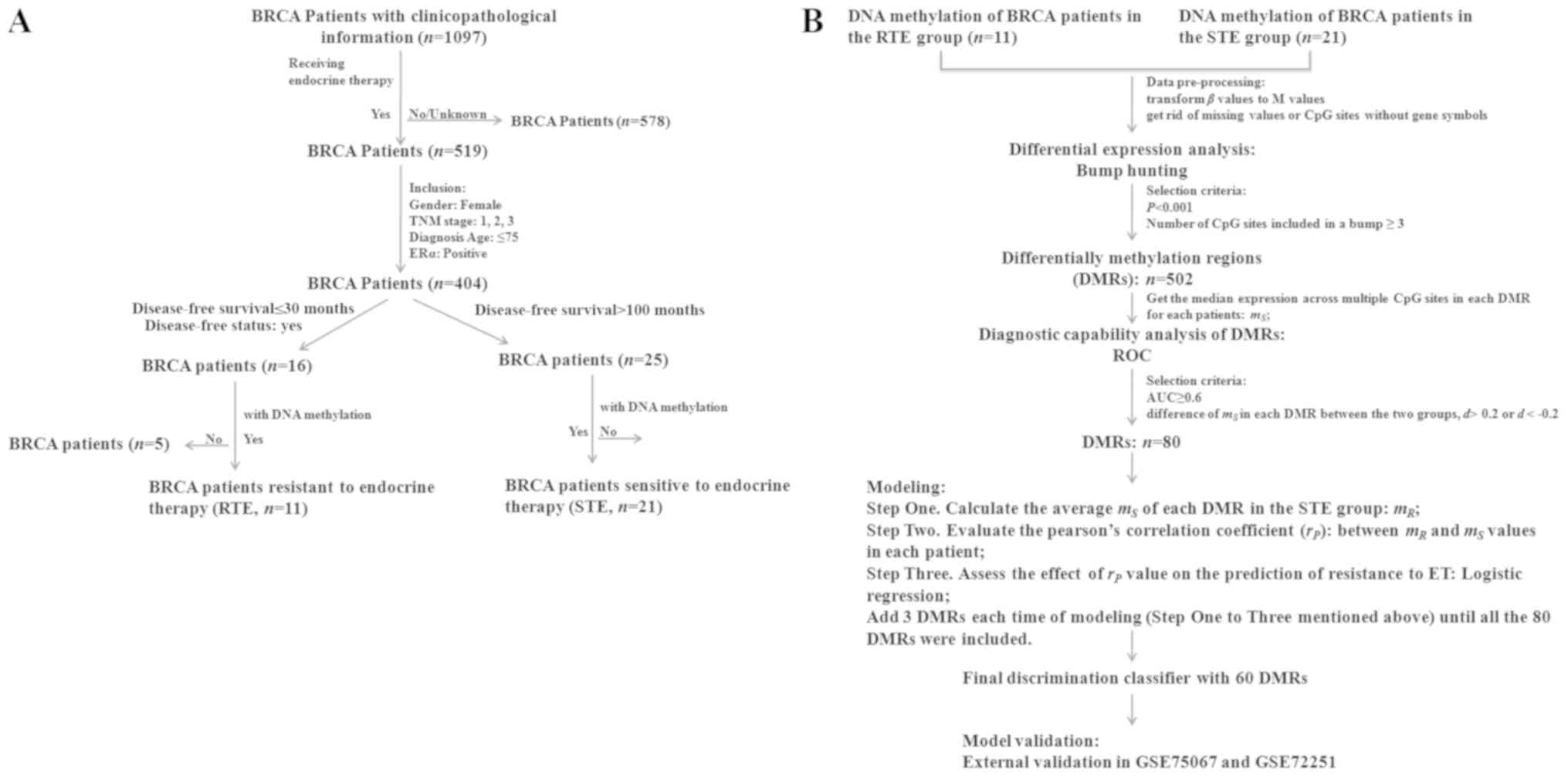
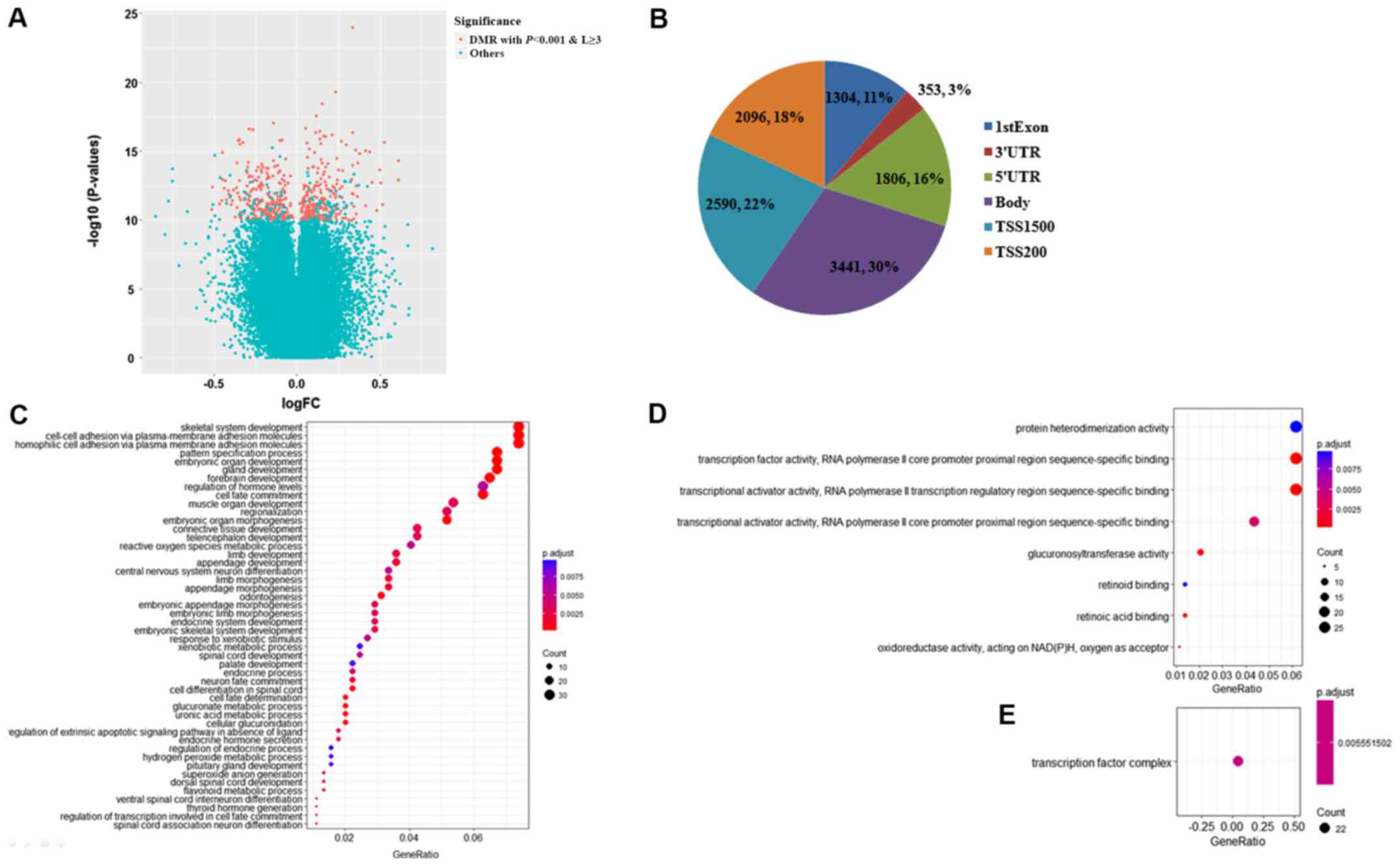
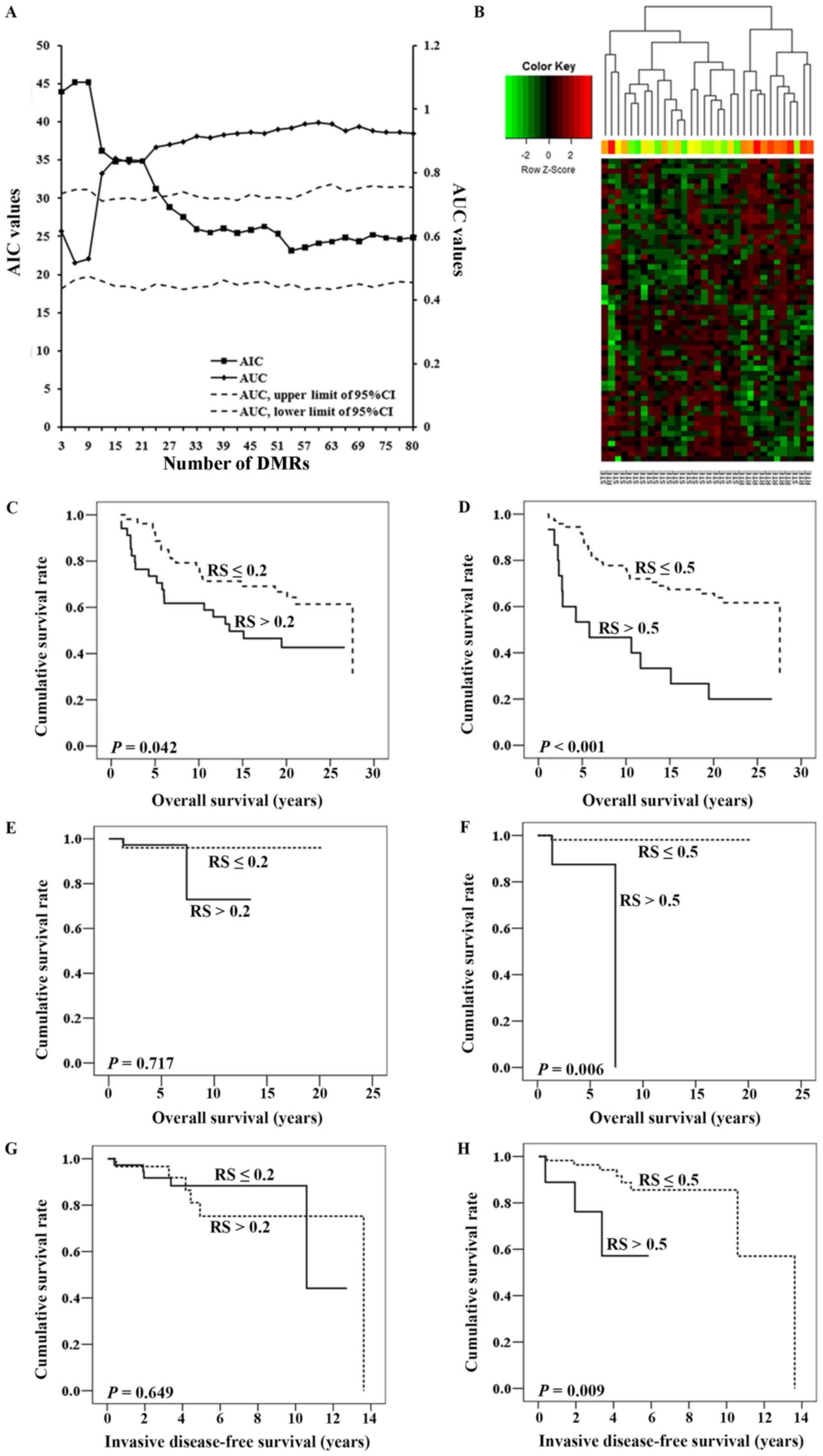
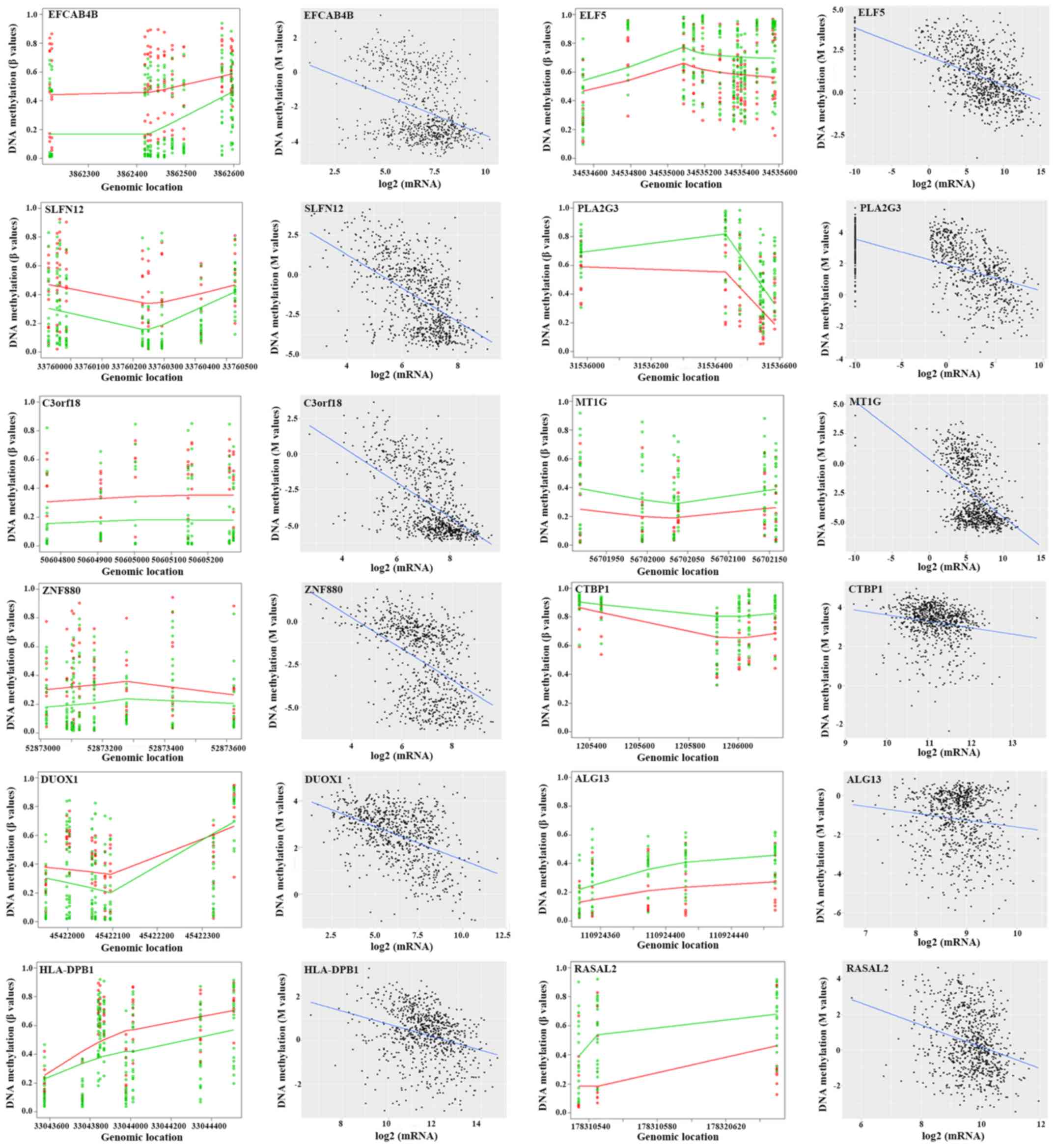
|
1
|
Jiang X, Tang H and Chen T: Epidemiology
of gynecologic cancers in China. J Gynecol Oncol. 29:e72018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Campbell LL and Polyak K: Breast tumor
heterogeneity: Cancer stem cells or clonal evolution? Cell Cycle.
6:2332–2338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nicolini A, Ferrari P and Duffy MJ:
Prognostic and predictive biomarkers in breast cancer: Past,
present and future. Semin Cancer Biol. 52:56–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer (unabridged version). Arch Pathol Lab Med.
134:e48–e72. 2010.PubMed/NCBI
|
|
5
|
Mendes TF, Kluskens LD and Rodrigues LR:
Triple negative breast cancer: Nanosolutions for a big challenge.
Adv Sci (Weinh). 2:15000532015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Badia E, Oliva J, Balaguer P and Cavaillès
V: Tamoxifen resistance and epigenetic modifications in breast
cancer cell lines. Curr Med Chem. 14:3035–3045. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abdel-Hafiz HA: Epigenetic mechanisms of
tamoxifen resistance in luminal breast cancer. Diseases. 5(pii):
E162017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harbeck N and Rody A: Lost in translation?
