Introduction
Head and neck squamous cell carcinoma (HNSCC),
arising in the oral cavity, oropharynx, hypopharynx and larynx, is
the sixth most common type of cancer worldwide, with ~635,000 new
cases diagnosed annually and >12% of these cases occurring in
China (1). Advances in early
diagnosis and surgical techniques combined with radiotherapy and
chemotherapy have improved the survival rate of patients with HNSCC
in the last 20 years, and the overall 5-year relative survival rate
has increased from 54.7% (1992–1996) to 65.9% (2002–2006) (2). However, even among patients with HNSCC
of the same classification, the prognosis may vary (3). Therefore, there is a requirement to
identify novel molecular biomarkers of aggressive tumor
behavior.
The human genome encodes ~20,000 protein-coding
genes, accounting for <2% of the human genome, as the majority
of the human genome is actively transcribed into non-coding RNAs
(ncRNAs) (4). These ncRNAs are
divided into two categories based on their sequence length: Short
ncRNAs, including microRNAs (miRNAs), and long ncRNAs (lncRNAs).
lncRNAs are often defined as transcripts >200 nucleotides in
length that lack protein-coding capacity (5). lncRNAs function in regulation of gene
expression and cellular activity through diverse mechanisms
(6). Previous studies have suggested
that lncRNA expression is frequently dysregulated in cancer and
that aberrant expression is associated with cancer diagnosis and
prognosis, suggesting that lncRNAs may be promising molecular
biomarkers (7–10). Certain lncRNAs have been implicated
in HNSCC, including H19 imprinted maternally expressed transcript
(11), HOX transcript antisense RNA
(12) and cytoskeleton regulator RNA
(13). However, the understanding of
lncRNA expression patterns and their prognostic roles in HNSCC
remains limited.
The aim of the current study was to determine the
prognostic value of lncRNA expression profiles and to identify
novel lncRNA biomarkers closely associated with the OS of patients
with HNSCC using a large cohort of >400 patients with HNSCC.
Materials and methods
HNSCC dataset
The clinical features of the patients with HNSCC
used in the present study were obtained from The Cancer Genome
Atlas (TCGA; tcga-data.nci.nih.gov/). The lncRNA expression data
were downloaded from the The Atlas of Noncoding RNAs in Cancer
(bioinformatics.mdanderson.org/main/TANRIC:Overview),
in which lncRNA expression was quantified and normalized using
reads per kilobase per million mapped values (14). To investigate the association between
lncRNA expression and OS of patients with HNSCC, only patients with
available survival data and lncRNA expression profiles were
selected. Thus, 425 patients were selected and randomly divided
into a training cohort (n=213) and testing cohort (n=212) for
identifying and validating survival-associated lncRNA
biomarkers.
Identification of survival-associated
lncRNA biomarkers
Univariate Cox regression analysis was used to
assess the association between lncRNA expression and OS time in the
training cohort. lncRNAs achieving significance of P<0.01 were
considered as candidate survival-associated lncRNAs. These
candidate survival-associated lncRNAs were then analyzed using
multivariate Cox regression analysis, and those achieving P<0.01
in this analysis were identified as independent survival-associated
lncRNAs. All survival-associated lncRNAs were combined to construct
a lncRNA expression signature using a risk scoring method. A lncRNA
expression signature was constructed using a risk-scoring method as
previously described (7,8,15–17): The
lncRNA expression signature was established by including the
expression values of each selected lncRNA, weighted by their
estimated regression coefficients from the multivariate Cox
regression analysis. A risk score was calculated for each patient
using the lncRNA expression signature. Patients were further
divided into low-risk and high-risk groups using the median score
of all patients of the training cohort as the cut-off point.
Statistical analysis
The Kaplan-Meier method and a log-rank test were
used to compare the difference in OS time between the high- and
low-risk groups. Univariate and multivariate Cox regression
analyses for OS time were performed for individual clinical
features, with and without consideration of the lncRNA expression
signature in each cohort. Hazard ratios (HR) and 95% confidence
intervals (CI) were calculated. Time-dependent receiver operating
characteristic (ROC) curve analysis for the 3-year OS time was
performed to assess the prognostic value of the lncRNA expression
signature using the timeROC package (version 0.3) in R (18). The survival analysis, univariate and
multivariate Cox regression analyses was performed using the
survival package in R (https://github.com/therneau/survival) (19). The correlations between
protein-coding genes and the lncRNA biomarkers were identified by
Pearson's correlation coefficient using the entire TCGA data cohort
of 425 patients with HNSCC. All statistical analyses were performed
using R/Bioconductor (version 3.0.2; bioconductor.org/).
Function enrichment analysis
To investigate the potential biological role of the
identified lncRNA biomarkers, Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.genome.jp/kegg/) functional enrichment
analysis of protein-coding genes associated with the identified
lncRNA biomarkers was performed using The Database for Annotation,
Visualization and Integrated Discovery (DAVID) (v.6.8; david.ncifcrf.gov/).
