Introduction
Liver cancer is a common cancer worldwide, ranking
sixth highest in incidence, with a fatality rate that is the third
highest in highly lethal tumors globally. It is thus known as ‘the
king of cancers’. Among these tumors, hepatocellular carcinoma
(HCC) accounts for 85–90% of deaths, and China is currently one of
the countries with the most HCC deaths worldwide. HCC has a high
degree of malignancy. Once the disease occurs, it progresses
rapidly and the patient survival time is relatively short. The
survival rate of HCC patients in the early stage in China is
approximately 60%. According to statistics the 5-year survival rate
of HCC patients is approximately 33%, which is mostly due to the
occult onset of liver cancer and the lack of specific clinical
manifestations in the early stage. In addition, patients were in
the middle and advanced stage when diagnosed, and more importantly,
there are few effective treatments (1–3).
At present, surgical resection remains the first
choice of treatment. However, the vast majority of patients have
already had near invasion or distant metastasis at the time of
definite diagnosis; thus, surgical resection does not constitute a
viable option. In addition, the recurrence rate of patients
undergoing early surgical resection is high. Therefore, the
identification of more effective methods for clinical diagnosis,
treatment and prognosis monitoring is imperative (4,5). Long
intergenic non-coding RNAs (lincRNAs) refer to a class of long
non-coding RNAs (lncRNAs) located between two coding genes.
Currently, an increasing number of lincRNAs have been identified,
such as lincRNA-p21, and an increasing number of studies have shown
that lincRNA-p21 is involved in the development of a variety of
human diseases, especially cancer (6,7). At the
same time, lincRNA-p21 is downregulated in many types of tumors
compared with that in adjacent normal tissues, suggesting that
lincRNA-p21 plays a key role as a tumor suppressor gene (8,9).
In this study, gene knockout technique was used to
intervene in the expression of lincRNA-p21 in HepG2 cells, and the
effects of lincRNA-p21 on the proliferation, migration and invasion
ability of HepG2 cell line were studied.
Materials and methods
Experimental materials
The HepG2 liver cancer cell line was purchased from
ATCC and preserved in our laboratory.. The cells were identified as
hepatocellular carcinoma cell lines by STR analysis, and there was
no contamination. The eukaryotic expression plasmids containing
lincRNA-p21 small interfering RNA (siRNA) sequence were constructed
and prepared by GenScript Biotech Co., Ltd. (Nanjing, China).
Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum
were purchased from HyClone (HyClone: GE Healthcare, Logan, UT,
USA). TRIzol reagent kit, messenger RNA (mRNA) extraction kit,
complementary DNA (cDNA) synthesis kit and reverse
transcription-polymerase chain reaction (RT-PCR) amplification kit
were purchased from Invitrogen (Invitrogen: Thermo Fisher
Scientific, Inc., Carlsbad, CA, USA). Cell Counting kit-8 (CCK-8)
and apoptosis detection kit were purchased from Beyotime Institute
of Biotechnology (Haimen, China). Transwell chamber and Matrigel
were purchased from Corning, Inc. (Corning, NY, USA). Fluorescent
quantitative PCR instrument was purchased from Applied Biosystems
(Applied Biosystems: Thermo Fisher Scientific, Inc., Foster City,
CA, USA). Flow cytometer was purchased from BD Biosciences
(Franklin Lakes, NJ, USA). Fluorescence microscope was purchased
from Olympus Corp. (Tokyo, Japan).
The study was approved by the Ethics Committee of
Jiangsu Provincial People's Hospital Affiliated to Nanjing Medical
University (Nanjing, China).
Construction and transfection of
lincRNA-p21 siRNA lentivirus vector
The constructed recombinant plasmids containing
lincRNA-p21 siRNA were transfected into 293T cells. After culture,
cell supernatants rich in virus particles were collected, filtered
and stored at −80°C for standby application. After 40–60% of the
cells were fused, HepG2 cells in the logarithmic growth phase were
added with the recombinant lentivirus vector containing lincRNA-p21
siRNA (experimental group) and empty virus vector (control group),
respectively. The cells were incubated in an incubator with 5%
CO2 at 37°C for 24 h, and then the medium was replaced.
Stable cells were obtained for expansion culture by G418
screening.
