Introduction
Uterine leiomyosarcomas (uLMS) are relatively rare
but highly malignant gynecological mesenchymal tumors. uLMS are
derived from smooth muscle of the myometrium, and are the most
common subtype of uterine sarcomas (1). Although uLMS account for only about 1%
of uterine malignancies, they account for about 70% of all uterine
cancer deaths (2). Hysterectomy is
the standard treatment for uLMS. However, even for uLMS patients
that received complete surgical treatments, the risk of recurrence
after surgical treatments was high (50–71%) because of the
aggressive nature of uLMS (3–9). The
poor prognosis is reflected by 5-year disease-specific survival
rates of less than 30% (10–12). Therefore, there is a need for an
effective therapeutic strategy for uLMS.
Carbonyl reductase 1 (CBR1) is a nicotinamide
adenine dinucleotide phosphate-dependent, mostly monomeric,
cytosolic enzyme with a broad substrate specifically for carbonyl
compounds (13,14). CBR1 is present in a variety of organs
including liver, kidney, breast, ovary, and vascular endothelial
cells, and its primary function is considered to control fatty acid
metabolism (15). Interestingly,
CBR1 has been reported to regulate malignant behaviors of cancer
cells: Decreasing the expression of CBR1 induced cell proliferation
and tumorigenesis in vitro and in vivo experiments
accompanied by a decreased expression of E-cadherin, and activated
matrix metalloproteinases (MMPs) in ovarian, uterine cervical, or
uterine endometrial cancers (16–21).
This suggests that the decreased expression of CBR1 promotes tumor
growth and invasion activities. Furthermore, low-expression of CBR1
is closely associated with lymph node metastasis and a poor
prognosis in ovarian, uterine cervical, or endometrial cancers
(17,18,20). In
contrast, increasing the expression of CBR1 repressed the
activities of cell proliferation and invasion, and tumorigenesis in
ovarian, uterine cervical, and endometrial cancers (17–20,22).
Additionally, CBR1 overexpression increased E-cadherin expression
and decreased N-cadherin expression, indicating that CBR1
expression inhibits the epithelial mesenchymal transition (EMT)
(17,18).
EMT is a well-known phenomenon that is associated
with the progression of malignant behaviors in epithelial tumors
(23,24). In the EMT process, cell-to-cell
adhesion becomes weak with the decreased expression of E-cadherin,
and invasive and metastatic activities are promoted (23–26).
Although EMT has been studied in epithelial malignant tumors
(cancers), it is of interest to note that EMT is involved in
malignant behaviors even in mesenchymal malignant tumors. Several
reports have indicated the potential existence of EMT-related
processes in sarcomas, suggesting that sarcomas can undergo
phenotypic changes reminiscent of EMT (27–32). EMT
leads to a gain in mesenchymal activities and promotes malignant
behaviors of mesenchymal tumors (27,30,32,33),
which may be associated with aggressive clinical behaviors.
Therefore, it is important to examine the role of EMT in uLMS as
well as in cancers.
Regarding the mechanism of EMT, transforming growth
factor (TGF)-β, which has roles in cell proliferation,
differentiation, and tumorigenesis, has been reported to be closely
associated with EMT in many epithelial and mesenchymal tumors
(34,35). We previously analyzed pathways that
are regulated by CBR1 in a uterine cervical cancer cell line
overexpressing CBR1 (21). Of 15
pathways that were analyzed, the TGF-β signaling pathway was found
to be the most important. This suggests that TGF-β signaling is
closely associated with EMT, which is regulated by CBR1.
We previously showed that CBR1 inhibited malignant
behaviors by suppressing EMT in uterine cervical squamous cell
carcinomas (21). It remained
unclear whether this also was the case in uLMS. Therefore, we
investigated whether an increase in CBR1 expression inhibits
malignant behaviors and EMT through TGF-β signaling in uLMS.
Materials and methods
Cell culture
SKN, which is a human uLMS cell line, and MES-SA,
which is human uterine sarcoma cell line, were used in this study.
SKN was purchased from Health Science Research Resources Bank
(HSRRB, Osaka, Japan), and MES-SA was purchased from American type
culture collection (ATCC, Virginia, USA). SKN cells were cultured
in Ham's F 12 (Sigma-Aldrich Japan K.K., Tokyo, Japan), and MES-SA
cells were cultured in McCoy's 5a medium (ATCC). Both media were
supplemented with 10% heat-incubated fetal bovine serum (FBS). The
cells were seeded at a density of 5×104 cells/well in a
six-well plate, and incubated at 37°C in a humidified 5%
CO2 incubator for 5 days. The cells were trypsinized and
counted by a cell counter (Vi-CELL XR; Beckman Coulter, Tokyo,
Japan) at each time point, as reported previously (36).
For TGF-β treatment, cells were cultured in Ham's
F12 and McCoy's 5a medium supplemented with 10% FBS containing 100
or 500 pg/ml TGF-β1 (Sigma-Aldrich Japan K.K.) for 24 h. In order
to block TGF-β signaling, SB431542 (WAKO, Tokyo, Japan), which is a
TGF-β type I receptor-selective blocker, was dissolved at a
concentration of 10 µM in dimethylsulfoxide (DMSO). Cells were
seeded at a density of 5×104 cells/well in a six-well
plate and cultured for 24 h and then cultured with new medium
supplemented with 10 µM SB431542 for more 48 h.
Gene transfection
To analyze the function of CBR1, stable clones were
established in which CBR1 expression was increased. The construct
of CBR1 was transfected into SKN and MES-SA by the lipofection
method as previously reported (37,38). To
improve the efficacy of transfection, we used the pEF1a-IRES-AcGFP
vector (Clontech Laboratories, Inc., Mountain View, CA, USA) that
encodes human CBR1, green fluorescent protein (GFP) and a neomycin
resistance gene. Transfected cells were cultured in Ham's F12 and
McCoy's 5a medium for 24 h, and then in medium supplemented with
300 µg/ml neomycin for one month. The overexpression of CBR1 was
confirmed by Western blot analysis. A clone transfected with the
empty vector was used as a control. In the preliminary experiments,
the reagent that was used for transfection did not influence cell
mobility or CBR1 expression.
In vitro migration and invasion
assays
Cell migration and invasion assays were assayed with
a BioCoat Matrigel Invasion Chamber (cat. no. 354480; Corning Life
Sciences Inc., Tewksbury, MA, USA) according to the manufacturer's
protocol as previously reported (17). This kit is used to measure how well
cells can migrate from the upper side of a membrane to the lower
side in a 12-well transwell system. Cells were trypsinized, and
5×104 cells in serum-free medium were seeded into
collagen-coated and uncoated (control) upper insert chambers of the
transwell system. The lower chamber was filled with 750 µl medium
containing 10% FBS as a chemo-attractant. After incubation for 20
h, the upper surfaces of the membranes were wiped with cotton swabs
to completely remove the cells. The membranes were fixed and
stained with Diff-Quick (cat. no. 24606-500; Sysmex, Kobe, Japan).
The cells on the lower surfaces of the membranes were counted at
200× magnification in five randomized field views, and the mean was
calculated. Migration activities were evaluated by the numbers of
cells that migrated to the control chamber. Invasion activities
were expressed as the number of cells in the coated chamber divided
by the number of cells that migrated to the uncoated chamber. Each
experiment was performed in triplicate, and the mean percentage was
obtained from three independent experiments. In the experiments
with MES-SA cells, we could not see the effects of CBR1
overexpression on migration and invasion activities because MES-SA
cells did not move.
Wound healing assay
Cells were cultured in a six-well plate until they
reached confluence. Linear scratch wounds were created in the
center of each well with a 1,000-µl sterile pipette tip. After 24
h, images were obtained to observe the wounds at the same fields
under the microscope, and the separation distance between wound
sides was measured as previously reported (36). Each experiment was performed in
triplicate, and the mean was obtained from three independent
experiments.
Reverse transcription-PCR
(RT-PCR)
The mRNA expressions of CDH1, SNAIL, SLUG,
TGF-β1 and TGF-β2 were examined by real-time RT-PCR.