Results
Identification of prognostic lncRNA
biomarkers in HNSCC
To identify lncRNAs associated with the OS time of
patients with HNSCC, univariate Cox regression analysis was used to
assess the association between lncRNA expression and OS time in the
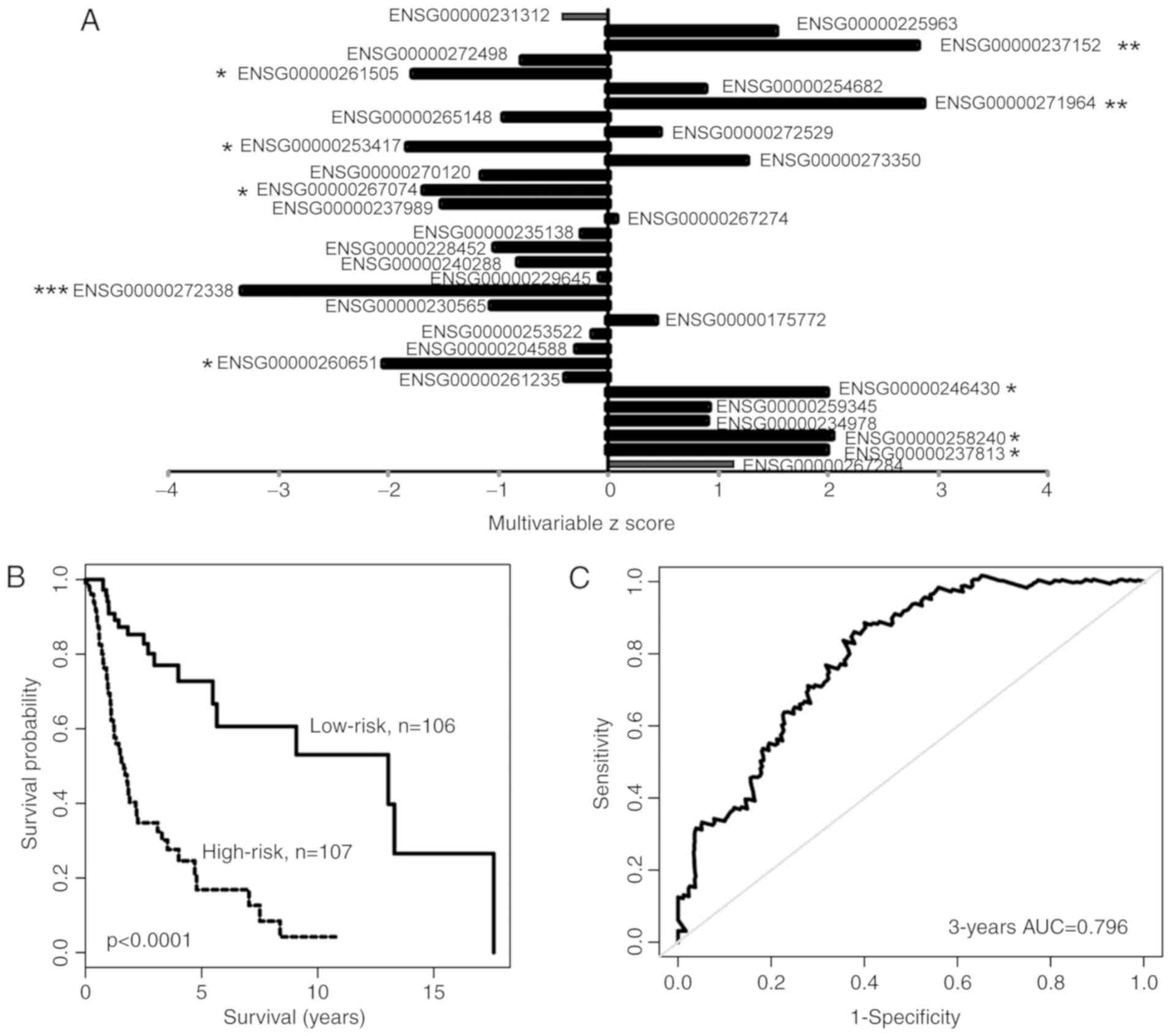
training cohort. A total of 32 lncRNAs were demonstrated to be
significantly associated with OS time for patients with HNSCC
(P<0.01) and were considered as candidate survival-associated
lncRNAs (Fig. 1A). By performing
multivariable Cox regression analysis on the 32 candidate
survival-associated lncRNAs, 10 candidate survival-associated
lncRNAs (P<0.01; Table I) were
identified as potential independent prognostic lncRNA biomarkers. A
total of four prognostic lncRNAs were identified as risk factors
and their overexpression was associated with a shorter OS time
(Table I). The remaining six
prognostic lncRNAs were protective factors and their overexpression
was associated with longer OS time (Table I).
 | Table I.The 10 independent prognostic long
non-coding RNA biomarkers in patients with head and neck squamous
cell carcinoma. |
Table I.
The 10 independent prognostic long
non-coding RNA biomarkers in patients with head and neck squamous
cell carcinoma.
| Ensembl ID | Gene symbol | Chromosomal
location | P-value | Hazard ratio | Cox multivariate
coefficient |
|---|
| ENSG00000237813 | AC002066.1 | Chromosome 7:
116,238,260–116,499,465 reverse strand | 0.0003 | 3.968 | 1.378 |
| ENSG00000258240 | AC002351.1 | Chromosome 12:
110,951,683–110,957,812 reverse strand | 0.0003 | 1.973 | 0.680 |
| ENSG00000246430 | LINC00968 | Chromosome 8:
56,496,246–56,559,823 reverse strand | 0.0017 | 10.549 | 2.356 |
| ENSG00000260651 | AF213884.3 | Chromosome 4:
102,500,841–102,501,319 reverse strand | 0.0020 | 0.00004 | 7.776 |
| ENSG00000272338 | AC067838.1 | Chromosome 8:
33,360,839–33,361,415 reverse strand | 0.0032 | 0.320 | 1.139 |
| ENSG00000267074 | AC015911.3 | Chromosome 17:
35,499,690–35,510,270 reverse strand | 0.0059 | 0.101 | 2.297 |
| ENSG00000253417 | LINC02159 | Chromosome 5:
160,931,778–160,938,626 reverse strand | 0.0067 | 0.469 | 0.758 |
| ENSG00000271964 | AC090948.1 | Chromosome 3:
16,314,439–16,314,987 forward strand | 0.0076 | 0.039 | 3.240 |
| ENSG00000261505 | AL031714.1 | Chromosome 16:
1,317,891–1,322,845 reverse strand | 0.0080 | 0.013 | 4.338 |
| ENSG00000237152 | DLEU7-AS1 | Chromosome 13:
50,807,856–50,849,905 forward strand | 0.0094 | 541.424 | 6.294 |
Construction and evaluation of the
lncRNA expression signature in predicting survival in the training
cohort
To construct an lncRNA-based prognostic model for
predicting the OS time of patients with HNSCC in the training
cohort, the ten lncRNA biomarkers were fitted into a multivariable
Cox regression model. A lncRNA expression signature was constructed
using a risk-scoring method as previously described (6,7,15–17):
lncRNA-based risk score=[(1.2680 × expression value of AC002066.1)
+ (0.7834 × expression value of AC002351.1) + (3.8306 × expression
value of LINC00968) + (7.3969 × expression value of AF213884.3) +
(1.7179 × expression value of AC067838.1) + (2.7346 × expression
value of AC015911.3) + (0.7579 × expression value of LINC02159) +
(3.5427 × expression value of AC090948.1) + (3.5687 × expression
value of AL031714.1) + (11.2941 × expression value of
DLEU7-AS1)].
A patient with HNSCC was classified as low-risk
(n=106) if their risk score was lower than the median risk score of
the training cohort (−1.0376) and as high-risk (n=107) if it was
higher. Patients with high-risk scores exhibited poorer OS times
compared with patients with low-risk scores (median OS time, 1.65
vs. 13.04 years; P<0.0001; Fig.
1B). In univariate analysis, the hazard ratios of the high-risk
group vs. the low-risk group for OS time were 5.142 (95% CI,
2.924–9.043; P<0.0001; Table
II). The 3- and 5-year survival rates of the high-risk group
were 34.8 and 16.8%, respectively, whereas those of the low-risk
group were 77 and 72.7%, respectively. The area under the curve
(AUC) of the time-dependent ROC curve for the lncRNA expression
signature was 0.796 for 3-year OS time (P<0.01; Fig. 1C).
 | Table II.Univariate and multivariate Cox
regression analysis of overall survival in each patient cohort. |
Table II.