Detection of lincRNA-p21 mRNA
expression via RT-PCR
The transfected HepG2 cells were lysed with TRIzol
reagent and the total cell RNA was extracted according to the
instructions of mRNA extraction kit to measure RNA concentration.
The total RNA was reversely transcribed into cDNA according to the
cDNA synthesis kit. lincRNA-p21 primers were designed, including
lincRNA-p21 forward, 5′-CCCGGGCTTGTCTTTTGTT-3′ and reverse,
5′-GAGTGGGTGGCTCACTCTTCTG-3′; glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-CTGGGCTACACTGAGCACC-3′ and
reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. According to the instructions
of RT-PCR amplification kit, SYBR-Green assay was used for
quantitative PCR detection. Reaction conditions: pre-denaturation
at 95°C for 15 min, denaturation at 95°C for 15 sec, annealing at
60°C for 30 sec, fluorescence collection, extension at 72°C for 30
sec, a total of 40 cycles, data reading and quantitative
analysis.
Detection of cell proliferation
ability via CCK-8 assay
Cells in experimental and control group were
digested with trypsin. The concentration of cells was adjusted to
5×104/ml with DMEM containing 10% fetal bovine serum. A
total of 100 µl of cell suspension was inoculated into a 96-well
plate and cultured at 37°C for 12, 24, 48, and 72 h, respectively.
A total of 10 µl CCK-8 was added and gently mixed well using an
oscillator, followed by cultivation in an incubator at 37°C for 4
h. Double-wavelength measurements were performed on a microplate
reader with a detection wavelength of 490 nm and a reference
wavelength of 650 nm. The cell proliferation curve was plotted
based on optical density (OD) values.
Detection of apoptosis by flow
cytometry
Logarithmic growth phase cells in experimental and
control group were digested with trypsin without
ethylenediaminetetraacetic acid (EDTA), collected, and washed with
phosphate-buffered saline (PBS) twice. A total of 5×105
cells were collected in each tube, and 500 µl binding buffer was
added to 100×101 dyeing buffer for cell resuspension.
Then, 5 µl Annexin V and 10 µl propidium iodide (PI) dyeing liquor
were added, respectively, and mixed well, followed by reaction at
room temperature in the dark for 15 min. Cell apoptosis was
detected by flow cytometry. Three parallel samples were set up in
each experimental group. Apoptosis rate (%) = (Annexin V + PI +
cell number + Annexin V + PI - cell number)/10,000 × 100%.
Transwell chamber migration
experiment
The Transwell chamber was placed in a 24-well plate.
HepG2 cells in the experimental and control groups were digested
with trypsin and pipetted into single cell suspension, which was
then washed with PBS twice. Cells were resuspended in serum-free
medium and the cell concentration was adjusted to
5×105/ml. A total of 100 ml suspension was taken and
inoculated in the Transwell chamber. In the lower chamber, 500 µl
DMEM containing 10% fetal bovine serum was added for cultivation in
an incubator at 37°C for 20–24 h. The Transwell chamber was taken
out and washed with PBS twice, followed by fixation with 5%
glutaraldehyde for 30 min, washing with PBS twice, staining with
0.1% crystal violet for 20 min and washing with PBS twice. The
unmigrated cells on the upper surface were erased with cotton ball
and the number of perforating cells was calculated by randomly
taking images of 8–10 visual fields by an optical microscope at a
magnification of ×100.
Transwell chamber invasion
experiment
The Transwell chamber was placed in a 24-well plate.
BD Matrigel was diluted with serum-free DMEM at 1:6 and 100 µl of
diluted BD Matrigel was added into the Transwell chamber, followed
by cultivation in an incubator at 37°C for 6 h. BD Matrigel was
washed with serum-free medium once. HepG2 cells in experimental and
control group were digested with trypsin and pipetted into single
cell suspension, which was then washed with PBS twice. Cells were
resuspended in serum-free medium and the cell concentration was
adjusted to 5×105/ml. A total of 100 ml suspension was
taken and inoculated into the Transwell chamber. In the lower
chamber, 500 µl DMEM containing 10% fetal bovine serum was added
for cultivation in an incubator at 37°C for 20–24 h. The Transwell
chamber was taken out and washed with PBS twice, followed by
fixation with 5% glutaraldehyde for 30 min, washing with PBS twice,
staining with 0.1% crystal violet for 20 min, and washing with PBS
twice. The upper layer cells of BD Matrigel and microporous
membrane were erased with cotton ball. The number of perforated
cells was calculated by randomly taking images of 8–10 visual
fields by an optical microscope at a magnification of ×100.
