Introduction
The incidence of papillary thyroid carcinoma (PTC)
is increasing rapidly worldwide (1).
Although the majority of patients with PTC have a good prognosis,
others face poor outcomes. Several organizations and experts have
tried to stratify patients with PTC into risk categories (2–4). As
different analysis methods were used in previous studies, the
incidence of multifocality is estimated with a wide range of 18–87%
(5–9). PTC foci may originate from the
intrathyroidal metastasis of the same clonal population of cells or
as a result of multiple tumors initiating independently. The origin
of these foci has not been determined. Multifocal tumors are often
correlated with a high risk of lymph-node metastasis, distant
metastasis and regional recurrence (10). Multifocality is not regarded as a
major factor in the classification of patients as low or high risk,
as studies have found that the cause-specific survival rate can
reach 100% in low-risk patients independently of multifocality
(11,12). However, a detailed understanding of
the nature of the clonal origin of multiple PTCs may aid in the
understanding of tumor pathogenesis and prognosis.
The examination of X-chromosome inactivation is one
approach to determine the clonal origin of multifocal PTCs. In the
somatic cells of females, one X-chromosome is inactivated through
the methylation of genes (13). The
inactivation, which occurs randomly in either the maternal or
paternal X-chromosome during embryogenesis, is restricted to a
single allele and is not changed during tumorigenesis (14). In the present study, the human
androgen receptor (HUMARA) gene assay, based on the polymerase
chain reaction (PCR), was applied to detect the presence of
X-chromosome inactivation in women (15). In the somatic cells of female
mammals, one X-chromosome is inactivated during embryogenesis. The
inactivation occurs randomly between one of the two chromosomes,
and it is somatically heritable. The methylation of cytosine
residues in the promoter regions of genes in the chromosome is one
of the main mechanisms of inactivation (12,13). The
HUMARA gene contains a highly polymorphic trinucleotide CAG short
tandem repeat (STR) in the first exon. The restriction endonuclease
HhaI has cleavage sites less than 100 base pairs away from
this polymorphic STR, which are accessible for cleavage only when
they are unmethylated. The PCR-based HUMARA assay takes advantage
of the highly polymorphic CAG STR region and nearby HhaI
sites in exon 1 of the HUMARA gene at the Xq13 region. Thus, the
HUMARA assay can be used to determine the clonality of multifocal
tumor tissues in cases where the patient is heterozygous for the
size of the STR region (15). The
aim of the present study was to investigate the X-chromosome
inactivation pattern of multifocal PTC, using a PCR-based assay to
detect a polymorphic CAG repeat locus in exon 1 of the HUMARA gene,
and use this to elucidate the clonal origin of multiple foci in PTC
cases.
Materials and methods
Tumor samples
A total of 89 female patients with a median age of
39±14 years (range, 18–60 years) who underwent a thyroidectomy for
the treatment of multiple distinct foci of PTC between January 2009
and December 2014 at the First Affiliated Hospital of Dalian
Medical University (Liaoning, China) were included in the present
study. All patients had PTC with 2 or 3 distinct foci in the
thyroid only. The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Dalian Medical
University. Histological slides were stained with hematoxylin and
eosin for 45 min at room temperature, and were reviewed by 2
independent pathologists who confirmed the diagnosis of classical
PTC. The diagnosis of classical PTC was based on the features of
classical nuclear morphology including: i) Enlarged and elongated
nuclei with crowding and overlap; ii) irregular nuclear contour;
iii) ground-glass nucleus; and iv) nuclear grooves.
Microdissection of tumor cells
Tissue samples were fixed in 10% formalin for 12 h
at room temperature and embedded in paraffin blocks preserved at
room temperature. Samples were cut into 10-µm sections that were
placed on clean slides. Large tumors where the tumor margins could
easily be recognized were microdissected by hand from the slides.