Univariate and multivariate Cox
regression analysis of overall survival in each patient cohort.
| A, Training cohort
(n=213) |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
| 10-lncRNA risk
score |
|
|
|
|
|
|
|
High/low | 5.1420 | 2.924–9.043 | <0.0001 | 16.0300 | 3.609–71.154 | 0.0003 |
| Age, years |
|
|
|
|
|
|
|
≥60/<60 | 1.2850 | 0.803–2.058 | 0.2960 | 1.9340 | 0.718–5.211 | 0.1921 |
| Sex |
|
|
|
|
|
|
|
Male/Female | 0.9770 | 0.588–1.622 | 0.9280 | 1.2660 | 0.39–4.113 | 0.6945 |
| Anatomic neoplasm
subdivision |
|
|
|
|
|
|
|
Larynx/Others | 1.7810 | 0.934–3.396 | 0.0798 | 1.8800 | 0.429–8.236 | 0.4021 |
| Oral
cavity/Others | 1.0140 | 0.488–2.108 | 0.9697 | 0.0801 | 0.007–0.919 | 0.0426 |
| Oral
tongue/Others | 1.2080 | 0.627–2.33 | 0.5721 | 0.9678 | 0.215–4.353 | 0.9660 |
| History of other
malignancy |
|
|
|
|
|
|
|
Yes/No | 0.3650 | 0.051–2.638 | 0.3180 | <0.0001 | 0-∞ | 0.9982 |
| Lymphovascular
invasion present |
|
|
|
|
|
|
|
Yes/No | 1.3080 | 0.675–2.533 | 0.4260 | 0.4456 | 0.132–1.506 | 0.1932 |
| Perineural invasion
present |
|
|
|
|
|
|
|
Yes/No | 1.4150 | 0.754–2.657 | 0.2800 | 2.1350 | 0.543–8.401 | 0.2776 |
| pN |
|
|
|
|
|
|
|
Positive/Negative | 0.7470 | 0.379–1.472 | 0.4000 | 0.0222 | 0.002–0.296 | 0.0040 |
| ECS |
|
|
|
|
|
|
|
Positive/Negative | 2.2550 | 1.236–4.114 | 0.0081 | 2.2020 | 0.593–8.174 | 0.2384 |
| Clinical stage |
|
|
|
|
|
|
|
III,IV/I,II | 1.2750 | 0.761–2.136 | 0.3567 | 2.6400 | 0.459–15.195 | 0.2769 |
| Pathological
stage |
|
|
|
|
|
|
|
III,IV/I,II | 1.7000 | 0.937–3.085 | 0.0810 | 0.7086 | 0.129–3.891 | 0.6918 |
| Alcohol
history |
|
|
|
|
|
|
|
Yes/No | 0.7990 | 0.496–1.289 | 0.3581 | 4.1760 | 0.955–18.265 | 0.0576 |
| Margin status |
|
|
|
|
|
|
|
Positive/Negative | 1.2310 | 0.64–2.365 | 0.5332 | 8.2830 | 0.954–71.921 | 0.0552 |
| Tumor grade |
|
|
|
|
|
|
|
G3,4/G1,2 | 0.7850 | 0.453–1.359 | 0.3867 | 3.6370 | 1.178–11.228 | 0.0247 |
|
| B, Test cohort
(n=212) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
| Variable | HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
|
| 10-lncRNA risk
score |
|
|
|
|
|
|
|
High/low | 1.9070 | 1.179–3.085 | 0.0085 | 4.3370 | 1.245–15.108 | 0.0212 |
| Age, years |
|
|
|
|
|
|
|
≥60/<60 | 1.2590 | 0.774–2.049 | 0.3529 | 6.0650 | 1.297–28.365 | 0.0220 |
| Sex |
|
|
|
|
|
|
|
Male/Female | 0.7420 | 0.45–1.224 | 0.2424 | 2.0370 | 0.442–9.386 | 0.3614 |
| Anatomic neoplasm
subdivision |
|
|
|
|
|
|
|
Larynx/Others | 0.6913 | 0.344–1.388 | 0.2990 | <0.0001 | 0-∞ | 0.9971 |
| Oral
cavity/Others | 1.3777 | 0.742–2.56 | 0.3110 | 1.3930 | 0.277–7.009 | 0.6873 |
| Oral
tongue/Others | 1.2292 | 0.631–2.395 | 0.5440 | 0.3850 | 0.095–1.552 | 0.1796 |
| History of other
malignancy |
|
|
|
|
|
|
|
Yes/No | 1.0080 | 0.404–2.516 | 0.9861 | 0.6159 | 0.061–6.203 | 0.6809 |
| Lymphovascular
invasion present |
|
|
|
|
|
|
|
Yes/No | 1.4140 | 0.746–2.683 | 0.2885 | 3.6560 | 0.829–16.13 | 0.0869 |
| Perineural invasion
present |
|
|
|
|
|
|
|
Yes/No | 1.8060 | 0.991–3.291 | 0.0535 | 4.0110 | 1.049–15.345 | 0.0424 |
| pN |
|
|
|
|
|
|
|
Positive/Negative | 0.5530 | 0.28–1.094 | 0.0889 | 0.0018 | 0.0002–0.1168 | 0.0030 |
| ECS |
|
|
|
|
|
|
|
Positive/Negative | 1.9730 | 1.105–3.523 | 0.0216 | 0.7748 | 0.196–3.071 | 0.7165 |
| Clinical stage |
|
|
|
|
|
|
|
III,IV/I,II | 0.9490 | 0.54–1.667 | 0.8554 | 16.1200 | 1.204–215.749 | 0.0357 |
| Pathologic
stage |
|
|
|
|
|
|
|