Statistical analysis
Statistical Product and Service Solutions (SPSS)
17.0 statistical software was used for statistical analysis.
Enumeration data are expressed as means ± SD. Analysis of variance
was used for multigroup comparison as well as least significant
difference (LSD) post hoc test. The t-test was used for comparison
between two groups. P<0.05 indicates that the difference was
statistically significant.
Results
Detection of knockout effects of
lincRNA-p21 mRNA via RT-PCR
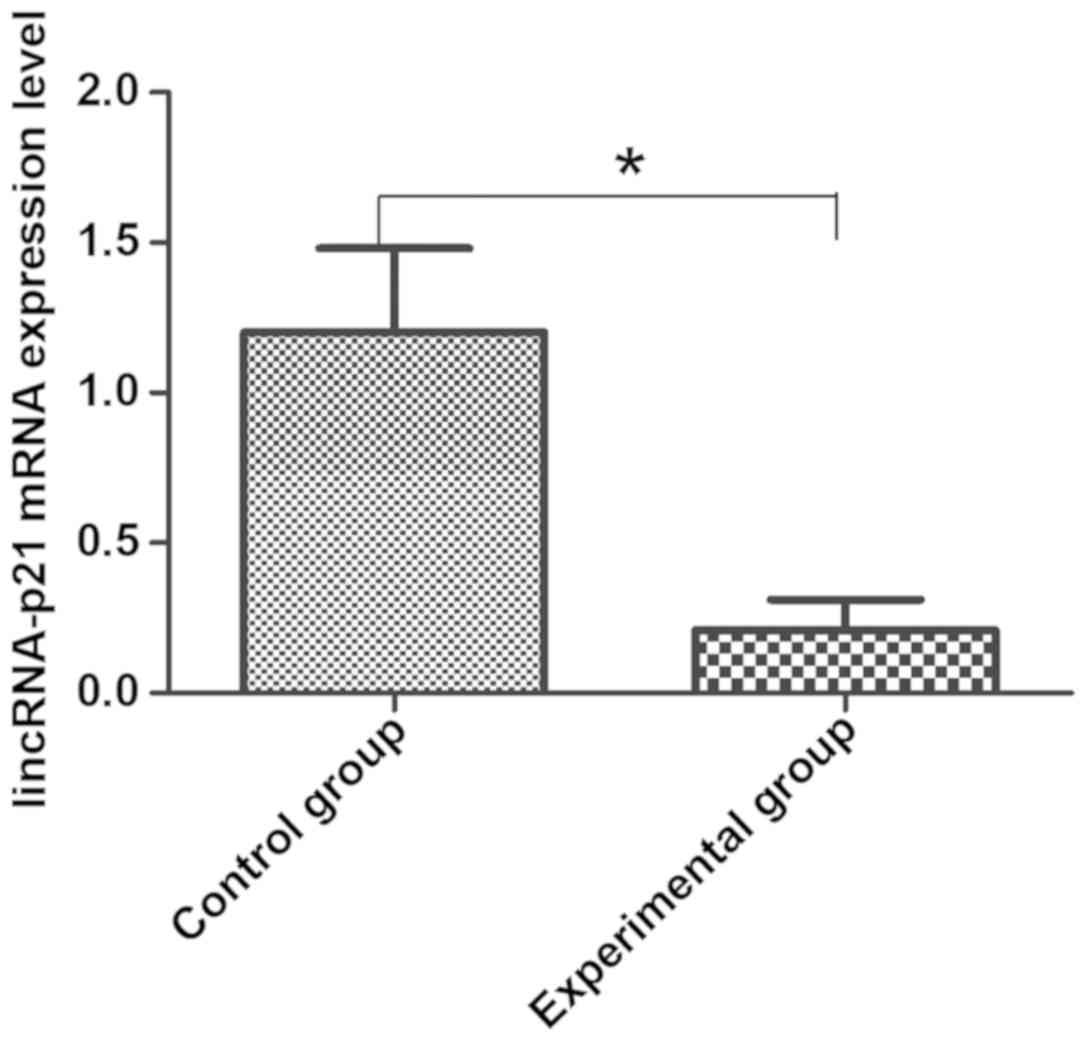
Compared with empty transfected HepG2 cells in
control group, the mRNA expression of lincRNA-p21 in cells in the
experimental group was significantly decreased, indicating that the
transfection of lincRNA-p21 siRNA can effectively interfere with
the expression level of lincRNA-p21 in HepG2 cells, which laid the
foundation for the next experiment (Fig.