Total RNA was isolated with an RNeasy mini kit (cat. no. 74104;
Qiagen, Tokyo, Japan). cDNA was synthesized from 1 µg of total RNA
using a Revertra Ace qPCR RT Master Mix (cat. no. FSQ-201; Toyobo
Life Science, Osaka, Japan). RT-PCR was performed using TB green
Primer Ex Taq II DNA Polymerase (cat. no. RR820S; Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's protocol with the
following amplifying primer pairs: SNAI1
(5′-ctccctgtcagatgaggacagt-3′ and 5′-tccttgttgcagtatttgcagt-3′),
SNAI2 (5′-cctgtcataccacaaccagag-3′ and
5′-cttcatcactaatggggctttc-3′), CDH1
(5′-cgggaatgcagttgaggatc-3′ and 5′-aggatggtgtaagcgatggc-3′),
TGF-β1 (5′-tggacaccaactattgcttcag-3′ and
5′-gtccaggctccaaatgtagg-3′), TGF-β2
(5′-ttgatggcacctccacatatac-3′ and 5′-agtggacgtaggcagcaatta-3′), and
glyceraldehyde 3-phospate dehydrogenase (GAPDH)
(5′-tgcaccaccaactgcttagc-3′ and 5′-ggcatggactgtggtcatgag-3′).
GAPDH served as an internal control. The thermal cycling
conditions were 45 cycles of 95°C for 5 sec, and 60°C for 20
sec.
Western blot analysis
Cultured cells were resuspended in RIPA buffer (Wako
Pure Chemical Industries, Ltd., Osaka, Japan) and mildly sonicated.
The insoluble materials were removed by centrifugation at 15,000
rpm for 10 min at 4°C. The supernatant was mixed with SDS sample
buffer (New England BioLabs, Tokyo, Japan), and the samples were
boiled for 5 min. Ten µg of proteins were electrophoresed on a 10%
SDS-polyacrylamide gel (PAGE). The proteins were then transferred
to a polyvinylidene difluoride membrane (New England BioLabs) with
a semi-dry type blotting system. The membrane was blocked with
blocking solution (5% skimmed milk with 0.1% tween 20 dissolved in
Tris buffered saline, pH 7.5), incubated with the appropriate first
antibodies (see below) diluted at 1:1,000 in blocking solution,
incubated with the peroxidase-conjugated secondary antibody (see
below) diluted at 1:2,000 in blocking solution, incubated in
ECL-Western blotting detection regents (GE Healthcare, Little
Chalfont, Buckinghamshire, UK) for 5 min and used to expose the
Hyperfilm-ECL (GE Healthcare). The first antibodies were goat
anti-human CBR1 polyclonal antibody (ab4148; Abcam, Tokyo, Japan),
rabbit anti-human E-cadherin monoclonal antibody (EP700Y; Abcam),
mouse anti-human cytokeratin monoclonal antibody (ab668; Abcam),
rabbit anti-human α-SMA monoclonal antibody (1184-1; Abcam), rabbit
anti-human fibronectin polyclonal antibody (ab23750; Abcam), rabbit
anti-human N-cadherin polyclonal antibody (ab76057; Abcam), rabbit
anti-human vimentin polyclonal antibody (EPR3776; Abcam), rabbit
anti-human snail polyclonal antibody (cat. no. 3879; CST, Tokyo,
Japan), rabbit anti-human smad 2 monoclonal antibody (cat. no.
3122; CST, Tokyo, Japan), rabbit anti-human Smad 3 monoclonal
antibody (cat. no. 9523; CST), rabbit anti-human phospho-Smad 2
monoclonal antibody (cat. no. 3122; CST), rabbit anti-human
phospho-Smad 3 monoclonal antibody (cat. no. 3104S; CST), and mouse
anti-human β-tubulin monoclonal antibody (T4026; Sigma-Aldrich
Japan K.K.). The second antibodies were anti-rabbit IgG/HRP
(PO399), anti-mouse IgG/HRP (PO260), and anti-goat IgG/HRP (PO449;
all from Dako; Agilent Technologies Japan, Ltd., Hachioji-shi,
Tokyo).
ELISA
After cells were cultured in the medium without FBS
for 48 h, TGF-β1 concentrations in the cell culture medium were
measured by a Quantikine ELISA Human TGF-β1 immunoassay kit (cat.
no. DB100B; R&D Systems Inc., Minneapolis, MN, USA), according
to the manufacturer's protocols. The absorbance (OD) was measured
at 450 nm.
Gelatin zymography
In order to analyze the effect of CBR1
overexpression on the production of activated MMPs, extracellular
MMPs in the medium were measured by gelatin zymography as
previously reported (17). Cells
were cultured at a density of 1×105 cells/ml in a
six-well microtiter plate at 37°C for 24 h under 5% CO2
in a humidified atmosphere. The medium was replaced with new medium
without FBS, and then cells were cultured for another 24 h. After
the culture, MMPs were detected with gelatin zymography. Twenty µl
of culture medium with 10 ml Red loading buffer (New England
Biolabs) without regent buffer was electrophoresed using detector
gels containing 0.9 mg/ml gelatin. As a positive control of MMPs, a
standard mixture of pro MMP-9 (92 kDa), pro MMP-2 (72kDa), and
MMP-2 (62kDa; Funakoshi, Tokyo, Japan) was electrophoresed on the
same gel. The gels were washed for 1 h in 2.5% w/v aqueous Triton
X-100 to remove SDS and renature the activity of MMPs, and then
incubated in activation buffer (50 mM Tris-HCl, 200 mM NaCl, 5 mM
CaCl2, pH 7.5) at 37°C for 72 h. The activities of MMPs
were detected as transparent gelatinolytic bands by staining with
CBB R-250. The band intensities were measured with Image J
software. In the experiments with MES-SA cells, we could not see
the effects of CBR1 overexpression on MMP secretion because MES-SA
cells did not secret MMP due to the character of the cells.
Statistical analysis
Unpaired Student's t-test was used to compare the
two groups. To analyze differences between groups, one-way ANOVA
followed by Tukey's test was used. P<0.05 was considered to
indicate a statistically significant difference. All statistics
were analyzed with SPSS 5.0 J for Windows (SAS Institute Inc.,
Cary, NC, USA).
Results
Establishment of a clone
overexpressing CBR1
To evaluate the roles of CBR1 in malignant behaviors
of uLMS cells, clones overexpressing CBR1 were established by
transfecting sense cDNA to CBR1 into SKN cells and MES-SA cells
(CBR1). Cells that were transfected with empty vector were used as
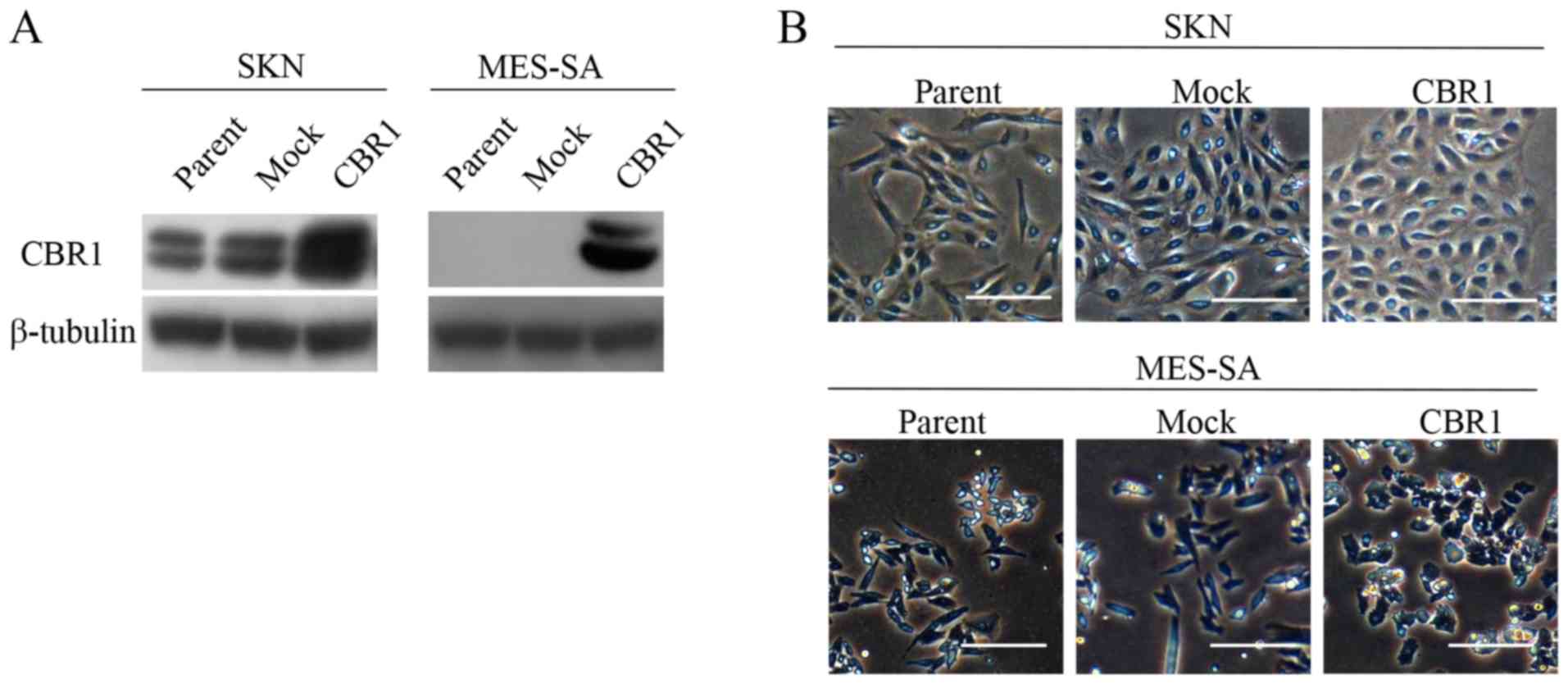
a control (Mock). Western blot analysis confirmed that CBR1 is
overexpressed in both CBR1 clones (Fig.