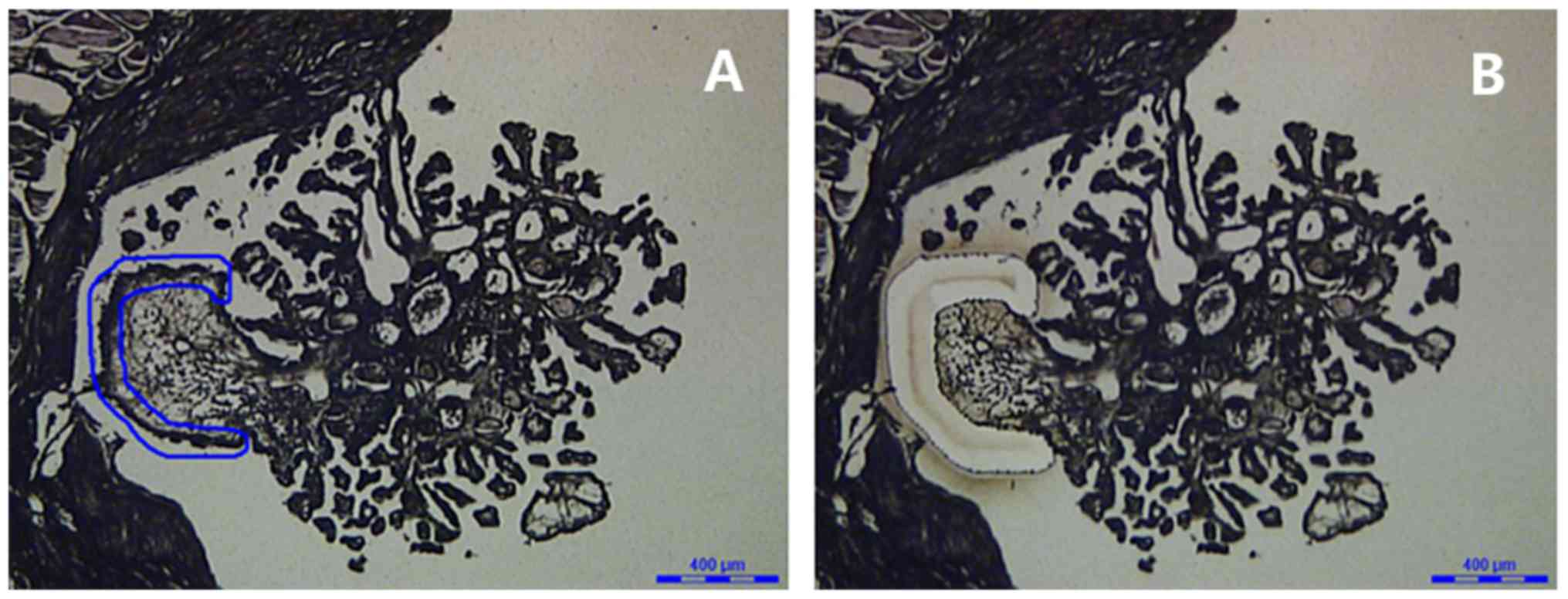
Small tumors or those with extensive inflammatory or stromal
components were dissecting using laser-capture microdissection
(Leica LMD6000; Leica Microsystems GmbH, Wetzlar, Germany) to
isolate the neoplastic cells (Fig.
1). Normal tissues at distances >1 cm from the tumors
obtained from each patient were microdissected as controls.
DNA extraction
DNA was extracted from formalin-fixed,
paraffin-embedded (FFPE) sections using the QiagenQIAmp FFPE kit
(Qiagen, Valencia, CA, USA) following the manufacturer's protocol.
Briefly, FFPE tissue was incubated in xylene to remove the
paraffin. Proteinase K was added and the samples were incubated at
90°C. DNA was bound to the membranes in the kit and contaminants
were washed through. DNA was eluted from the membrane and
quantified for use. An ultraviolet spectrophotometer was used to
quantify the extracted DNA (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
HhaI enzyme digestion and PCR-based
HUMARA analysis
A PCR-based method for assessing X-chromosome
inactivation according to the status of the X-linked HUMARA gene
was performed. DNA samples (5 µl) were incubated overnight at 37°C
with 1 µl HhaI (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a 10-µl reaction volume. The following PCR
primers were used to amplify the HUMARA exon 1 (16): Forward,
5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ and reverse,
5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′. The 30-µl PCR mixture included 3 µl
10X PCR buffer, 1.5 µl dNTP, 0.3 µl Taq DNA polymerase (Beijing
BLKW Biotechnology Co., Ltd., Beijing, China), 0.5 µl primers, 2 µl
DNA template and 22.7 µl ddH2O. The thermocycling
conditions were as follows: Denaturation at 94°C for 8 min, then 38
cycles at 95°C for 40 sec, 63°C for 40 sec and 72°C for 59 sec,
followed by a final extension at 72°C for 10 min. The PCR products
were separated by electrophoresis using an agarose gel (25 g/l with
1% ethidium bromide) at 100V for 40 min. X-chromosome inactivation
patterns were subsequently revealed by imaging the gels.