1).
Effects of lincRNA-p21 knockout on the
proliferation of HepG2 cells
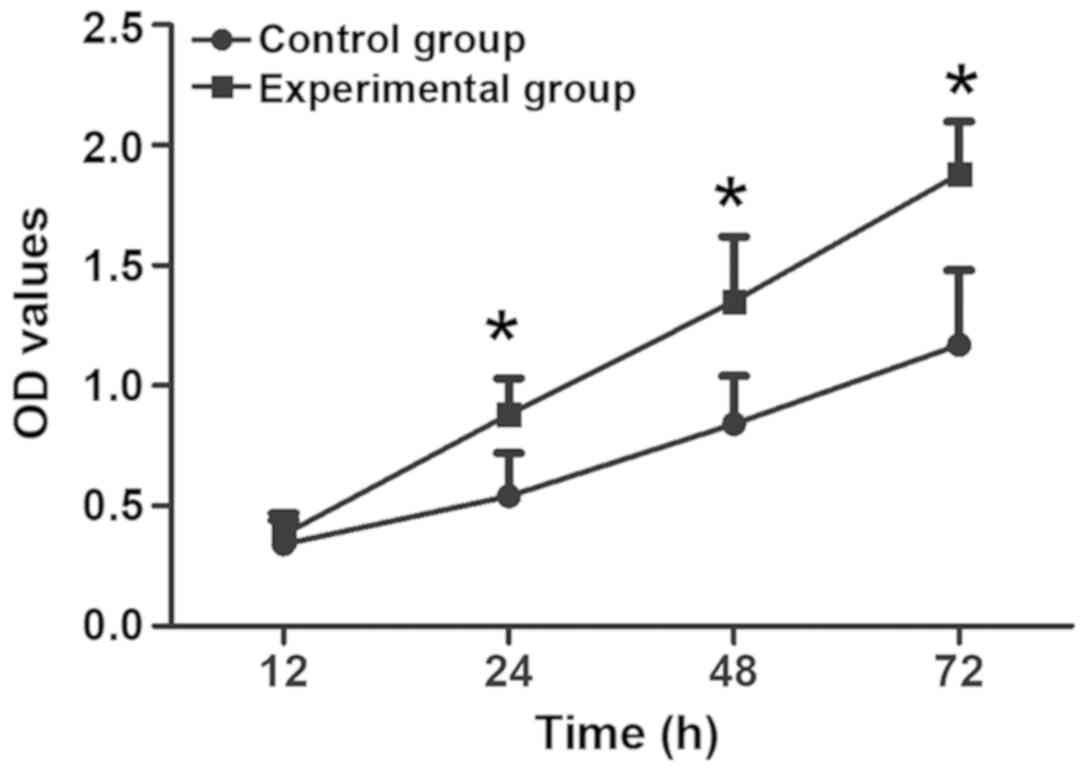
CCK-8 assay was used to detect the relationship
between the downregulation of lincRNA-p21 expression and HepG2 cell
proliferation. The results are shown in Fig. 2 and Table
I. Compared with that in control group, the cell proliferation
ability in the experimental group was obviously enhanced, and the
difference was obvious from 24 h (p<0.05), indicating that the
downregulation of lincRNA-p21 expression can promote the
proliferation of HepG2 cells.
 | Table I.Effects of lincRNA-p21 knockout on the
proliferation of HepG2 cells. |
Table I.