1A). Parent cells and mock cells appeared as spindle-shaped
fibroblasts while both types of CBR1 cells appeared as cobble
stone-like epithelial cells (Fig.
1B).
Cell proliferation, cell migration,
and wound healing assays
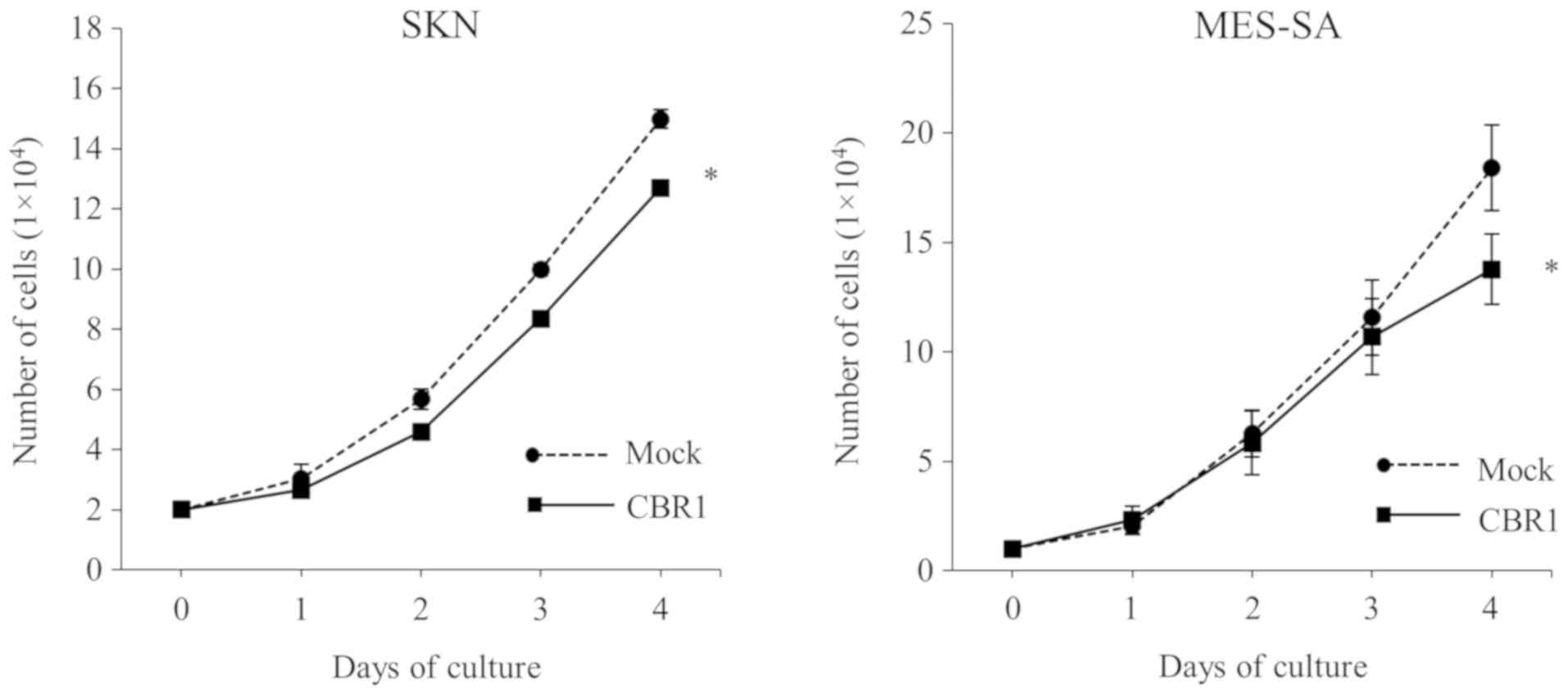
After four days of culture, the number of cells was
significantly lower in the CBR1 clones than in the mock clones for
both SKN and MES-SA cells (Fig. 2),
suggesting that CBR1 overexpression decreased cell
proliferation.
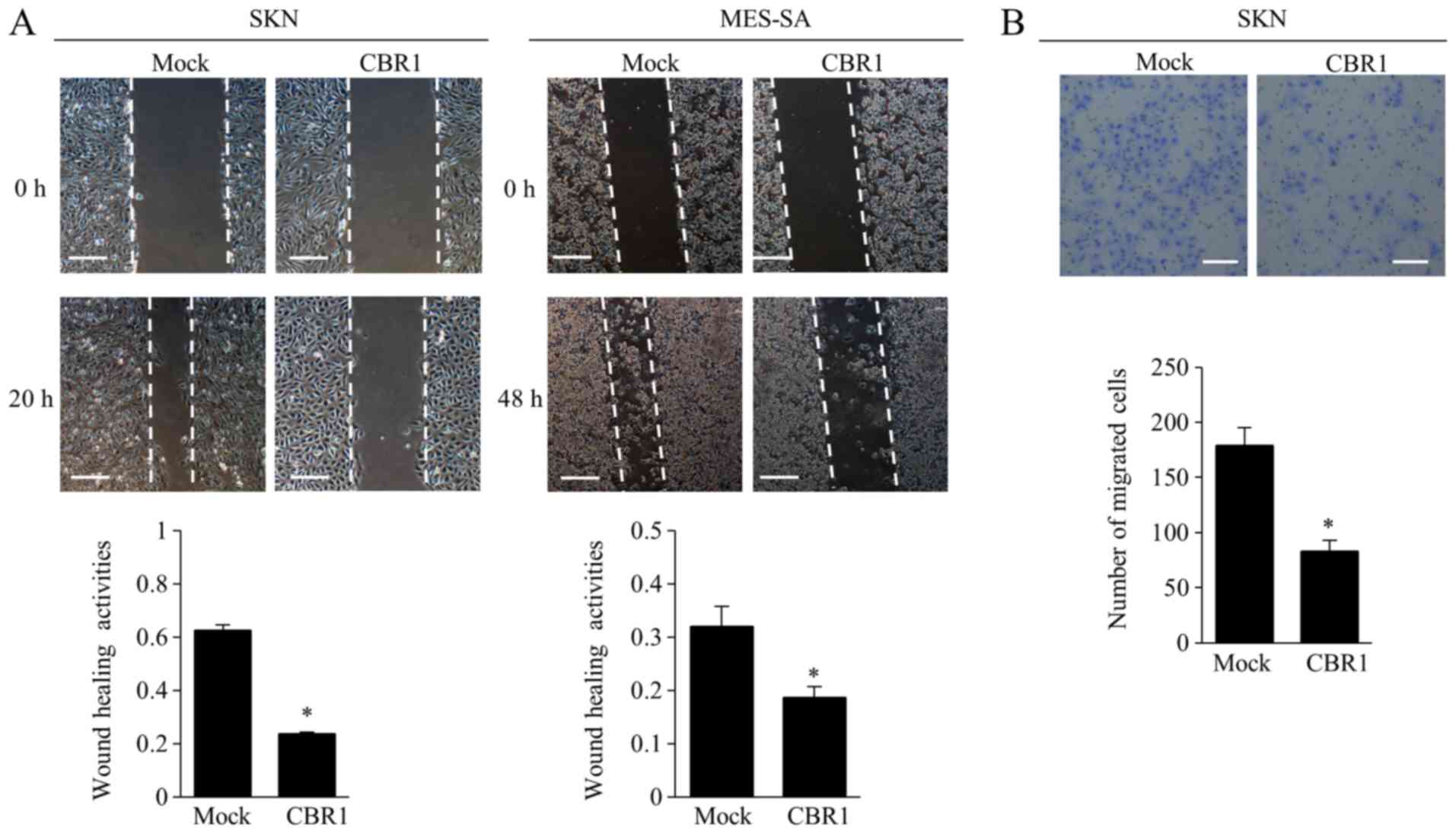
The wound healing assay showed that the cells in the
CBR1 clone filled the wounded area significantly more slowly than
did the cells in the mock clone for both cell types (Fig. 3A). In the migration assay, the number
of SKN cells that migrated to the lower side of the membrane was
significantly lower in the CBR1 clone than that in the mock clone
(Fig. 3B).