Examination of X-chromosome
inactivation
The cases were informative if there were 2 bands in
the electrophoresis gel of normal control tissue samples without
treatment with HhaI. Only informative cases were selected
for evaluation of X-chromosome inactivation. In tumor tissues
following HhaI digestion, the presence of 2 bands was
considered to indicate that the tumor was of polyclonal origin, and
the presence of 1 band indicated a monoclonal origin for the tumor.
Foci were considered to be of the same clonal origin if the same
X-chromosome inactivation status was revealed in the separate
tumors. Foci were considered to be of independent origin if
X-chromosome inactivation patterns were different in each tumor
(17,18).
Results
In the present study, multifocal PTC
samples from 89 female patients were analyzed
Heterozygosity for the HUMARA polymorphism,
indicated by the appearance of 2 bands in the electrophoresis gel
of normal control tissue, was identified in 10 out of 89 (11%)
cases. Of these 10 cases, 5 were considered to be informative, as
they had 2 bands for the HUMARA gene following amplification using
DNA from normal tissue without treatment with HhaI. The foci
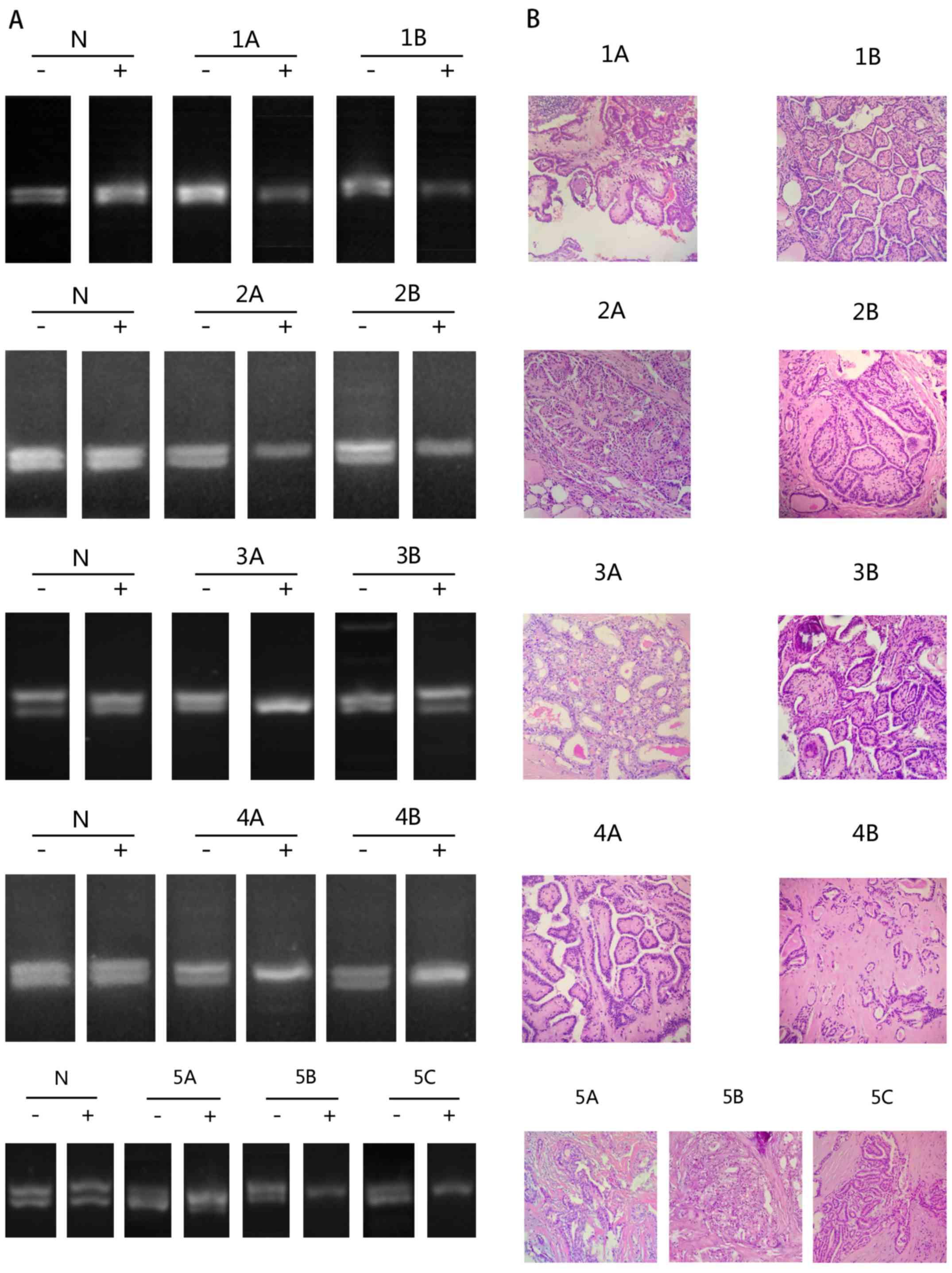
from cases 1, 2 and 4 had concordant patterns of X-chromosome
inactivation, with the 2 foci in each case exhibiting the same
pattern of bands following enzyme digestion. In cases 3 and 5,
contralateral tumor foci showed discordant patterns of X-chromosome
inactivation, with tumors from one lobe of the thyroid having a
different number of bands than tumors from the other lobe (Table I; Fig.