Effects of lincRNA-p21 knockout on the
proliferation of HepG2 cells.
| Group | 12 h | 24 h | 48 h | 72 h |
|---|
| Control | 0.34±0.10 | 0.54±0.18 | 0.84±0.20 | 1.17±0.31 |
| Experimental | 0.38±0.09 | 0.88±0.15 | 1.35±0.27 | 1.88±0.22 |
| P-value | 0.109 | 0.043 | 0.031 | 0.0102 |
Effects of lincRNA-p21 knockout on
apoptosis of HepG2 cells
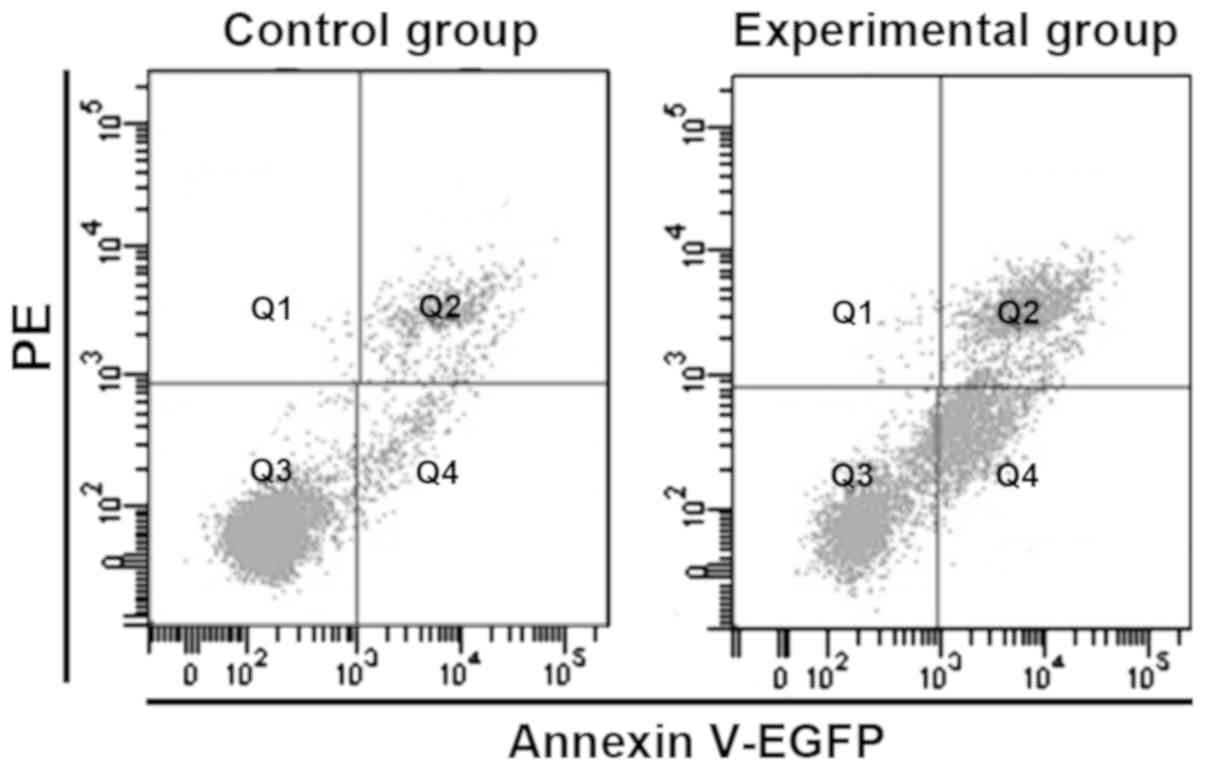
The apoptosis rate of HepG2 cells in logarithmic
growth phase in experimental and control group was detected via
flow cytometry. As shown in Fig. 3,
the apoptotic rates in the experimental and control group were
5.21±1.43 and 26.33±5.13, respectively. Compared with that in
control group, the apoptosis rate in experimental group was
significantly decreased (p<0.05), indicating that knockout
lincRNA-p21 can inhibit HepG2 cell apoptosis.
Effects of lincRNA-p21 knockout on the
migration and invasion ability of HepG2 cells
The relationship between the downregulation of
lincRNA-p21 expression and the migration and invasion of HepG2
cells was detected via Transwell assay. The results revealed that
after crystal violet staining, cells in control and experimental
group passing through chamber filter membrane were observed in both
migration and invasion experiments. The number of cells that passed
through the membrane in experimental group was significantly higher
than that in control group (p<0.05), indicating that knockout of
lincRNA-p21 can effectively improve the migration and invasion
ability of HepG2 cells (Table
II).
 | Table II.Comparisons of cell migration and
invasion number between two groups (cells/HP). |
Table II.