MMPs secretions and cell invasion
assay
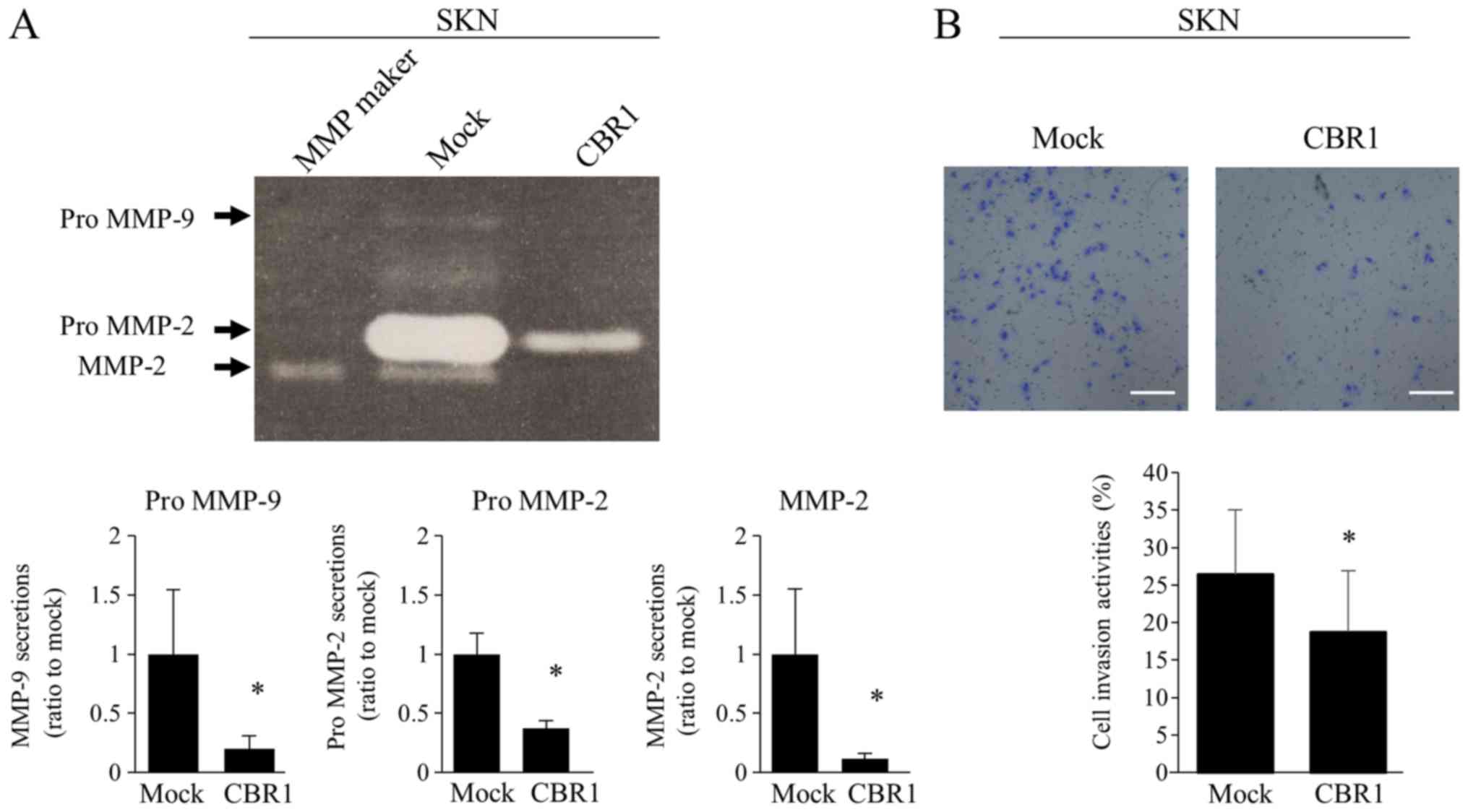
SKN cells were examined in these assays, in which
MMP-2 and MMP-9 secretions were determined by gelatin zymography.
The band intensities of pro-MMP-9, pro-MMP-2, and MMP-2 were
significantly lower in the CBR1 clone than those in the mock clone
(Fig. 4A). The CBR1 clone showed
significantly lower invasion activities than the mock clone
(Fig. 4B).
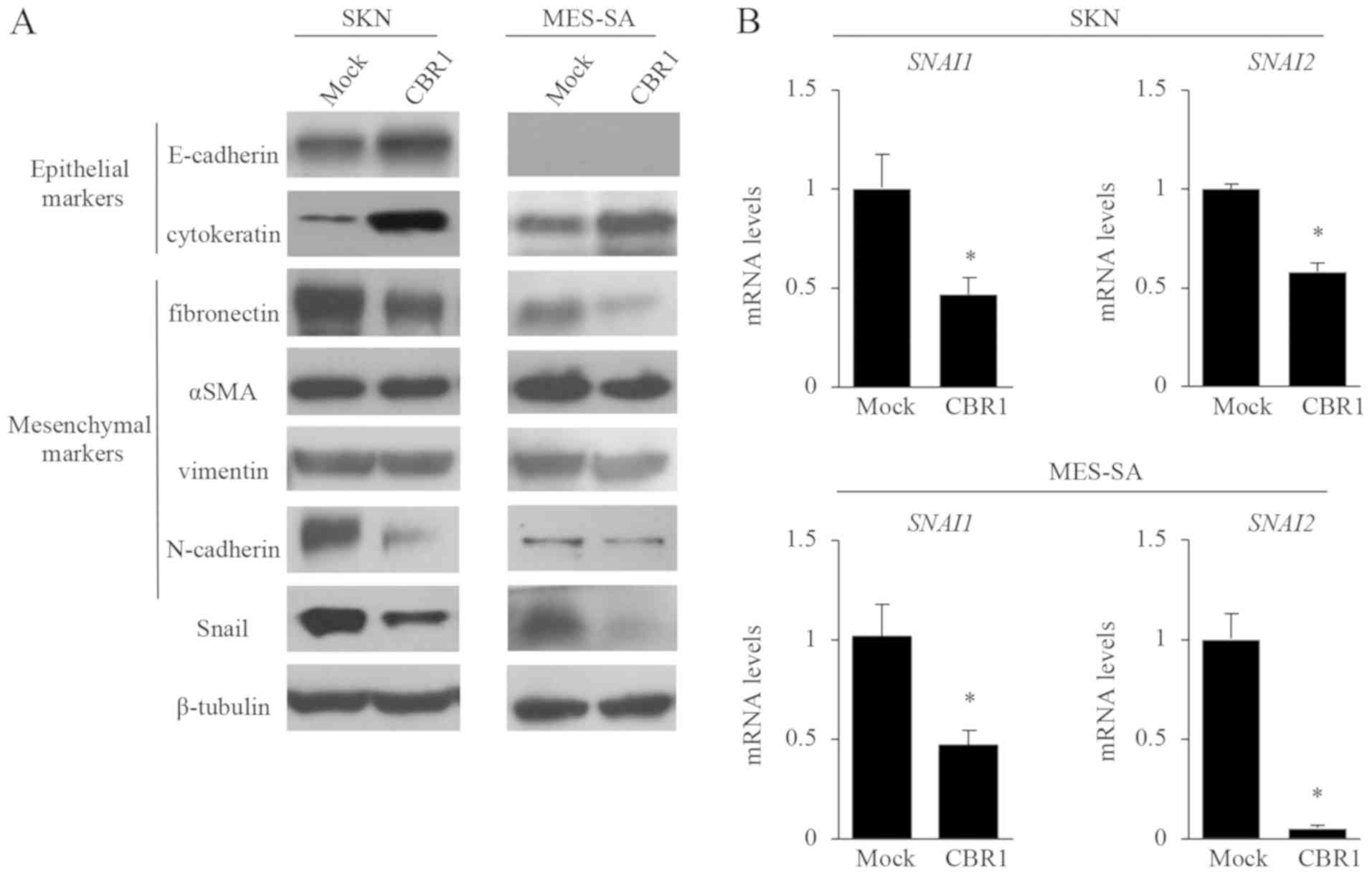
Effects of CBR1 overexpression on
EMT-related markers
In SKN cells, the epithelial markers E-cadherin and
cytokeratin were expressed more strongly in the CBR1 clone than in
the mock clone (Fig. 5A). In MES-SA
cells, cytokeratin was also expressed more strongly in the CBR1
clone (Fig. 5A). Since MEA-SA cells
do not express E-cadherin, we did not observe the effect of CBR1
overexpression on E-cadherin. On the other hand, in SKN cells, the
expression levels of the mesenchymal markers fibronectin and
N-cadherin were lower in the CBR1 clone than in the mock clone,
while the expression levels of αSMA and vimentin did not differ
between the two clones (Fig. 5A). In
MES-SA cells, the expression levels of fibronectin, αSMA, and
N-cadherin were lower in the CBR1 clone than in the mock clone
(Fig. 5A).