2).
 | Table I.X-chromosome inactivation pattern
identified by HUMARA assay in 5 cases of multifocal papillary
thyroid carcinoma. |
Table I.
X-chromosome inactivation pattern
identified by HUMARA assay in 5 cases of multifocal papillary
thyroid carcinoma.
| Case no. | Age, years | Location | Tumor | Tumor size, cm | HUMARA result |
|---|
| 1 | 57 | Left | 1A | 1.5 | Monoclonal |
|
|
| Right | 1B | 0.8 | Monoclonal |
| 2 | 57 | Left | 2A | 0.6 | Monoclonal |
|
|
| Right | 2B | 0.8 | Monoclonal |
| 3 | 29 | Left | 3A | 1.8 | Monoclonal |
|
|
| Right | 3B | 0.8 | Polyclonal |
| 4 | 43 | Left | 4A | 2.0 | Monoclonal |
|
|
| Right | 4B | 0.7 | Monoclonal |
| 5 | 60 | Left | 5A | 1.7 | Polyclonal |
|
|
| Right | 5B | 2.7 | Monoclonal |
|
|
| Right | 5C | 0.7 | Monoclonal |
Discussion
PTC is one of the most common types of malignant
tumor of the endocrine system (1).
Patients with PTC have a good prognosis, with a 95% survival rate
at 10 years. However, up to 20% of patients relapse following the
initial treatment (19). The high
occurrence and low mortality rates of PTC have led to differing
opinions on the optimal management of individual patients.
Independent factors that influence the survival rate
of patients with PTC are the presence of extra-thyroidal extension
of the tumor, the age of the patient and the presence of distant
metastasis. Although the occurrence of multiple foci in PTC is a
common clinical finding, the origin of the foci is undetermined
(20). Clarifying the origin of the
foci may be valuable for determining the treatment of patients with
multifocal PTC and their prognosis.
Previous genetic studies have revealed that a high
frequency (70%) of cases of PTC are associated with
mitogen-activated protein kinase signaling pathway activation via
point mutations in BRAF or RAS, or chromosomal fusions, including
translocations resulting in activated RET proto-oncogene or the TRK
fusion oncogene (21–26). Mutations in the phosphoinositide
3-kinase pathway, including protein kinase B, phosphatidylinositol
4,5-bisphosphate 3-kinase subunit A, and phosphatase and tensin
homolog, have also been reported in cases of PTC (27). Analysis of the clonal origin of PTC
with the unique clonal genetic alterations has been reported in
several studies (28,29). Gene alterations may not be an early
event during PTC progression. Thus, investigation into whether
cancer cells originate from a single precursor or multiple
precursors with a unique clonal genetic alteration is not
conclusive. Loss of heterozygosity and microRNA profiling have also
been used to identify the clonality of multifocal PTC (16,17,30).
Definite loss of heterozygosity or microRNA-specific signatures may
require refining to allow for clonal analysis of different
foci.
According to a hypothesis by Lyon (13), one X-chromosome is randomly
inactivated during embryogenesis in somatic cells. Methylation of
cytosine residues in the promoter regions of genes in the
chromosome leads to the inactivation of the X-chromosome (13). Inactivation of the X-chromosome
happens prior to cell transformation and therefore is irrelevant to
tumor formation. Consequently, detecting the pattern of
X-chromosome inactivation in different tumor foci can accurately
determine the clonal origin of multiple foci in PTC (31,32). The
HUMARA gene resides on the X-chromosome and the first exon of the
HUMARA gene includes a highly polymorphic trinucleotide CAG repeat.