Estrogen receptor status and endocrine responsiveness in breast
cancer. J Clin Oncol. 30:686–689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gururaj AE, Rayala SK, Vadlamudi RK and
Kumar R: Novel mechanisms of resistance to endocrine therapy:
Genomic and nongenomic considerations. Clin Cancer Res.
12:1001s–1007s. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ali S and Coombes RC: Endocrine-responsive
breast cancer and strategies for combating resistance. Nat Rev
Cancer. 2:101–112. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smith IE, Walsh G, Skene A, Llombart A,
Mayordomo JI, Detre S, Salter J, Clark E, Magill P and Dowsett M: A
phase II placebo-controlled trial of neoadjuvant anastrozole alone
or with gefitinib in early breast cancer. J Clin Oncol.
25:3816–3822. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johnston SR, Martin LA, Leary A, Head J
and Dowsett M: Clinical strategies for rationale combinations of
aromatase inhibitors with novel therapies for breast cancer. J
Steroid Biochem Mol Biol. 106:180–186. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paik S, Shak S, Tang G, Kim C, Baker J,
Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al: A
multigene assay to predict recurrence of tamoxifen-treated,
node-negative breast cancer. N Engl J Med. 351:2817–2826. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cardoso F, van't Veer LJ, Bogaerts J,
Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S,
DeLorenzi M, et al: 70-Gene signature as an aid to treatment
decisions in early-stage breast cancer. N Engl J Med. 375:717–729.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stone A, Valdes-Mora F and Clark SJ:
Exploring and exploiting the aberrant DNA methylation profile of
endocrine-resistant breast cancer. Epigenomics. 5:595–598. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Glasspool RM, Teodoridis JM and Brown R:
Epigenetics as a mechanism driving polygenic clinical drug
resistance. Br J Cancer. 94:1087–1092. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ward AK, Mellor P, Smith SE, Kendall S,
Just NA, Vizeacoumar FS, Sarker S, Phillips Z, Alvi R, Saxena A, et
al: Epigenetic silencing of CREB3L1 by DNA methylation is
associated with high-grade metastatic breast cancers with poor
prognosis and is prevalent in triple negative breast cancers.
Breast Cancer Res. 18:122016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lubecka K, Kurzava L, Flower K, Buvala H,
Zhang H, Teegarden D, Camarillo I, Suderman M, Kuang S, Andrisani
O, et al: Stilbenoids remodel the DNA methylation patterns in
breast cancer cells and inhibit oncogenic NOTCH signaling through
epigenetic regulation of MAML2 transcriptional activity.
Carcinogenesis. 37:656–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Looi ML, Zakaria H, Osman J and Jamal R:
Quantity and quality assessment of DNA extracted from saliva and
blood. Clin Lab. 58:307–312. 2012.PubMed/NCBI
|
|
21
|
Bediaga NG, Acha-Sagredo A, Guerra I,
Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J,
Montaner D, et al: DNA methylation epigenotypes in breast cancer
molecular subtypes. Breast Cancer Res. 12:R772010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng W, Shen L, Wen S, Rosen DG, Jelinek
J, Hu X, Huan S, Huang M, Liu J, Sahin AA, et al: Correlation
between CpG methylation profiles and hormone receptor status in
breast cancers. Breast Cancer Res. 9:R572007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Holm K, Hegardt C, Staaf J,
Vallon-Christersson J, Jonsson G, Olsson H, Borg A and Ringnér M:
Molecular subtypes of breast cancer are associated with
characteristic DNA methylation patterns. Breast Cancer Res.
12:R362010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stone A, Valdés-Mora F, Gee JM, Farrow L,
McClelland RA, Fiegl H, Dutkowski C, McCloy RA, Sutherland RL,
Musgrove EA and Nicholson RI: Tamoxifen-induced epigenetic
silencing of oestrogen-regulated genes in anti-hormone resistant
breast cancer. PLoS One. 7:e404662012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Magnani L, Stoeck A, Zhang X, Lanczky A,
Mirabella AC, Wang TL, Gyorffy B and Lupien M: Genome-wide
reprogramming of the chromatin landscape underlies endocrine
therapy resistance in breast cancer. Proc Natl Acad Sci USA.