III,IV/I,II | 1.2590 | 0.667–2.376 | 0.4771 | 1.7120 | 0.136–21.539 | 0.6772 |
| Alcohol
history |
|
|
|
|
|
|
|
Yes/No | 0.9900 | 0.601–1.631 | 0.9696 | 3.4710 | 0.684–17.617 | 0.1332 |
| Margin status |
|
|
|
|
|
|
|
Positive/Negative | 1.8580 | 0.954–3.617 | 0.0685 | 0.7339 | 0.095–5.688 | 0.7671 |
| Tumor grade |
|
|
|
|
|
|
|
G3,4/G1,2 | 0.8440 | 0.504–1.412 | 0.5178 | 2.6290 | 0.589–11.728 | 0.2052 |
|
| C, Entire The
Cancer Genome Atlas cohort (n=425) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variable | HR | 95% CI of
HR | P-value | HR | 95% CI of
HR | P-value |
|
| 10-lncRNA risk
score |
|
|
|
|
|
|
|
High/low | 3.0140 | 2.111–4.304 | <0.0001 | 4.5375 | 2.169–9.491 | 0.0001 |
| Age, years |
|
|
|
|
|
|
|
≥60/<60 | 1.2860 | 0.918–1.802 | 0.1440 | 1.8743 | 0.947–3.709 | 0.0712 |
| Sex |
|
|
|
|
|
|
|
Male/Female | 0.8370 | 0.588–1.192 | 0.3250 | 1.1553 | 0.519–2.573 | 0.7239 |
| Anatomic neoplasm
subdivision |
|
|
|
|
|
|
|
Larynx/Others | 1.1600 | 0.733–1.836 | 0.5270 | 0.3415 | 0.105–1.112 | 0.0745 |
| Oral
cavity/Others | 1.2170 | 0.762–1.944 | 0.4120 | 0.5533 | 0.178–1.719 | 0.3062 |
| Oral
tongue/Others | 1.1780 | 0.745–1.864 | 0.4840 | 0.9037 | 0.418–1.956 | 0.7972 |
| History of other
malignancy |
|
|
|
|
|
|
|
Yes/No | 0.7650 | 0.337–1.737 | 0.5216 | 0.5668 | 0.069–4.659 | 0.5973 |
| Lymphovascular
invasion present |
|
|
|
|
|
|
|
Yes/No | 1.3320 | 0.844–2.1 | 0.2183 | 1.0942 | 0.512–2.339 | 0.8165 |
| Perineural invasion
present |
|
|
|
|
|
|
|
Yes/No | 1.6360 | 1.06–2.526 | 0.0263 | 1.8780 | 0.894–3.947 | 0.0963 |
| pN |
|
|
|
|
|
|
|
Positive/Negative | 0.6400 | 0.396–1.034 | 0.0680 | 0.0264 | 0.003–0.2 | 0.0004 |
| ECS |
|
|
|
|
|
|
|
Positive/Negative | 2.0310 | 1.342–3.074 | 0.0008 | 1.4968 | 0.692–3.237 | 0.3055 |
| Clinical stage |
|
|
|
|
|
|
|
III,IV/I,II | 1.1240 | 0.771–1.638 | 0.5433 | 2.7613 | 0.729–10.464 | 0.1351 |
| Pathological
stage |
|
|
|
|
|
|
|
III,IV/I,II | 1.4940 | 0.969–2.305 | 0.0694 | 1.0189 | 0.278–3.732 | 0.9775 |
| Alcohol
history |
|
|
|
|
|
|
|
Yes/No | 0.8930 | 0.633–1.258 | 0.5160 | 1.9261 | 0.854–4.342 | 0.1140 |
| Margin status |
|
|
|
|
|
|
|
Positive/Negative | 1.5120 | 0.95–2.406 | 0.0809 | 1.9081 | 0.59–6.173 | 0.2808 |
| Tumor grade |
|
|
|
|
|
|
|
G3,4/G1,2 | 0.8280 | 0.571–1.199 | 0.3174 | 1.0951 | 0.511–2.346 | 0.8153 |
Independent validation of the lncRNA
expression signature in the testing and entire TCGA cohorts
To test the predictive performance of the lncRNA
expression signature, the lncRNA expression signature was validated
in the test cohort. By using the aforementioned risk score model,
212 patients from the testing cohort were classified into high-risk
(n=85) and low-risk (n=127) groups using the risk score cut-off
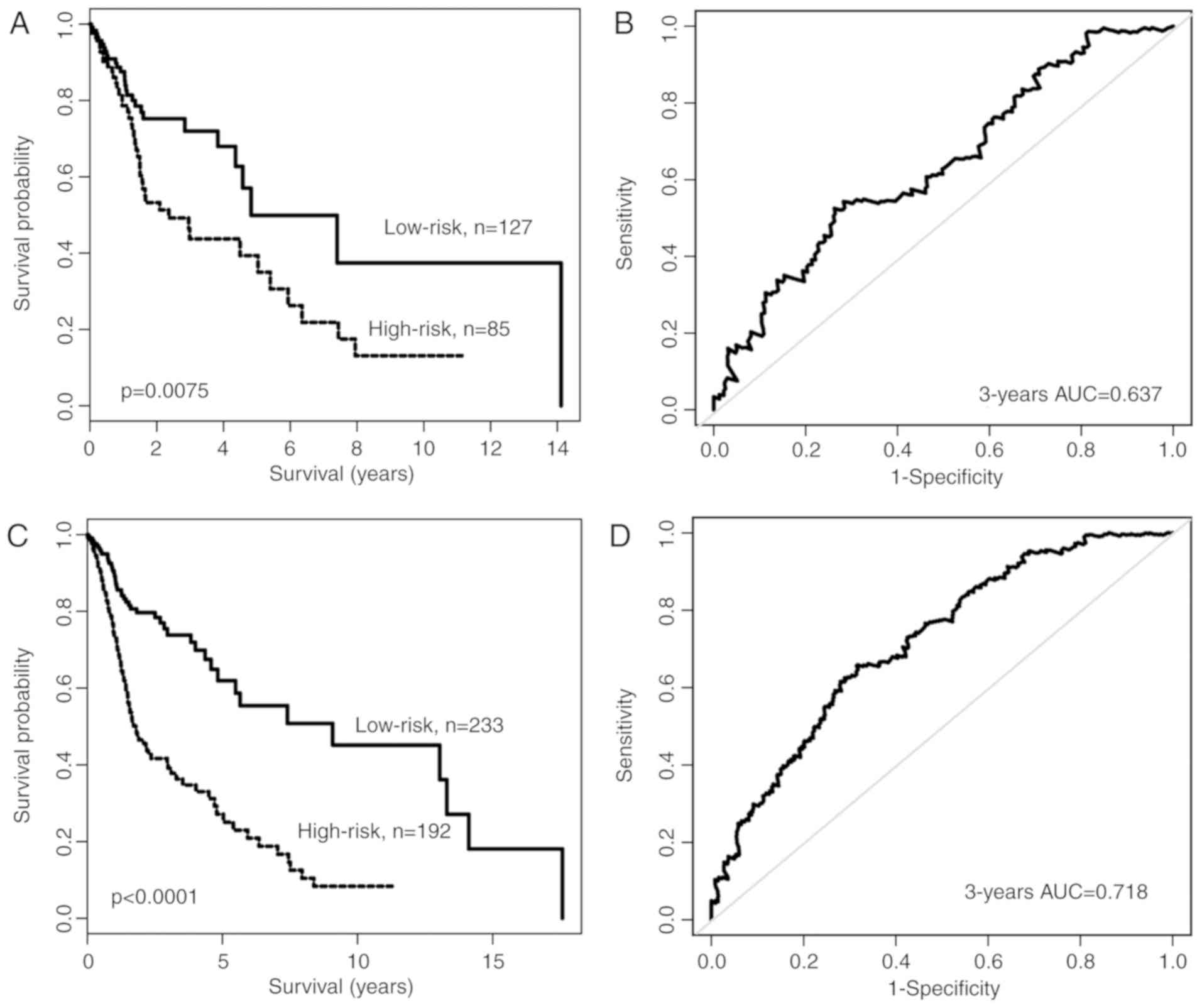
derived from training cohort (−1.0376). As observed in the training
cohort, the OS time of patients in the high-risk group was
significantly shorter compared with that of patients in the
low-risk group (median OS time, 2.36 vs. 4.83 years, respectively;
P=0.0075; Fig. 2A). In univariate
analysis, the HR of the high-risk vs. low-risk group for OS time
was 1.907 (95% CI, 1.179–3.085; P=0.0085; Table II). The 3- and 5-year survival rates
of the high-risk group were 43.7 and 39.3%, respectively, whereas
those of the low-risk group were 71.9 and 49.9%, respectively. The
AUC of the time-dependent ROC curve for the lncRNA expression
signature was 0.637 for 3-year OS time (P<0.01; Fig. 2B).