The presence of an HhaI restriction site makes it possible
to detect the inactivation of this chromosome with a PCR-based
method. Previous studies have used a HUMARA-based assay to detect
the presence of non-random X-chromosome inactivation for
determining the clonal origins of multiple PTC foci (16,31–35).
However, the results of these studies are contradictory. Among the
6 reports, McCarthy et al (16) identified a concordant non-random
X-chromosome inactivation pattern in all 13 informative cases
studied. A more recent study revealed a high frequency of the same
inactivation pattern in separate foci, and therefore, the study
concluded that individual tumors from multifocal PTCs arise from a
single clone (33). Wang et
al (34) also suggested that
multifocal tumors arise from the same clone. By contrast, Shattuck
et al (32) collected 10
informative cases suitable for analysis and observed discordant
X-chromosome inactivation patterns in 5 samples, concluding that
that individual foci often arise as independent tumors. Moniz et
al (31) identified 3 out of 8
informative cases of multifocal PTC that had a polyclonal pattern
of X-chromosome inactivation, agreeing with the findings of
Shattuck et al (32).
Additionally, Kuhn et al (35) suggested that certain cases of
multifocal PTC are the result of true multicentricity, but that
others are the result of intra-thyroid spread by an original single
tumor mass. The explanation of the data may be an important factor
when considering the disparate results in these studies. However,
the results of this assay may occasionally be uninterpretable due
to complicated patterns (36,37).
Heterozygosity for the HUMARA polymorphism was
identified in 10 out of 89 (11%) cases in the present study. Of the
10 cases, 5 were considered to be informative, as the normal
control samples did not show 2 bands in the electrophoresis gel
following treatment with HhaI. Of the informative cases, 3
demonstrated evidence of monoclonality and 2 revealed patterns
consistent with multifocality.
Normal female tissue is a mosaic consisting of a
roughly equal mixture of 2 types of cells: Cells that contain an
active maternal X-chromosome, and others that possess an active
paternal X-chromosome. Once X-chromosome inactivation is
established during embryogenesis, it is fixed for all future cell
divisions. Therefore, determination of clonality by X-chromosome
inactivation analysis has the advantage that the inactivated
X-chromosome does not change with the formation of a tumor. The
main limitation of this method is that it can only be performed on
females. In the present study, 5 heterozygous cases were excluded
from the group of informative cases. This is due to findings by
Jovanovic et al (38) that
indicate that monoclonality in the thyroid is not restricted to
neoplastic tissue, but that large portions of normal tissue are
also monoclonally-derived, as embryonic patch size areas of normal
thyroid follicular epithelium display non-random patterns of
X-chromosome inactivation. Therefore, it was necessary to assess
adjacent normal thyroid tissue as a control in the present study,
in order to remove the possibility of false results due to
embryonic patch size.
In conclusion, the results of the present study
indicate that multifocal PTC may arise from the same clonal tumor
cell or independently. The largest sample population in any
previous study contained only 13 cases suitable for analysis.
Therefore, investigation in a larger cohort is required to verify
these results.
Acknowledgements
Not applicable.
Funding
Funding was provided by the Liaoning Provincial
Natural Science Foundation of China (grant no. 20180550036).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HG and LW designed the study. DC and SX collected
the data. LF, WQ and JW performed the PCR and electrophoresis gel
analysis. DC drafted the manuscript. HG and LW revised and
proofread the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients in this study provided informed consent
prior to undergoing thyroidectomy. All procedures in this study
were approved and performed in accordance with the principles of
the Research Ethics Committee of the First Affiliated Hospital of
Dalian Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kilfoy BA, Zheng T, Holford TR, Han X,
Ward MH, Sjodin A, Zhang Y, Bai Y, Zhu C, Guo GL, et al:
International patterns and trends in thyroid cancer incidence,
1973–2002. Cancer Causes Control. 20:525–531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
American Thyroid Association (ATA)
Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid
Cancer, ; Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL,
Mandel SJ, Mazzaferri EL, McIver B, Pacini F, Schlumberger M, et
al: Revised American Thyroid Association management guidelines for
patients with thyroid nodules and differentiated thyroid cancer.