Snail (SNAI1) and Slug (SNAI2) are
transcription factors that repress E-cadherin expression (30). The protein expression of Snail
(Fig. 5A) and the mRNA levels of
SNAI1 and SNAI2 (Fig.
5B) were all significantly lower in the CBR1 clone than in the
mock clone in both cell lines. These results suggest that EMT is
suppressed in the CBR1 clone overexpressing CBR1.
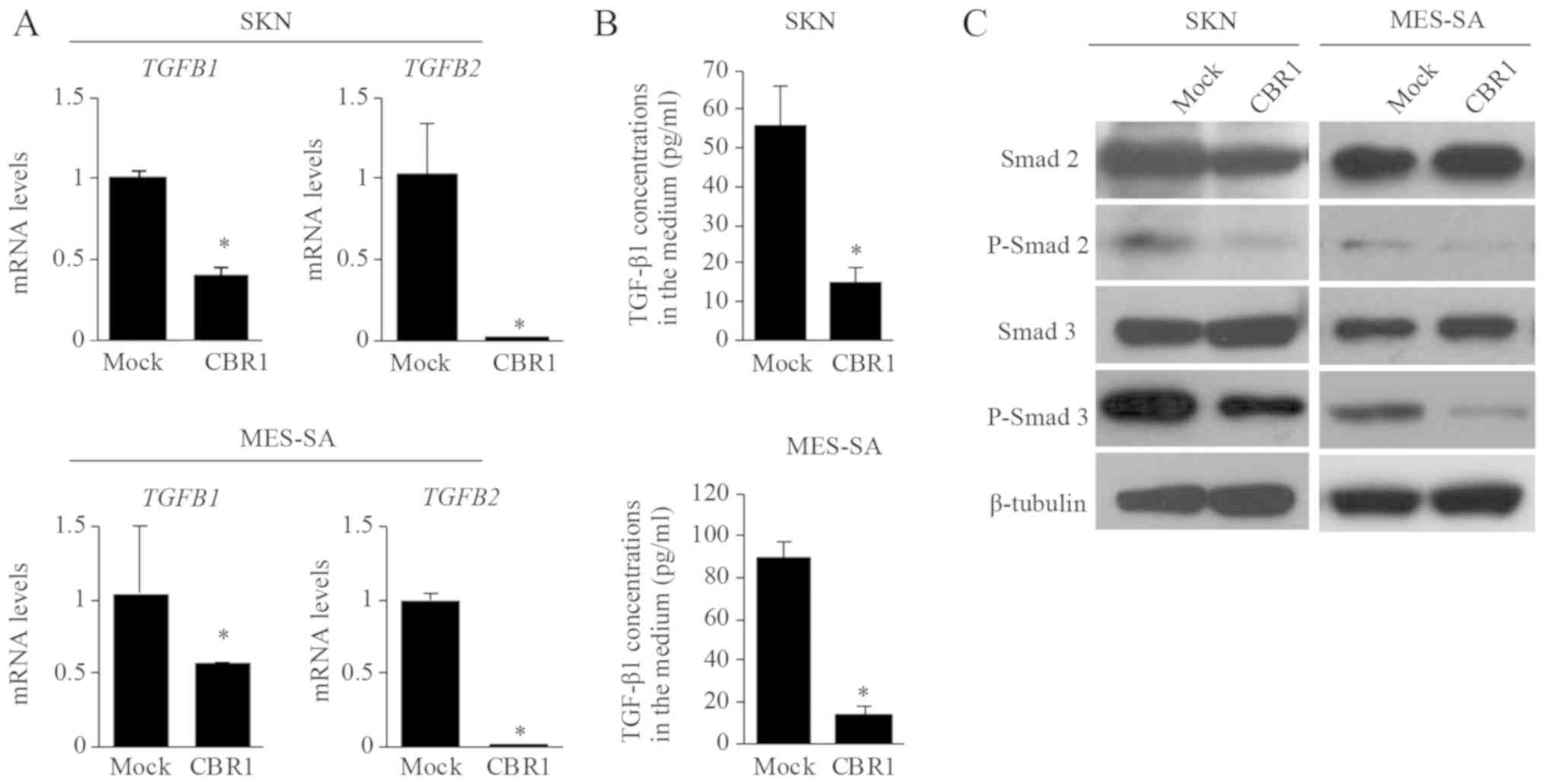
TGF-β production and signaling in CBR1
overexpressing clone
We investigated whether CBR1 overexpression affects
TGF-βs production. TGFB1 and TGFB2 mRNA levels were
significantly lower in the CBR1 clone than in the mock clone in
both SKN and MES-SA cells (Fig. 6A).
The TGF-β1 concentrations in the medium were also significantly
lower in the CBR1 clone than in the mock clone in both cell lines
(Fig. 6B). These results indicate
that TGF-βs production is suppressed in the CBR1 clone
overexpressing CBR1.
We further investigated whether CBR1 overexpression
suppresses TGFβ/Smad signaling. The expression levels of
phosphorylated-Smad2 (P-Smad2) and phosphorylated-Smad3 (P-Smad3)
as measured by Western blotting were significantly lower in the
CBR1 clone than in the mock clone in both SKN and MES-SA cells
(Fig. 6C). This result suggests that
the TGF-β signaling pathway is suppressed in the CBR1 clone.