The prognostic value of the lncRNA signature was
subsequently analyzed in the entire TCGA cohort of 425 patients.
Using the aforementioned risk score model and cut-off value of the
training cohort, patients were segregated into high-risk (n=179)
and low-risk (n=233) groups with significantly different OS times
(median OS time, 1.79 vs. 9.08 years; P<0.0001; Fig. 2C). In univariate analysis, the HR of
the high-risk vs. low-risk group for OS time was 3.014 (95% CI,
2.111–4.304; P<0.0001l Table
II). The 3- and 5-year survival rates of the high-risk group
were 39.1 and 27.1%, respectively, whereas those of the low-risk
group were 73.8 and 61.9%, respectively. The AUC of time-dependent
ROC curve for the lncRNA expression signature was 0.718 for 3-year
OS time (P<0.01; Fig. 2D).
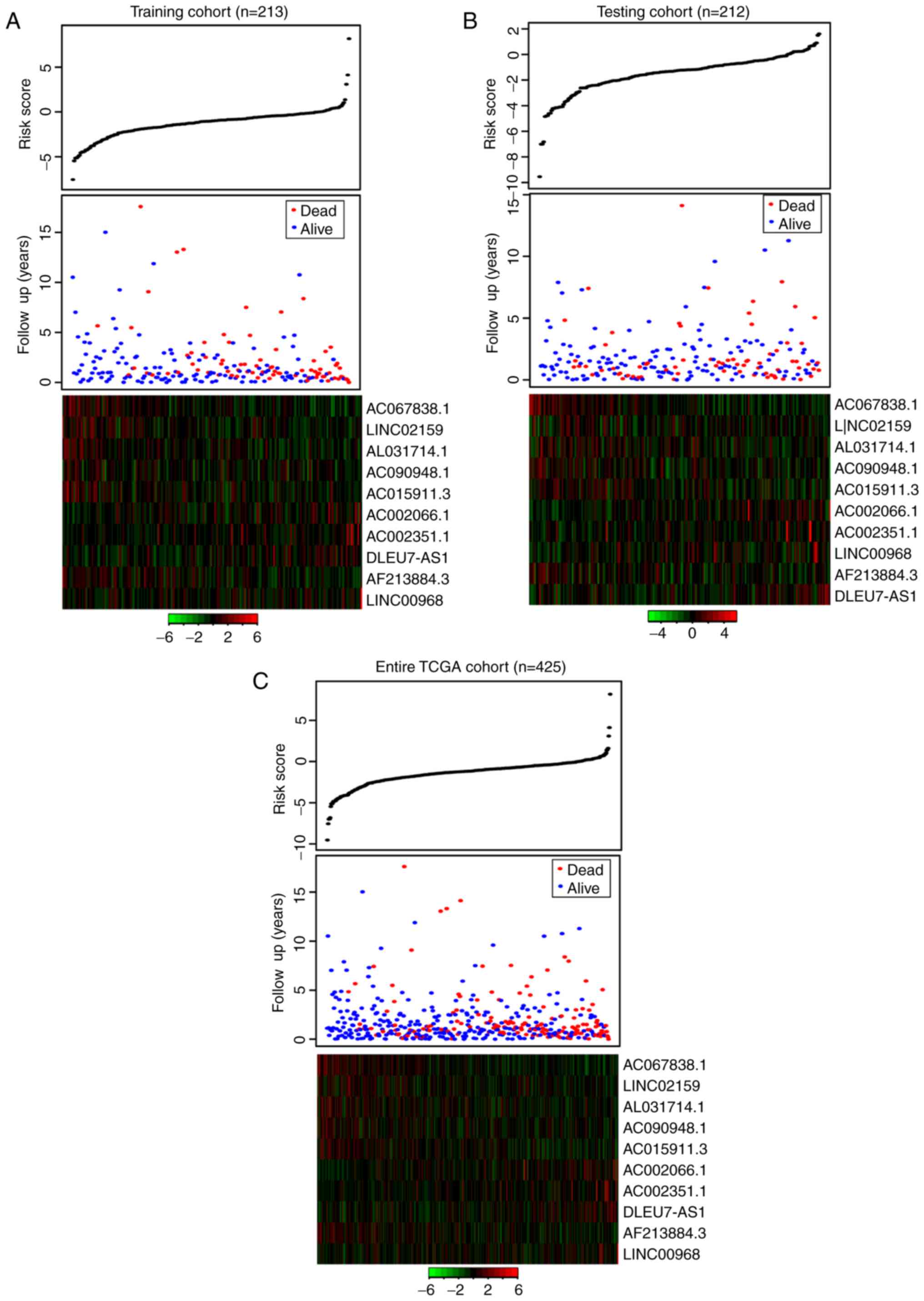
The distribution of risk score, the survival status
of the patients with HNSCC and the expression pattern of the lncRNA
biomarkers in the training cohort, testing cohort and entire TCGA
cohort are presented in Fig. 3.