Comparisons of cell migration and
invasion number between two groups (cells/HP).
| Group | Cell migration
no. | Cell invasion
no. |
|---|
| Control | 90.7±14.8 | 30.6±8.6 |
| Experimental |
215.3±18.9a |
87.5±10.3a |
Discussion
Liver cancer is the most common malignant tumor in
China, whose morbidity and mortality accounts for ~50% of those in
the whole world, making China one of the regions with the highest
incidence of liver cancer in the world. Since liver cancer patients
often do not have specific symptoms or signs, it is difficult to
diagnose early. Therefore, in clinic, many patients with liver
cancer have entered the middle or late stages when they are
diagnosed. For patients with middle or late liver cancer, there is
still no effective clinical treatment (10,11). It
is noteworthy that liver cancer is characterized by high
invasiveness, early metastasis, high recurrence rate, poor
prognosis, insensitivity to radiotherapy, low effective rate of
chemotherapy, and extreme resistance to drugs. Its mortality rate
has remained high, and metastasis is the main reason for the high
mortality rate. Therefore, it is of great significance to study the
mechanism of tumor metastasis and explore effective treatment
methods to control the metastasis and recurrence of liver cancer
and improve the survival rate and quality of life of liver cancer
patients (12–14). lncRNAs and their subtype lincRNAs are
biologically functional non-coding RNAs discovered in recent years.
lncRNA is a type of RNA molecule that does not encode proteins and
has a transcript of >200 nt in length. It regulates gene
expression at multiple levels in the form of RNA (15,16).
Research has manifested that lncRNAs play an important role in the
formation and development of tumors. Therefore, in-depth study of
the function and mechanism of lncRNA in tumors will help find
potential target genes for the treatment of tumors.
p53 is a key molecule that regulates a variety of
biological behaviors of cells, and plays an important role in the
repair of damaged DNA, apoptosis, proliferation and other
regulatory aspects. lincRNA-p21 is found to be a kind of lncRNA
most closely related to p53 expression. p53 binds directly to the
upstream regulatory region of lincRNA-p21 and regulates its
transcriptional level (17,18). Clinical studies have proven that the
low expression of lincRNA-p21 predicts a lower survival rate of
cancer patients, and it can be used in the future as a new
molecular marker for the diagnosis and prognosis to stimulate new
research directions and treatment options.
In order to further clarify the role of lincRNA-p21
in the progression of liver cancer, RNA interference (RNAi)
technology was used to interfere with the expression of lincRNA-p21
in HCC HepG2 cells in this study. The results revealed that after
the expression of lincRNA-p21 was successfully inhibited, the
proliferation ability of HCC cell line HepG2 was obviously
enhanced, flow cytometry showed a significant decrease in the
apoptosis ability, and the results of Transwell chamber experiment
demonstrated that the invasion and migration ability were
remarkably increased. These results further confirmed that
lincRNA-p21 promotes apoptosis and inhibits the proliferation,
invasion and metastasis ability of HCC cells, pointing out a
direction of further study on lincRNA-p21 transcriptional
regulation mechanism and new drugs for liver cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW, JL, SL, ZY and XH were responsible for the
conception and study design. TW, JL, SL, ZY and XH were responsible
for the collection and assembly of data. TW, JL and SL were
responsible for data analysis and interpretation. TW, JL, SL, ZY
and XH were responsible for the writing and final approval of
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Jiangsu Provincial People's Hospital Affiliated to Nanjing Medical
University (Nanjing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun T, Liu H and Ming L: Multiple roles of
autophagy in the sorafenib resistance of hepatocellular carcinoma.
Cell Physiol Biochem. 44:716–727. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pinter M, Weinmann A, Wörns MA, Hucke F,
Bota S, Marquardt JU, Duda DG, Jain RK, Galle PR, Trauner M, et al:
Use of inhibitors of the renin-angiotensin system is associated
with longer survival in patients with hepatocellular carcinoma.
