and Tang P: p53
inhibits the upregulation of sirtuin 1 expression induced by c-Myc.
Oncol Lett. 14:4396–4402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Voulgari A and Pintzas A:
Epithelial-mesenchymal transition in cancer metastasis: Mechanisms,
markers and strategies to overcome drug resistance in the clinic.
Biochim Biophys Acta. 1796:75–90. 2009.PubMed/NCBI
|
|
41
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kudo-Saito C, Shirako H, Takeuchi T and
Kawakami Y: Cancer metastasis is accelerated through
immunosuppression during Snail-induced EMT of cancer cells. Cancer
Cell. 15:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu ZJ, Liu HL, Zhou HC and Wang GC: TIPE2
Inhibits Hypoxia-Induced Wnt/β-catenin pathway activation and EMT
in glioma cells. Oncol Res. 24:255–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Z, Wu Y, Wang Y, Jin Y, Ma X, Zhang Y
and Ren H: Matrine inhibits the invasive properties of human glioma
cells by regulating epithelial-to-mesenchymal transition. Mol Med
Rep. 11:3682–3686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Byles V, Zhu L, Lovaas JD, Chmilewski LK,
Wang J, Faller DV and Dai Y: SIRT1 induces EMT by cooperating with
EMT transcription factors and enhances prostate cancer cell
migration and metastasis. Oncogene. 31:4619–4629. 2012. View Article : Google Scholar : PubMed/NCBI
|