Thyroid. 19:1167–1214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pitoia F, Ward L, Wohllk N, Friguglietti
C, Tomimori E, Gauna A, Camargo R, Vaisman M, Harach R, Munizaga F,
et al: Recommendations of the Latin American Thyroid Society on
diagnosis and management of differentiated thyroid cancer. Arq Bras
Endocrinol Metabol. 53:884–887. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pacini F, Schlumberger M, Dralle H, Elisei
R, Smit JW and Wiersinga W; European Thyroid Cancer Taskforce, :
European consensus for the management of patients with
differentiated thyroid carcinoma of the follicular epithelium. Eur
J Endocrino. 154:787–803. 2006. View Article : Google Scholar
|
|
5
|
Iida F, Yonekura M and Miyakawa M: Study
of intragladular dissemination of thyroid cancer. Cancer.
24:764–771. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tscholl-Ducommun J and Hedinger CE:
Papillary thyroid carcinomas. Morphology and prognosis. Virchows
Arch A Pathol Anat Histol. 396:19–39. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katoh R, Sasaki J, Kurihara H, Suzuki K,
Iida Y and Kawaoi A: Multiple thyroid involvement (intraglandular
metastasis) in papillary thyroid carcinoma. A clinicopathologic
study of 105 consecutive patients. Cancer. 70:1585–1590. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pitt SC, Sippel RS and Chen H:
Contralateral papillary thyroid cancer: Does size matter? Am J Sur.
197:342–347. 2009. View Article : Google Scholar
|
|
9
|
Hawk WA and Hazard JB: The many
appearances of papillary carcinoma of the thyroid. Cleve Clin Q.
43:207–215. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mazzaferri EL and Jhiang SM: Long-term
impact of initial surgical and medical therapy on papillary and
follicular thyroid cancer. Am J Med. 97:418–428. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hay ID, Thompson GB, Grant CS, Bergstralh
EJ, Dvorak CE, Gorman CA, Maurer MS, McIver B, Mullan BP, Oberg AL,
et al: Papillary thyroid carcinoma managed at the Mayo Clinic
during six decades (1940–1999): Temporal trends in initial therapy
and long-term outcome in 2444 consecutively treated patients. World
J Surg. 26:879–885. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gemsenjager E, Perren A, Seifert B,
Schuler G, Schweizer I and Heitz PU: Lymph node surgery in
papillary thyroid carcinoma. J Am Coll Surg. 197:182–190. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lyon MF: X-chromosome inactivation: A
repeat hypothesis. Cytogenet Cell Genet. 80:133–137. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lyon MF: X-chromosome inactivation and
developmental patterns in mammals. Biol Rev Camb Philos Soc.
47:1–35. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allen RC, Zoghbi HY, Moseley AB,
Rosenblatt HM and Belmont JW: Methylation of HpaII and HhaI
sites near the polymorphic CAG repeat in the human
androgen-receptor gene correlates with X chromosome inactivation.
Am J Hum Genet. 51:1229–1239. 1992.PubMed/NCBI
|
|
16
|
McCarthy RP, Wang M, Jones TD, Strate RW
and Cheng L: Molecular evidence for the same clonal origin of
multifocal papillary thyroid carcinomas. Clin Cancer Res.
12:2414–2418. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng L, Gu J, Eble JN, Bostwick DG,
Younger C, MacLennan GT, Abdul-Karim FW, Geary WA, Koch MO, Zhang S
and Ulbright TM: Molecular genetic evidence for different clonal
origin of components of human renal angiomyolipomas. Am J Surg
Pathol. 25:1231–1236. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng L, Gu J, Ulbright TM, MacLennan GT,
Sweeney CJ, Zhang S, Sanchez K, Koch MO and Eble JN: Precise
microdissection of human bladder carcinomas reveals divergent tumor
subclones in the same tumor. Cancer. 94:104–110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davies L and Welch HG: Current thyroid
cancer trends in the United States. JAMA Otolaryngol Head Neck
Surg. 140:317–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pacini F and De Groot LJ: Thyroid cancer.
endotext.org, version of July 1, 2016, published by. simpleMDText.comInc.(South Dartmouth, MA).