United European Gastroenterol J. 5:987–996. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanduzzi Zamparelli M, Rocco A, Compare D
and Nardone G: The gut microbiota: A new potential driving force in
liver cirrhosis and hepatocellular carcinoma. United European
Gastroenterol J. 5:944–953. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
An C, Hu ZL, Liang P, Cheng ZG, Han ZY, Yu
J and Liu FY: Ultrasound-guided percutaneous microwave ablation vs.
surgical resection for thoracoabdominal wall implants from
hepatocellular carcinoma: Intermediate-term results. Int J
Hyperthermia. 21:1–10. 2017.
|
|
5
|
Chen X, Jiang W, Yue C, Zhang W, Tong C,
Dai D, Cheng B, Huang C and Lu L: Heparanase contributes to
trans-endothelial migration of hepatocellular carcinoma cells. J
Cancer. 8:3309–3317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen S, Liang H, Yang H, Zhou K, Xu L, Liu
J, Lai B, Song L, Luo H, Peng J, et al: LincRNa-p21: Function and
mechanism in cancer. Med Oncol. 34:982017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chillón I and Pyle AM: Inverted repeat Alu
elements in the human lincRNA-p21 adopt a conserved secondary
structure that regulates RNA function. Nucleic Acids Res.
44:9462–9471. 2016.PubMed/NCBI
|
|
8
|
Shen Y, Liu Y, Sun T and Yang W:
LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor
cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway.
Exp Cell Res. 358:188–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen Y, Wei G, Xia H, Yu H, Tang Q and Bi
F: Down regulation of lincRNA-p21 contributes to gastric cancer
development through Hippo-independent activation of YAP.
Oncotarget. 8:63813–63824. 2017.PubMed/NCBI
|
|
10
|
Wang JY, Fang M, Boye A, Wu C, Wu JJ, Ma
Y, Hou S, Kan Y and Yang Y: Interaction of microRNA-21/145 and
Smad3 domain-specific phosphorylation in hepatocellular carcinoma.
Oncotarget. 8:84958–84973. 2017.PubMed/NCBI
|
|
11
|
Feng LH, Wang H, Dong H, Zhu YY and Cong
WM: The stromal morphological changes for differential diagnosis of
uninodular high-grade dysplastic nodule and well-differentiated
small hepatocellular carcinoma. Oncotarget. 8:87329–87339. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ye JZ, Chen JZ, Li ZH, Bai T, Chen J, Zhu
SL, Li LQ and Wu FX: Efficacy of postoperative adjuvant
transcatheter arterial chemoembolization in hepatocellular
carcinoma patients with microvascular invasion. World J
Gastroenterol. 23:7415–7424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sim HW and Knox J: Hepatocellular
carcinoma in the era of immunotherapy. Curr Probl Cancer. 42:40–48.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Honda H, Takamura M, Yamagiwa S, Genda T,
Horigome R, Kimura N, Setsu T, Tominaga K, Kamimura H, Matsuda Y,
et al: Overexpression of a disintegrin and metalloproteinase 21 is
associated with motility, metastasis, and poor prognosis in
hepatocellular carcinoma. Sci Rep. 7:154852017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding G, Peng Z, Shang J, Kang Y, Ning H
and Mao C: LincRNA-p21 inhibits invasion and metastasis of
hepatocellular carcinoma through miR-9/E-cadherin cascade signaling
pathway molecular mechanism. OncoTargets Ther. 10:3241–3247. 2017.
View Article : Google Scholar
|
|
16
|
Jia M, Jiang L, Wang YD, Huang JZ, Yu M
and Xue HZ: lincRNA-p21 inhibits invasion and metastasis of
hepatocellular carcinoma through Notch signaling-induced
epithelial-mesenchymal transition. Hepatol Res. 46:1137–1144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu F, Guo Y, Chen B, Shi L, Dong P, Zhou M
and Zheng J: LincRNA-p21 Inhibits the Wnt/β-catenin pathway in
activated hepatic stellate cells via sponging microRNA-17-5p. Cell
Physiol Biochem. 41:1970–1980. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Castellano JJ, Navarro A, Viñolas N,
Marrades RM, Moises J, Cordeiro A, Saco A, Muñoz C, Fuster D,
Molins L, et al: LincRNA-p21 impacts prognosis in resected
non-small cell lung cancer patients through angiogenesis
regulation. J Thorac Oncol. 11:2173–2182. 2016. View Article : Google Scholar : PubMed/NCBI
|