110:E1490–E1499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aran D and Hellman A: DNA methylation of
transcriptional enhancers and cancer predisposition. Cell.
154:11–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: How the genome integrates intrinsic
and environmental signals. Nat Genet. 33 (Suppl):S245–S254. 2003.
View Article : Google Scholar
|
|
28
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jaffe AE, Murakami P, Lee H, Leek JT,
Fallin MD, Feinberg AP and Irizarry RA: Bump hunting to identify
differentially methylated regions in epigenetic epidemiology
studies. Int J Epidemiol. 41:200–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Holm K, Staaf J, Lauss M, Aine M, Lindgren
D, Bendahl PO, Vallon-Christersson J, Barkardottir RB, Höglund M,
Borg Å, et al: An integrated genomics analysis of epigenetic
subtypes in human breast tumors links DNA methylation patterns to
chromatin states in normal mammary cells. Breast Cancer Res.
18:272016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeschke J, Bizet M, Desmedt C, Calonne E,
Dedeurwaerder S, Garaud S, Koch A, Larsimont D, Salgado R, Van den
Eynden G, et al: DNA methylation-based immune response signature
improves patient diagnosis in multiple cancers. J Clin Invest.
127:3090–3102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim SI, Park BW and Lee KS: Comparison of
stage-specific outcome of breast cancer based on 5th and 6th AJCC
staging system. J Surg Oncol. 93:221–227. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
The Gene Ontology Consortium: The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
40
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival- associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tuo YL and Ye YF: MGP is downregulated due
to promoter methylation in chemoresistant ER+ breast cancer and
high MGP expression predicts better survival outcomes. Eur Rev Med
Pharmacol Sci. 21:3871–3878. 2017.PubMed/NCBI
|
|
43
|
Sano K, Tanihara H, Heimark RL, Obata S,
Davidson M, St John T, Taketani S and Suzuki S: Protocadherins: A
large family of cadherin-related molecules in central nervous
system. EMBO J. 12:2249–2256. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yagi T and Takeichi M: Cadherin
superfamily genes: Functions, genomic organization, and neurologic
diversity. Genes Dev. 14:1169–1180. 2000.PubMed/NCBI
|
|
45
|
Kawaguchi M, Toyama T, Kaneko R, Hirayama
T, Kawamura Y and Yagi T: Relationship between DNA methylation
states and transcription of individual isoforms encoded by the
protocadherin-alpha gene cluster. J Biol Chem. 283:12064–12075.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu JS, Koujak S, Nagase S, Li CM, Su T,
Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, et al: PCDH8,
the human homolog of PAPC, is a candidate tumor suppressor of
breast cancer. Oncogene. 27:4657–4665. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li AM, Tian AX, Zhang RX, Ge J, Sun X and
Cao XC: Protocadherin-7 induces bone metastasis of breast cancer.
Biochem Biophys Res Commun. 436:486–490. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu P, Sun M, Jiang W, Zhao J, Liang C and
Zhang H: Identification of targets of miRNA-221 and miRNA-222 in
fulvestrant-resistant breast cancer. Oncol Lett. 12:3882–3888.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu H, Wang G, Yang L, Qu J, Yang Z and
Zhou X: Knockdown of long non-coding RNA UCA1 increases the
tamoxifen sensitivity of breast cancer cells through inhibition of
Wnt/β-catenin pathway. PLoS One. 11:e01684062016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mansouri S, Naghavi-Al-Hosseini F,
Farahmand L and Majidzadeh AK: MED1 may explain the interaction
between receptor tyrosine kinases and ERα66 in the complicated
network of Tamoxifen resistance. Eur J Pharmacol. 804:78–81. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bhatlekar S, Fields JZ and Boman BM: Role
of HOX genes in stem cell differentiation and cancer. Stem Cells
Int 2018. 35694932018.