02748https://www.ncbi.nlm.nih.gov/books/NBK285543/
|
|
21
|
Bongarzone I, Butti MG, Coronelli S,
Borrello MG, Santoro M, Mondellini P, Pilotti S, Fusco A, Della
Porta G and Pierotti MA: Frequent activation of ret protooncogene
by fusion with a new activating gene in papillary thyroid
carcinomas. Cancer Res. 54:2979–2985. 1994.PubMed/NCBI
|
|
22
|
Cohen Y, Xing M, Mambo E, Guo Z, Wu G,
Trink B, Beller U, Westra WH, Ladenson PW and Sidransky D: BRAF
mutation in papillary thyroid carcinoma. J Natl Cancer Inst.
95:625–627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fagin JA: Challenging dogma in thyroid
cancer molecular genetics-role of RET/PTC and BRAF in tumor
initiation. J Clin Endocrinol Metab. 89:4264–4266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sozzi G, Bongarzone I, Miozzo M, Borrello
MG, Blutti MG, Pilotti S, Della Porta G and Pierotti MA: A t(10;17)
translocation creates the RET/PTC2 chimeric transforming sequence
in papillary thyroid carcinoma. Genes Chromosomes Cancer.
9:244–250. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA,
Nikiforov YE and Fagin JA: High prevalence of BRAF mutations in
thyroid cancer: genetic evidence for constitutive activation of the
RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma.
Cancer Res. 63:1454–1457. 2003.PubMed/NCBI
|
|
26
|
Cancer Genome Atlas Research Network, .
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xing M: Molecular pathogenesis and
mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park SY, Park YJ, Lee YJ, Lee HS, Choi SH,
Choe G, Jang HC, Park SH, Park DJ and Cho BY: Analysis of
differential BRAF(V600E) mutational status in multifocal papillary
thyroid carcinoma: evidence of independent clonal origin in
distinct tumor foci. Cancer. 107:1831–1838. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Z, Ciampi R, Nikiforova MN, Gandhi M
and Nikiforov YE: Prevalence of RET/PTC rearrangements in thyroid
papillary carcinomas: effects of the detection methods and genetic
heterogeneity. J Clin Endocrinol Metab. 91:3603–3610. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aherne ST, Smyth PC, Flavin RJ, Russell
SM, Denning KM, Li JH, Guenther SM, O'Leary JJ and Sheils OM:
Geographical mapping of a multifocal thyroid tumour using genetic
alteration analysis and miRNA profiling. Mol Cancer. 7:892008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moniz S, Catarino AL, Marques AR, Cavaco
B, Sobrinho L and Leite V: Clonal origin of non-medullary thyroid
tumours assessed by non-random X-chromosome inactivation. Eur J
Endocrinol. 146:27–33. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shattuck TM, Westra WH, Ladenson PW and
Arnold A: Independent clonal origins of distinct tumor foci in
multifocal papillary thyroid carcinoma. N Engl J Med.
352:2406–2412. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakazawa T, Kondo T, Tahara I, Kasai K,
Inoue T, Oishi N, Mochizuki K, Kubota T and Katoh R: Multicentric
occurrence of multiple papillary thyroid carcinomas-HUMARA and BRAF
mutation analysis. Cancer Med. 4:1272–1280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang W, Wang H, Teng X, Wang H, Mao C,
Teng R, Zhao W, Cao J, Fahey TJ III and Teng L: Clonal analysis of
bilateral, recurrent, and metastatic papillary thyroid carcinomas.
Hum Pathol. 41:1299–1309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuhn E, Teller L, Piana S, Rosai J and
Merino MJ: Different clonal origin of bilateral papillary thyroid
carcinoma, with a review of the literature. Endocr Pathol.
23:101–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Siu IM, Robinson DR, Schwartz S, Kung HJ,
Pretlow TG, Petersen RB and Pretlow TP: The identification of
monoclonality in human aberrant crypt foci. Cancer Res. 59:63–66.
1999.PubMed/NCBI
|
|
37
|
El Kassar N, Hetet G, Brière J and
Grandchamp B: X-chromosome inactivation in healthy females:
incidence of excessive lyonization with age and comparison of
assays involving DNA methylation and transcript polymorphisms. Clin
Chem. 44:61–67. 1998.PubMed/NCBI
|
|
38
|
Jovanovic L, Delahunt B, McIver B,
Eberhardt NL and Grebe SK: Thyroid gland clonality revisited: The
embryonal patch size of the normal human thyroid gland is very
large, suggesting X-chromosome inactivation tumor clonality studies
of thyroid tumors have to be interpreted with caution. J Clin
Endocrinol Metab. 88:3284–3291. 2003. View Article : Google Scholar : PubMed/NCBI
|