Patients in the high-risk group exhibited higher expression of the
four lncRNAs associated with poor prognosis compared with patients
in the low-risk group, whereas patients in the low-risk group
expressed higher levels of the six protective prognostic lncRNAs
compared with the high-risk group.
lncRNA expression signature is
independent of clinical features
To assess whether the survival-prediction ability of
the lncRNA expression signature was independent of clinical
features, multivariate Cox regression analysis was performed using
the following variables: Risk score, age, sex, anatomic neoplasm
subdivision, history of other malignancies, lymphovascular
invasion, perineural invasion, pathological lymph node status (pN),
extracapsular spread, clinical stage, pathological stage,
alcohol-consumption history, margin status and tumor grade. The
results demonstrated that the lncRNA expression signature was
significantly associated with OS time in the training cohort (HR,
16.03; 95% CI, 3.609–71.154; P=0.0003), testing cohort (HR, 4.337;
95% CI, 1.245–15.108; P=0.0212) and entire TCGA cohort (HR, 4.5375;
95% CI, 2.169–9.491; P=0.0001; Table
II).
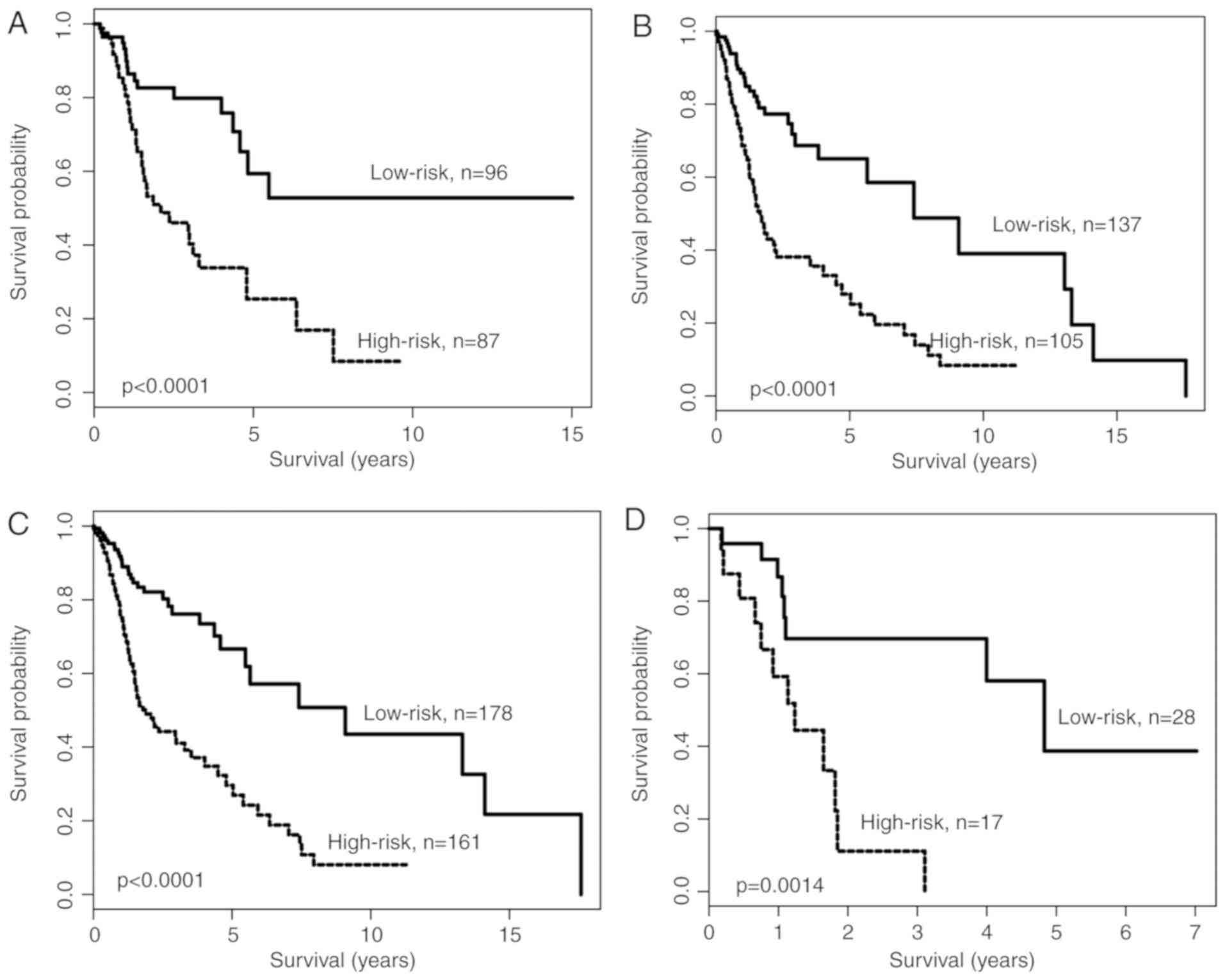
Data stratification analysis was performed for age
and pN status, as these two variables were significant in the
multivariate analysis. The patients were divided into two cohorts:
Younger (<60 years; n=183) and older (≥60 years; n=242). Using
the lncRNA expression signature, patients in the younger cohort
were further divided into high-risk and low-risk groups (Fig. 4A). Similar results were observed when
the lncRNA expression signature was applied to the older cohort, in
which patients were further divided into high-risk and low-risk
groups (Fig. 4B). All patients were
subsequently divided into two patient cohorts according to pN
status: pN-positive cohort (n=339) and pN-negative cohort (n=45).
The lncRNA expression signature was applied to classify patients of
the pN-positive cohort and the pN-negative cohort into high-risk
and low-risk groups. Kaplan-Meier survival analysis indicated that
the OS time of patients in the high-risk group was significantly
shorter compared with that of patients in the low-risk group,
despite having the same pN status (P<0.0001 for pN-positive
cohort and P=0.0014 for pN-negative cohort). These results
indicated that the predictive ability of the lncRNA expression
signature was independent of commonly used clinical features for
predicting the survival of patients with HNSCC.
Functional characteristics of the
identified lncRNA biomarkers
To investigate the potential biological role of the
identified lncRNA biomarkers, GO and KEGG functional enrichment
analysis of protein-coding genes associated with the identified
lncRNA biomarkers was performed. The correlations between
protein-coding genes and the lncRNA biomarkers were identified by
Pearson's correlation coefficient using the entire TCGA data cohort
of 425 patients with HNSCC. The protein-coding genes ranked in the
top 1% were used for GO and KEGG functional enrichment analysis.