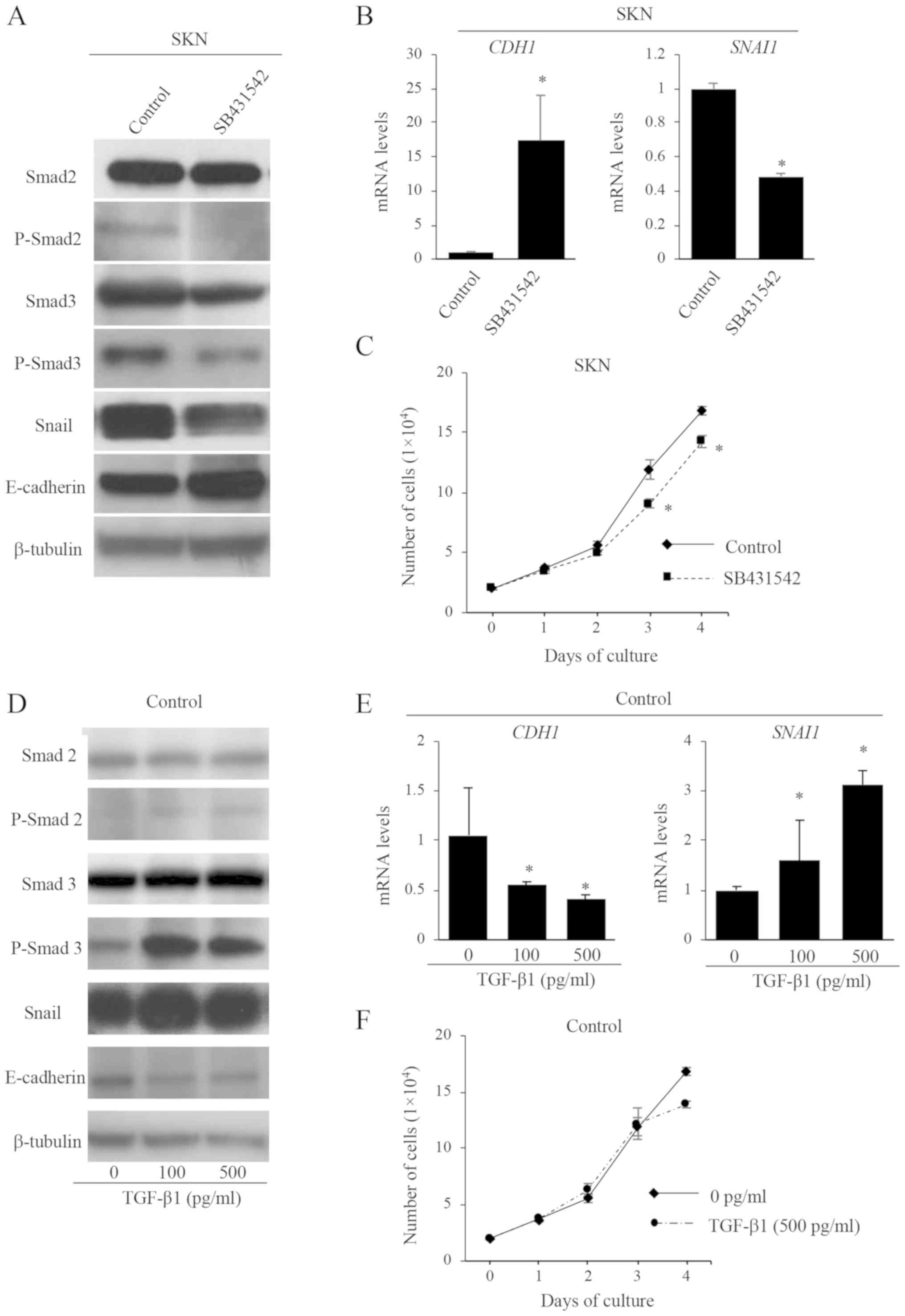
TGF-β signaling and the EMT
To investigate whether TGF-β signaling is associated
with the promotion of EMT in uLMS cells, SKN parent cells were
treated with a TGF-β receptor blocker, SB431542. The expression
levels of P-Smad2 and P-Smad3 proteins were confirmed to be
decreased by SB431542 (Fig. 7A). The
protein and mRNA levels of E-cadherin (CDH1) expression were
increased while those of Snail (SNAI1) were decreased by
SB431542 treatment (Fig. 7A and B).
In addition, SB431542 treatment significantly decreased cell
proliferation activities (Fig. 7C),
but did not affect cell morphology (data not shown). These results
suggest that TGF-β signaling is associated with the promotion of
EMT.
We further examined whether TGF-β stimulation
promotes EMT in uLMS cells. Parent cells (control) and
CBR1-overexpressing SKN cells were treated with TGF-β1. The
expression levels of P-Smad2 and P-Smad3 were confirmed to be
increased by TGF-β stimulation in control cells (Fig. 7D). The protein and mRNA levels of
E-cadherin (CDH1) were decreased while those of Snail
(SNAI1) were increased by TGF-β treatment in control cells
(Fig. 7D and E). TGF-β treatment did
not affect the cell proliferation activity (Fig. 7F) or cell morphology (data not
shown). These results provide additional evidence that TGF-β
signaling is partly associated with the promotion of EMT in uLMS
cells.
On the other hand, the effects by TGF-β treatment on
the CBR1 cells were similar to those observed in the control cells
(Fig. S1), indicating that CBR1
overexpression did not affect TGF-β signaling stimulated by
exogenous TGF-β. This could be due to either (1) the suppressive effect of CBR1 on TGF-β
signaling and EMT may have been overcome by exogenous TGF-β or
(2) CBR1 inhibits endogenous TGF-β
expression, but does not directly interrupt the TFG-β signaling
pathway.
Discussion
The present results show that CBR1 overexpression
inhibited cell proliferation, migration, and invasion activities
and that these changes were accompanied by increased expressions of
epithelial markers (E-cadherin and cytokeratin), and decreased
expressions of mesenchymal markers (N-cadherin and fibronectin) in
uLMS. These results suggest that the increased CBR1 expression
represses malignant behaviors of uLMS cells by inhibiting EMT. We
previously reported that increased CBR1 expression repressed
migration and invasion activities by inhibiting EMT in uterine
cervical and endometrial cancers (17,18).
Therefore, it is likely that CBR1 controls malignant behaviors of
tumor cells in uLMS as well as in uterine cancers. This the first
report suggesting the anti-tumor effect of CBR1 in uLMS.
The mechanism by which CBR1 inhibits EMT in uLMS
cells remains unclear. Our previous microarray and IPA pathway
analyses showed that suppression of CBR1 expression activates the
intracellular signaling pathway of TGF-β in uterine cervical cancer
(21). TGF-β is a multifunctional
cytokine, and has been shown to be a modulator of EMT in many types
of cancers (24,34,39–44).
However, it is still controversial how TGF-β acts on sarcomas. Qi
et al (35,45) reported that the TGF-β/Smad/Snail
signaling pathway is closely associated with EMT, which promotes
cell proliferation, migration, and invasion in synovial sarcomas.
Dwivedi et al (46) showed
that TGF-β promotes cell migration by phosphorylating Smad2 and
Smad3 in uterine carcinosarcomas. These findings suggest that TGF-β
signaling is associated with EMT in certain sarcomas as well as in
cancers. Our results clearly show that TGF-β treatment activates
EMT, that the blockage of TGF-β signaling inhibits EMT in uLMS
cells (Fig. 7), and that CBR1
overexpression inhibits TGF-β production and signaling (Fig. 6). Therefore, these results suggest
that CBR1 inhibits EMT through suppressing TGF-β production and the
subsequent signaling pathway in uLMS cells. This is also the first
report showing that that the suppressive effect of CBR1 is mediated
through TGF-β signaling.
In this study, the blockage of TGF-β signaling by
SB431542 (a TGF-β type 1 receptor-selective blocker) decreased cell
proliferation activities while exogenous TGF-β treatment had no
effects. TGF-β is a multifunctional cytokine and exogenous
treatment of TGF-β stimulates several intracellular signaling
pathways (47). Therefore, the
effect of a TGF-β receptor blocker may more correctly reflect the
involvement of TGF-β signaling pathway. It is unclear why the
exogenous TGF-β treatment and TGF-β-blocker did not influence cell
morphology. We speculate that the experimental conditions used in
this study changed the levels of EMT markers, but did not affect
cellular phenotypes such as cell morphology. The findings that
exogenous TGF-β treatment had no effects on cell proliferation and
that TGF-β-blocker did not influence cell morphology do not
contradict our hypothesis that TGF-β signaling is involved in the
suppressive effect of CBR1 on EMT. Since CBR1 is involved in a
number of intracellular signaling pathways other than TGF-β
signaling pathways (21), CBR1 may
regulate cell proliferation activities or cell morphology through
several intracellular signaling pathways.