|
|
52
|
Shah M, Cardenas R, Wang B, Persson J,
Mongan NP, Grabowska A and Allegrucci C: HOXC8 regulates
self-renewal, differentiation and transformation of breast cancer
stem cells. Mol Cancer. 16:382017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee JY, Hur H, Yun HJ, Kim Y, Yang S, Kim
SI and Kim MH: HOXB5 promotes the proliferation and invasion of
breast cancer cells. Int J Biol Sci. 11:701–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kamalakaran S, Varadan V, Giercksky
Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N,
Synnestvedt M, Schlichting E, et al: DNA methylation patterns in
luminal breast cancers differ from non-luminal subtypes and can
identify relapse risk independent of other clinical variables. Mol
Oncol. 5:77–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Q, Seo JH, Stranger B, McKenna A, Pe'er
I, Laframboise T, Brown M, Tyekucheva S and Freedman ML:
Integrative eQTL-based analyses reveal the biology of breast cancer
risk loci. Cell. 152:633–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Maston GA, Evans SK and Green MR:
Transcriptional regulatory elements in the human genome. Annu Rev
Genomics Hum Genet. 7:29–59. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cardoso F, Costa A, Norton L, Senkus E,
Aapro M, André F, Barrios CH, Bergh J, Biganzoli L, Blackwell KL,
et al: ESO-ESMO 2nd international consensus guidelines for advanced
breast cancer (ABC2)†. Ann Oncol. 25:1871–1888. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ali S, Rasool M, Chaoudhry H, N Pushparaj
P, Jha P, Hafiz A, Mahfooz M, Abdus Sami G, Azhar Kamal M, Bashir
S, et al: Molecular mechanisms and mode of tamoxifen resistance in
breast cancer. Bioinformation. 12:135–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kalyuga M, Gallego-Ortega D, Lee HJ, Roden
DL, Cowley MJ, Caldon CE, Stone A, Allerdice SL, Valdes-Mora F,
Launchbury R, et al: ELF5 suppresses estrogen sensitivity and
underpins the acquisition of antiestrogen resistance in luminal
breast cancer. PLoS Biol. 10:e10014612012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mittal MK, Singh K, Misra S and Chaudhuri
G: SLUG-induced elevation of D1 cyclin in breast cancer cells
through the inhibition of its ubiquitination. J Biol Chem.
286:469–479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ingle JN, Liu M, Wickerham DL, Schaid DJ,
Wang L, Mushiroda T, Kubo M, Costantino JP, Vogel VG, Paik S, et
al: Selective estrogen receptor modulators and pharmacogenomic
variation in ZNF423 regulation of BRCA1 expression: Individualized
breast cancer prevention. Cancer Discov. 3:812–825. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Marino N, Woditschka S, Reed LT, Nakayama
J, Mayer M, Wetzel M and Steeg PS: Breast cancer metastasis: Issues
for the personalization of its prevention and treatment. Am J
Pathol. 183:1084–1095. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mehrotra J, Vali M, McVeigh M, Kominsky
SL, Fackler MJ, Lahti-Domenici J, Polyak K, Sacchi N, Garrett-Mayer
E, Argani P and Sukumar S: Very high frequency of hypermethylated
genes in breast cancer metastasis to the bone, brain, and lung.
Clin Cancer Res. 10:3104–3109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ostrakhovitch EA and Li SS: NIP1/DUOXA1
expression in epithelial breast cancer cells: Regulation of cell
adhesion and actin dynamics. Breast Cancer Res Treat. 119:773–786.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Park SY, Kwon HJ, Choi Y, Lee HE, Kim SW,
Kim JH, Kim IA, Jung N, Cho NY and Kang GH: Distinct patterns of
promoter CpG island methylation of breast cancer subtypes are
associated with stem cell phenotypes. Mod Pathol. 25:185–196. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Feske S: Calcium signalling in lymphocyte
activation and disease. Nat Rev Immunol. 7:690–702. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Svendsen AJ, Gervin K, Lyle R,
Christiansen L, Kyvik K, Junker P, Nielsen C, Houen G and Tan Q:
Differentially methylated DNA regions in monozygotic twin pairs
discordant for rheumatoid arthritis: An epigenome-wide study. Front
Immunol. 7:5102016. View Article : Google Scholar : PubMed/NCBI
|