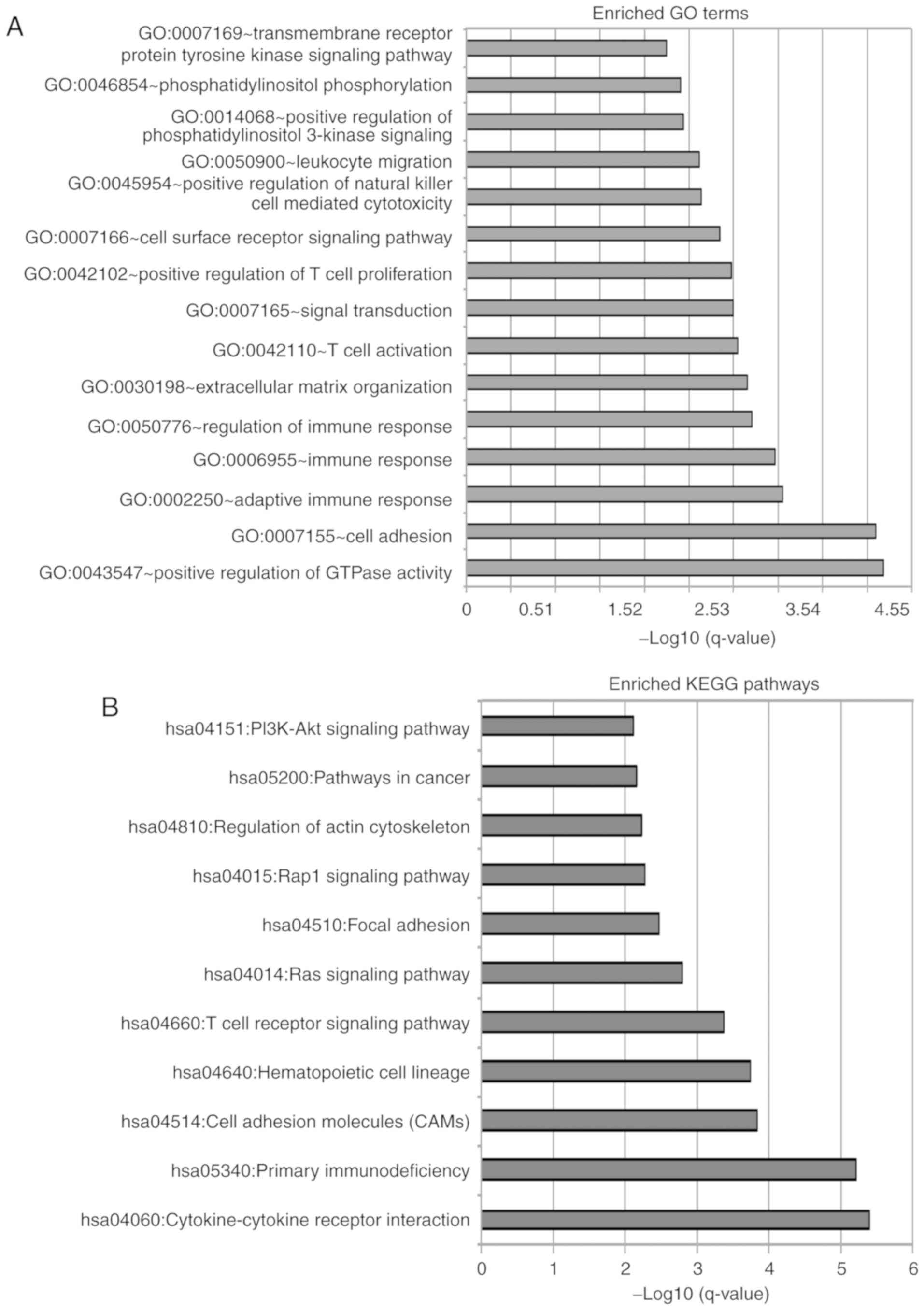
The results of GO analysis suggested that the protein-coding genes
were enriched in 14 GO terms, which could be categorized into two
functional clusters: ‘Cell-adhesion’ and ‘immune response’
(Fig. 5A). The results of GO
analysis suggested that protein-coding genes correlated with the
identified lncRNA biomarkers were enriched in 11 KEGG biological
pathways, including ‘Cytokine-cytokine receptor interaction’,
‘Primary immunodeficiency’, ‘Cell adhesion molecules’,
‘Hematopoietic cell lineage’, ‘T cell receptor signaling pathway’,
‘Ras signaling pathway’, ‘Focal adhesion’, ‘Rap1 signaling
pathway’, ‘Regulation of actin cytoskeleton’, ‘Pathways in cancer’,
and ‘PI3K-Akt signaling pathway’ (Fig.
5B).
Discussion
HNSCC is a heterogeneous disease characterized by
distinct clinical and molecular features (20). Traditional staging diagnosis,
treatment options and prognostic prediction of HNSCC do not allow
for precision medicine, due to the diverse molecular features
between patients with identical American Joint Committee on Cancer
TNM staging (21). Molecular
profiles of patients with HNSCC have been investigated in previous
reports, which demonstrated the potential of molecular profiles as
novel biomarkers to predict treatment outcome and to guide
treatment strategies (21–24). However, these studies are restricted
to protein-coding gene data and miRNA data. An improved
understanding of the role of lncRNAs in HNSCC may result in lncRNA
expression emerging as a promising molecular biomarker for
predicting the prognosis of patients with HNSCC (25,26).
The current study investigated the prognostic values
of lncRNA expression profiles in predicting the OS time of patients
with HNSCC by integrating clinical and profiling data from TCGA. A
total of ten novel lncRNAs were identified as potential prognostic
markers for patients with HNSCC. These were used to develop a
prognostic signature using a risk scoring method, which classified
the patients into 2 groups with significantly different OS times.
The lncRNA signature identified in the training cohort demonstrated
a similar prognostic value in the testing and the entire TCGA
cohorts. Multivariable Cox regression analysis indicated that the
signature was an independent prognostic factor for patients with
HSNCC. Thus, the prognostic value of the lncRNA signature may have
clinical potential for patients with HNSCC.
Furthermore, functional enrichment analysis was
performed to investigate the biological roles of the lncRNA
signature in HNSCC. Protein-coding genes, whose expression values
were positively associated with the lncRNA signature, were enriched
in 14 GO biological terms and 11 KEGG biological pathways. These
enriched GO biological processes and KEGG pathways were categorized
into ‘cell-adhesion’ and ‘immune response’. Thus, the ten lncRNAs
associated with the survival of patients with HNSCC may be involved
in cell-adhesion and the immune response. A number of studies have
indicated that dysfunction of cell-adhesion and cell-migration
serves an important role in the processes of invasion and
metastasis in HNSCC (27,28). HNSCC is an immunosuppressive disease
characterized by dysregulation of immunocompetent cells and
impaired cytokine secretion (29).