Much is known about the role of EMT in epithelial
malignant tumors (cancers) whereas little is known about its role
in mesenchymal malignant tumors. Since sarcomas originally have
mesenchymal features, they do not need to undergo EMT. However,
there is accumulating evidence that EMT is involved in malignant
behaviors in a variety of mesenchymal malignant tumors (28,30–33,48).
Yang et al (48) reported
that suppression of EMT with an increase in E-cadherin expression
was associated with inhibition of cell proliferation and invasion
activities in leiomyosarcomas, which is consistent with our
results. EMT enhances mesenchymal activities and promotes malignant
behaviors such as cell proliferation and invasion activities in
mesenchymal malignant tumor cells (27,30,49–53),
which may be associated with aggressive clinical features.
Therefore, controlling EMT may be a key to controlling the
aggressive malignant behaviors of sarcomas. Interestingly, the
reverse process of EMT, termed the mesenchymal to epithelial
transition (MET), has been reported to occur in sarcomas, which
results in more favorable tumor behaviors (27,49,54,55). Our
results show that CBR1 inhibits EMT. In other words, CBR1 may
induce MET in uLMS cells.
It is unclear how CBR1 suppresses TGF-β expression.
Miura et al (22) reported
that CBR1 represses c-jun expression in ovarian cancer cells. c-jun
combines with AP-1, and the complex stimulates TGF-β-induced EMT
and prevents apoptosis (56).
Therefore, there is a possibility that CBR1 inhibits TGF-β
expression and its subsequent signaling by decreasing c-jun
expression. However, further studies are needed to test these
ideas.
In this study, only one cell line (SKN) derived from
uLMS cells was used because we were unable to find other uLMS cell
lines that are commercially available. This is a limitation of this
study although the data using a uterine sarcoma cell line (MEA-SA)
were very similar to those observed in SKN cells.
In summary, the present study shows that increased
CBR1 expression inhibits malignant behaviors and EMT by suppressing
TGF-β signaling in uLMS cells. uLMS is a highly malignant
mesenchymal tumor, and there is no efficient therapy for it except
surgery at present. Our findings suggest that CBR1 can be a
promising new target for therapy of uLMS. A therapeutic shifting of
uLMSs to a more epithelial-like status could attenuate their
aggressive features and improve patient prognosis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported in part by JSPS
KAKENHI (grant nos. 23592425, 23791845, 26462525, 15K10719 and
16H07009) for Scientific Research from the Ministry of Education,
Science and Culture, Japan.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
TK and NS designed the study. TK, AM, MO, KN and KS
performed the experiments. TK and SS analyzed and interpreted the
data. TK drafted the first manuscript. NS directed the research and
drafted the final manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zang Y, Gu L, Zhang Y, Wang Y and Xue F:
Identification of key genes and pathways in uterine leiomyosarcoma
through bioinformatics analysis. Oncol Lett. 15:9361–9368.
2018.PubMed/NCBI
|
|
2
|
Ricci S, Stone RL and Fader AN: Uterine
leiomyosarcoma: Epidemiology, contemporary treatment strategies and
the impact of uterine morcellation. Gynecol Oncol. 145:208–216.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
George S, Barysauskas C, Serrano C,
Oduyebo T, Rauh-Hain JA, Del Carmen MG, Demetri GD and Muto MG:
Retrospective cohort study evaluating the impact of intraperitoneal
morcellation on outcomes of localized uterine leiomyosarcoma.
Cancer. 120:3154–3158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raine-Bennett T, Tucker LY, Zaritsky E,
Littell RD, Palen T, Neugebauer R, Axtell A, Schultze PM, Kronbach
DW, Embry-Schubert J, et al: Occult uterine sarcoma and
leiomyosarcoma: Incidence of and survival associated with
morcellation. Obstet Gynecol. 127:29–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Einstein MH, Barakat RR, Chi DS, Sonoda Y,
Alektiar KM, Hensley ML and Abu-Rustum NR: Management of uterine
malignancy found incidentally after supracervical hysterectomy or
uterine morcellation for presumed benign disease. Int J Gynecol
Cancer. 18:1065–1070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oduyebo T, Rauh-Hain AJ, Meserve EE,
Seidman MA, Hinchcliff E, George S, Quade B, Nucci MR, Del Carmen
MG and Muto MG: The value of re-exploration in patients with
inadvertently morcellated uterine sarcoma. Gynecol Oncol.
132:360–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parker WH, Fu YS and Berek JS: Uterine
sarcoma in patients operated on for presumed leiomyoma and rapidly
growing leiomyoma. Obstet Gynecol. 83:414–418. 1994.PubMed/NCBI
|
|
8
|
Harris JA, Swenson CW, Uppal S, Kamdar N,
Mahnert N, As-Sanie S and Morgan DM: Practice patterns and
postoperative complications before and after US Food and Drug
Administration safety communication on power morcellation. Am J
Obstet Gynecol. 214:e1–98.e13. 2016. View Article : Google Scholar
|
|
9
|
Ducie JA and Leitao MM Jr: The role of
adjuvant therapy in uterine leiomyosarcoma. Expert Rev Anticancer
Ther. 16:45–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Amant F, Lorusso D, Mustea A, Duffaud F
and Pautier P: Management strategies in advanced uterine
leiomyosarcoma: Focus on trabectedin. Sarcoma. 2015:7041242015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kapp DS, Shin JY and Chan JK: Prognostic
factors and survival in 1396 patients with uterine leiomyosarcomas:
Emphasis on impact of lymphadenectomy and oophorectomy. Cancer.
112:820–830. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zivanovic O, Leitao MM, Iasonos A, Jacks
LM, Zhou Q, Abu-Rustum NR, Soslow RA, Juretzka MM, Chi DS, Barakat
RR, et al: Stage-specific outcomes of patients with uterine
leiomyosarcoma: A comparison of the international Federation of
gynecology and obstetrics and American joint committee on cancer
staging systems. J Clin Oncol. 27:2066–2072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Penning TM and Drury JE: Human aldo-keto
reductases: Function, gene regulation, and single nucleotide
polymorphisms. Arch Biochem Biophys. 464:241–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mindnich RD and Penning TM: Aldo-keto
reductase (AKR) superfamily: Genomics and annotation. Hum Genomics.
3:362–370. 2009.PubMed/NCBI
|
|
15
|
Wermuth B, Bohren KM, Heinemann G, von
Wartburg JP and Gabbay KH: Human carbonyl reductase. Nucleotide
sequence analysis of a cDNA and amino acid sequence of the encoded
protein. J Biol Chem. 263:16185–16188. 1988.PubMed/NCBI
|
|
16
|
Ismail E, Al-Mulla F, Tsuchida S, Suto K,
Motley P, Harrison PR and Birnie GD: Carbonyl reductase: A novel
metastasis-modulating function. Cancer Res. 60:1173–1176.
2000.PubMed/NCBI
|
|
17
|
Murakami A, Fukushima C, Yoshidomi K,
Sueoka K, Nawata S, Yokoyama Y, Tsuchida S, Ismail E, Al-Mulla F
and Sugino N: Suppression of carbonyl reductase expression enhances
malignant behaviour in uterine cervical squamous cell carcinoma:
Carbonyl reductase predicts prognosis and lymph node metastasis.
Cancer Lett. 311:77–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Murakami A, Yakabe K, Yoshidomi K, Sueoka
K, Nawata S, Yokoyama Y, Tsuchida S, Al-Mulla F and Sugino N:
Decreased carbonyl reductase 1 expression promotes malignant
behaviours by induction of epithelial mesenchymal transition and
its clinical significance. Cancer Lett. 323:69–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Osawa Y, Yokoyama Y, Shigeto T, Futagami M
and Mizunuma H: Decreased expression of carbonyl reductase 1
promotes ovarian cancer growth and proliferation. Int J Oncol.
46:1252–1258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Umemoto M, Yokoyama Y, Sato S, Tsuchida S,
Al-Mulla F and Saito Y: Carbonyl reductase as a significant
predictor of survival and lymph node metastasis in epithelial
ovarian cancer. Br J Cancer. 85:1032–1036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishimoto Y, Murakami A, Sato S, Kajimura
T, Nakashima K, Yakabe K, Sueoka K and Sugino N: Decreased carbonyl
reductase 1 expression promotes tumor growth via epithelial
mesenchymal transition in uterine cervical squamous cell
carcinomas. Reprod Med Biol. 17:173–181. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miura R, Yokoyama Y, Shigeto T, Futagami M
and Mizunuma H: Inhibitory effect of carbonyl reductase 1 on
ovarian cancer growth via tumor necrosis factor receptor signaling.
Int J Oncol. 47:2173–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Banyard J and Bielenberg DR: The role of
EMT and MET in cancer dissemination. Connect Tissue Res.
56:403–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kahlert UD, Joseph JV and Kruyt FAE: EMT-
and MET-related processes in nonepithelial tumors: Importance for
disease progression, prognosis, and therapeutic opportunities. Mol
Oncol. 11:860–877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song C, Liu W and Li J: USP17 is
upregulated in osteosarcoma and promotes cell proliferation,
metastasis, and epithelial-mesenchymal transition through
stabilizing SMAD4. Tumour Biol. 39:10104283177171382017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng SX, Cai QC, Guo CH, Zhi LQ, Dai X,
Zhang DF and Ma W: High expression of TRIM29 (ATDC) contributes to
poor prognosis and tumor metastasis by inducing
epithelial-mesenchymal transition in osteosarcoma. Oncol Rep.
38:1645–1654. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sannino G, Marchetto A, Kirchner T and
Grünewald TGP: Epithelial-to-Mesenchymal and
Mesenchymal-to-Epithelial transition in mesenchymal tumors: A
paradox in sarcomas? Cancer Res. 77:4556–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kong G, Jiang Y, Sun X, Cao Z, Zhang G,
Zhao Z, Zhao Y, Yu Q and Cheng G: Irisin reverses the IL-6 induced
epithelial-mesenchymal transition in osteosarcoma cell migration
and invasion through the STAT3/Snail signaling pathway. Oncol Rep.
38:2647–2656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peng C, Zhao H, Song Y, Chen W, Wang X,
Liu X, Zhang C, Zhao J, Li J, Cheng G, et al: SHCBP1 promotes
synovial sarcoma cell metastasis via targeting TGF-β1/Smad
signaling pathway and is associated with poor prognosis. J Exp Clin
Cancer Res. 36:1412017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang J, Shen L, Yang Q and Zhang C:
Overexpression of metadherin mediates metastasis of osteosarcoma by
regulating epithelial-mesenchymal transition. Cell Prolif.
47:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qi Y, Wang N, He Y, Zhang J, Zou H, Zhang
W, Gu W, Huang Y, Lian X, Hu J, et al: Transforming growth
factor-β1 signaling promotes epithelial-mesenchymal transition-like
phenomena, cell motility, and cell invasion in synovial sarcoma
cells. PLoS One. 12:e01826802017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yoshidomi K, Murakami A, Yakabe K, Sueoka
K, Nawata S and Sugino N: Heat shock protein 70 is involved in
malignant behaviors and chemosensitivities to cisplatin in cervical
squamous cell carcinoma cells. J Obstet Gynaecol Res. 40:1188–1196.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Murakami A, Nakagawa T, Kaneko M, Nawata
S, Takeda O, Kato H and Sugino N: Suppression of SCC antigen
promotes cancer cell invasion and migration through the decrease in
E-cadherin expression. Int J Oncol. 29:1231–1235. 2006.PubMed/NCBI
|
|
38
|
Nakagawa T, Murakami A, Torii M, Nawata S,
Takeda O and Sugino N: E-cadherin increases squamous cell carcinoma
antigen expression through phosphatidylinositol-3 kinase-Akt
pathway in squamous cell carcinoma cell lines. Oncol Rep.
18:175–179. 2007.PubMed/NCBI
|
|
39
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deckers M, van Dinther M, Buijs J, Que I,
Löwik C, van der Pluijm G and ten Dijke P: The tumor suppressor
Smad4 is required for transforming growth factor beta-induced
epithelial to mesenchymal transition and bone metastasis of breast
cancer cells. Cancer Res. 66:2202–2209. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Morizane R, Fujii S, Monkawa T, Hiratsuka
K, Yamaguchi S, Homma K and Itoh H: miR-34c attenuates
epithelial-mesenchymal transition and kidney fibrosis with ureteral
obstruction. Sci Rep. 4:45782014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Valcourt U, Kowanetz M, Niimi H, Heldin CH
and Moustakas A: TGF-beta and the Smad signaling pathway support
transcriptomic reprogramming during epithelial-mesenchymal cell
transition. Mol Biol Cell. 16:1987–2002. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ghahhari NM and Babashah S: Interplay
between microRNAs and WNT/β-catenin signalling pathway regulates
epithelial-mesenchymal transition in cancer. Eur J Cancer.
51:1638–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu QQ, Chen K, Ye Q, Jiang XH and Sun YW:
Oridonin inhibits pancreatic cancer cell migration and
epithelial-mesenchymal transition by suppressing Wnt/β-catenin
signaling pathway. Cancer Cell Int. 16:572016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qi Y, Wang CC, He YL, Zou H, Liu CX, Pang
LJ, Hu JM, Jiang JF, Zhang WJ and Li F: The correlation between
morphology and the expression of TGF-β signaling pathway proteins
and epithelial-mesenchymal transition-related proteins in synovial
sarcomas. Int J Clin Exp Pathol. 6:2787–2799. 2013.PubMed/NCBI
|
|
46
|
Dwivedi SK, McMeekin SD, Slaughter K and
Bhattacharya R: Role of TGF-β signaling in uterine carcinosarcoma.
Oncotarget. 6:14646–14655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Horbelt D, Denkis A and Knaus P: A
portrait of transforming growth factor β superfamily signalling:
Background matters. Int J Biochem Cell Biol. 44:469–474. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang J, Eddy JA, Pan Y, Hategan A, Tabus
I, Wang Y, Cogdell D, Price ND, Pollock RE, Lazar AJ, et al:
Integrated proteomics and genomics analysis reveals a novel
mesenchymal to epithelial reverting transition in leiomyosarcoma
through regulation of slug. Mol Cell Proteomics. 11:2405–2413.
2010. View Article : Google Scholar
|
|
49
|
Li Y, Shao G, Zhang M, Zhu F, Zhao B, He C
and Zhang Z: miR-124 represses the mesenchymal features and
suppresses metastasis in Ewing sarcoma. Oncotarget. 8:10274–10286.
2017.PubMed/NCBI
|
|
50
|
Zhang M, Wang D, Zhu T and Yin R: RASSF4
overexpression inhibits the proliferation, invasion, EMT, and Wnt
signaling pathway in osteosarcoma cells. Oncol Res. 25:83–91. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang D, Jiang F, Wang X and Li G:
Downregulation of Ubiquitin-Specific protease 22 inhibits
proliferation, invasion, and Epithelial-mesenchymal transition in
osteosarcoma cells. Oncol Res. 25:743–751. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Guo X, Zhang J, Pang J, He S, Li G, Chong
Y, Li C, Jiao Z, Zhang S and Shao M: MicroRNA-503 represses
epithelial-mesenchymal transition and inhibits metastasis of
osteosarcoma by targeting c-myb. Tumour Biol. 37:9181–9187. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang D, Wang S, Chen J, Liu H, Lu J,
Jiang H, Huang A and Chen Y: Fibulin-4 promotes osteosarcoma
invasion and metastasis by inducing epithelial to mesenchymal
transition via the PI3K/Akt/mTOR pathway. Int J Oncol.
50:1513–1530. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Somarelli JA, Shetler S, Jolly MK, Wang X,
Bartholf Dewitt S, Hish AJ, Gilja S, Eward WC, Ware KE, Levine H,
et al: Mesenchymal-epithelial transition in sarcomas is controlled
by the combinatorial expression of MicroRNA 200s and GRHL2. Mol
Cell Biol. 36:2503–2513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang J, Du X, Wang G, Sun Y, Chen K, Zhu
X, Lazar AJ, Hunt KK, Pollock RE and Zhang W: Mesenchymal to
epithelial transition in sarcomas. Eur J Cancer. 50:593–601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yi EY, Park SY, Jung SY, Jang WJ and Kim
Y: Mitochondrial dysfunction induces EMT through the
TGF-β/Smad/Snail signaling pathway in Hep3B hepatocellular
carcinoma cells. Int J Oncol. 47:1845–1853. 2015. View Article : Google Scholar : PubMed/NCBI
|