The immune system serves an important role in the occurrence and
progression of HNSCC, and the status of the immune system is likely
to be of prognostic value in HNSCC (30). The pathway ‘immune response’ was
significantly associated with the lncRNA signature, suggesting that
the lncRNA expression-based risk scoring system described in the
current study may reflect the basic status of the immune response
in the tumor microenvironment. However, several limitations of the
present study should be noted. Firstly, the ten lncRNA biomarkers
identified in the present study were only validated in TCGA
datasets. Further testing in other independent datasets is
required. Secondly, the functions of the ten lncRNA biomarkers were
only predicted using bioinformatics methods; therefore, these
require further investigation using experimental methods.
In conclusion, the present study identified ten
lncRNAs associated with the OS time of patients with HSNCC from a
large cohort. These ten lncRNA biomarkers were used to develop a
lncRNA signature which robustly predicted the survival of patients
with HSNCC in the training, testing and entire TCGA cohorts.
Further analysis revealed that the prognostic value was independent
of the clinical and pathological characteristics of patients with
HSNCC. While the results presented in the current study require
further validation, the current study indicates that lncRNA
expression profiles may be used as molecular markers to improve the
clinical prognosis for patients with HSNCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Hubei Province
Health and Family Planning Scientific Research Project (grant no.
WJ2017M145).
Availability of data and material
The datasets generated and/or analyzed during the
current study are available in the The Cancer Genome Atlas (TCGA;
tcga-data.nci.nih.gov/) and The Atlas of
Noncoding RNAs in Cancer (bioinformatics.mdanderson.org/main/TANRIC:Overview).
Authors' contributions
YL conceived and designed the experiments. JL, YHL,
XW performed the experiments and analyzed the data. YL wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Feng Z, Xu QS, Qin LZ, Li H, Huang X, Su M
and Han Z: Second primary cancer after index head and neck squamous
cell carcinoma in Northern China. Oral Surg Oral Med Oral Pathol
Oral Radiol. 123:95–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pulte D and Brenner H: Changes in survival
in head and neck cancers in the late 20th and early 21st century: A
period analysis. Oncologist. 15:994–1001. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grégoire V, Lefebvre JL, Licitra L and
Felip E; EHNS-ESMO-ESTRO Guidelines Working Group, : Squamous cell
carcinoma of the head and neck: EHNS-ESMO-ESTRO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 21
(Suppl 5):v184–v186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
ENCODE Project Consortium, Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hangauer MJ, Vaughn IW and McManus MT:
Pervasive transcription of the human genome produces thousands of
previously unidentified long intergenic noncoding RNAs. PLoS Genet.
9:e10035692013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L
and Sun J: An immune-related six-lncRNA signature to improve
prognosis prediction of glioblastoma multiforme. Mol Neurobiol.
55:3684–3697. 2018.PubMed/NCBI
|
|
8
|
Zhou M, Zhao H, Xu W, Bao S, Cheng L and
Sun J: Discovery and validation of immune-associated long
non-coding RNA biomarkers associated with clinically molecular
subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer.
16:162017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Zhang Z, Zhao H, Bao S and Sun J:
A novel lncRNA-focus expression signature for survival prediction
in endometrial carcinoma. BMC Cancer. 18:392018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou M, Zhao H, Wang X, Sun J and Su J:
Analysis of long noncoding RNAs highlights region-specific altered
expression patterns and diagnostic roles in Alzheimer's disease.
Brief Bioinform. Apr 17–2018.Doi: 10.1093/bib/bby021.
|
|
11
|
Guan GF, Zhang DJ, Wen LJ, Xin D, Liu Y,
Yu DJ, Su K, Zhu L, Guo YY and Wang K: Overexpression of lncRNA
H19/miR-675 promotes tumorigenesis in head and neck squamous cell
carcinoma. Int J Med Sci. 13:914–922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu B, Liu J, Wang B, Liao X, Cui Z and
Ding N: Association on polymorphisms in LncRNA HOTAIR and
susceptibility to HNSCC in Chinese population. Eur Rev Med
Pharmacol Sci. 22:702–706. 2018.PubMed/NCBI
|
|
13
|
Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang
Y, Yang L, He Y, Lian Y, Li X, et al: Upregulated long non-coding
RNA LINC00152 expression is associated with progression and poor
prognosis of tongue squamous cell carcinoma. J Cancer. 8:523–530.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Han L, Roebuck P, Diao L, Liu L,
Yuan Y, Weinstein JN and Liang H: TANRIC: An interactive open
platform to explore the function of lncRNAs in cancer. Cancer Res.
75:3728–3737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
17
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blanche P, Dartigues JF and Jacqmin-Gadda
H: Estimating and comparing time-dependent areas under receiver
operating characteristic curves for censored event times with
competing risks. Stat Med. 32:5381–5397. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moreno-Betancur M, Sadaoui H, Piffaretti C
and Rey G: Survival analysis with multiple causes of death:
Extending the competing risks model. Epidemiology. 28:12–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Network: Comprehensive
genomic characterization of head and neck squamous cell carcinomas.
Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chung CH, Parker JS, Karaca G, Wu J,
Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X,
et al: Molecular classification of head and neck squamous cell
carcinomas using patterns of gene expression. Cancer Cell.
5:489–500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feldman R, Gatalica Z, Knezetic J, Reddy
S, Nathan CA, Javadi N and Teknos T: Molecular profiling of head
and neck squamous cell carcinoma. Head Neck. 38 (Suppl
1):E1625–E1638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramdas L, Giri U, Ashorn CL, Coombes KR,
El-Naggar A, Ang KK and Story MD: miRNA expression profiles in head
and neck squamous cell carcinoma and adjacent normal tissue. Head
Neck. 31:642–654. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji
T, Chen WT and Zou X: A three-lncRNA signature derived from the
Atlas of ncRNA in cancer (TANRIC) database predicts the survival of
patients with head and neck squamous cell carcinoma. Oral Oncol.
65:94–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang ZL, Zhao LJ, Chai L, Zhou SH, Wang
F, Wei Y, Xu YP and Zhao P: Seven LncRNA-mRNA based risk score
predicts the survival of head and neck squamous cell carcinoma. Sci
Rep. 7:3092017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Behrens J: The role of cell adhesion
molecules in cancer invasion and metastasis. Breast Cancer Res
Treat. 24:175–184. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Howell GM and Grandis JR: Molecular
mediators of metastasis in head and neck squamous cell carcinoma.
Head Neck. 27:710–717. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Varilla V, Atienza J and Dasanu CA: Immune
alterations and immunotherapy prospects in head and neck cancer.
Expert Opin Biol Ther. 13:1241–1256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Economopoulou P, Perisanidis C, Giotakis
EI and Psyrri A: The emerging role of immunotherapy in head and
neck squamous cell carcinoma (HNSCC): Anti-tumor immunity and
clinical applications. Ann Transl Med. 4:1732016. View Article : Google Scholar : PubMed/NCBI
|